1. Introduction
Our broad objective is the extension of the Wiener-Physically-Informed-Neural-Network (
W-PINN) (see
Figure 1c) approach developed by [
1] to improve modeling effectiveness when
dynamic processes (i.e., systems) have highly nonlinear static behavior. This work is an extension and advancement of the modeling approach in [
2] that was applied to three types of freely existing data sets. The first data set consisted of four (4) nutrient inputs (
xi,
i = 1, …, 4) and modeled the change in weight (
y) over time using first-order dynamic structures (
vi) for each input
i, and a quadratic, multiple linear regression, static output structure,
where
is a vector of the
vi’s. The second one consisted of nine (9)
xi’s and modeled the top tray temperature (
y) of a pilot distillation column using second-order dynamic structures for the
vi’s and a first-order multiple linear regression, static structure for
Our goal, for the fitted correlation coefficient (
) of
y and
(the fitted
y), is
for test sets or validation sets when the test set is not possible, and this was met for these two cases in [
2].
The third case in [
2] consisted of individually modeling eleven (11), two-week, type 1 diabetes data sets, with twelve inputs (
xi’s), originally modeled in [
3]. For each data set, its first week is used as training data and its second week as validation data. The sensor glucose concentration (
SGC) sampling rate of five (5) minutes resulted in two very large data sets for each of the eleven modeling cases. The critical complexity of this case is that it requires forecast modeling for closed-loop forecast control, a future objective. Thus, all the inputs must have a model deadtime greater than or equal to the effective deadtime (
θMV) of the manipulated variable (
MV) unless it has a scheduled (known) change (e.g., a meal) and a deadtime less than
θMV, called an “announcement input”. These requirements were not followed in [
3] and it is, therefore, not applicable to the modeling objectives of this work. The modeling strategy in [
2] estimates
θMV first, then any announcement inputs, and then all other inputs, using a one-input simple linear regression structure, to obtain initial estimates of the dynamic parameters for each input separately. After completion of this step for all the inputs, a full second-order dynamic structure and first-order static structure strategy was used to obtain final parameter estimates. This approach resulted in an average validation set
of 0.68 and a maximum
of 0.77, considerably below the individual goal of
.
Our hypothesis is that [
2] used an effective dynamic structure and estimated the dynamic parameters sufficiently accurately for the eleven (11)
SGC cases. However, the first-order linear regression static structure did not, and cannot, accurately capture the complex static forecast nature of
SGC of these data sets. Thus, the overall goal of this work is the development of a two-stage modeling methodology for highly static and highly dynamic behavior that achieves the modeling goal of
, for one or more of the eleven
SGC data sets. While our process example is
SGC, it was selected because of its highly dynamic and highly complex static nature. Thus, it is not the objective of this work to focus on the issues related to advancing
SGC modeling but to present a general approach for modeling highly dynamic and highly complex static processes.
More specifically, the approach of this work is to use the
’s results in [
2], for each model
i, as the first stage in a two stage
W-PINN approach, where the second stage is a nonlinear static
ANN structure. Note that empirical dynamic
ANN modeling and
PINN modeling methods are outside this scope since they do not use
’s (i.e.,
ANN) or one for each input (i.e.,
PINN).
W-PINN is the only methodology that can directly use
’s results obtained in [
2] and in any context.
Moreover, the objective of this work is the development of an effective
W-PINN modeling approach for systems with significantly varying input dynamic behavior and complex nonlinear static behavior. More specifically, by using the Stage 1 dynamic modeling results obtained in [
2], the objective of this work is to significantly increase
using a novel, proposed, two-stage (
TS),
W-PINN modeling approach. Two types of
TS methodologies are proposed, one that uses JMP and one that uses Python coding to develop a novel input factor
W-PINN approach.
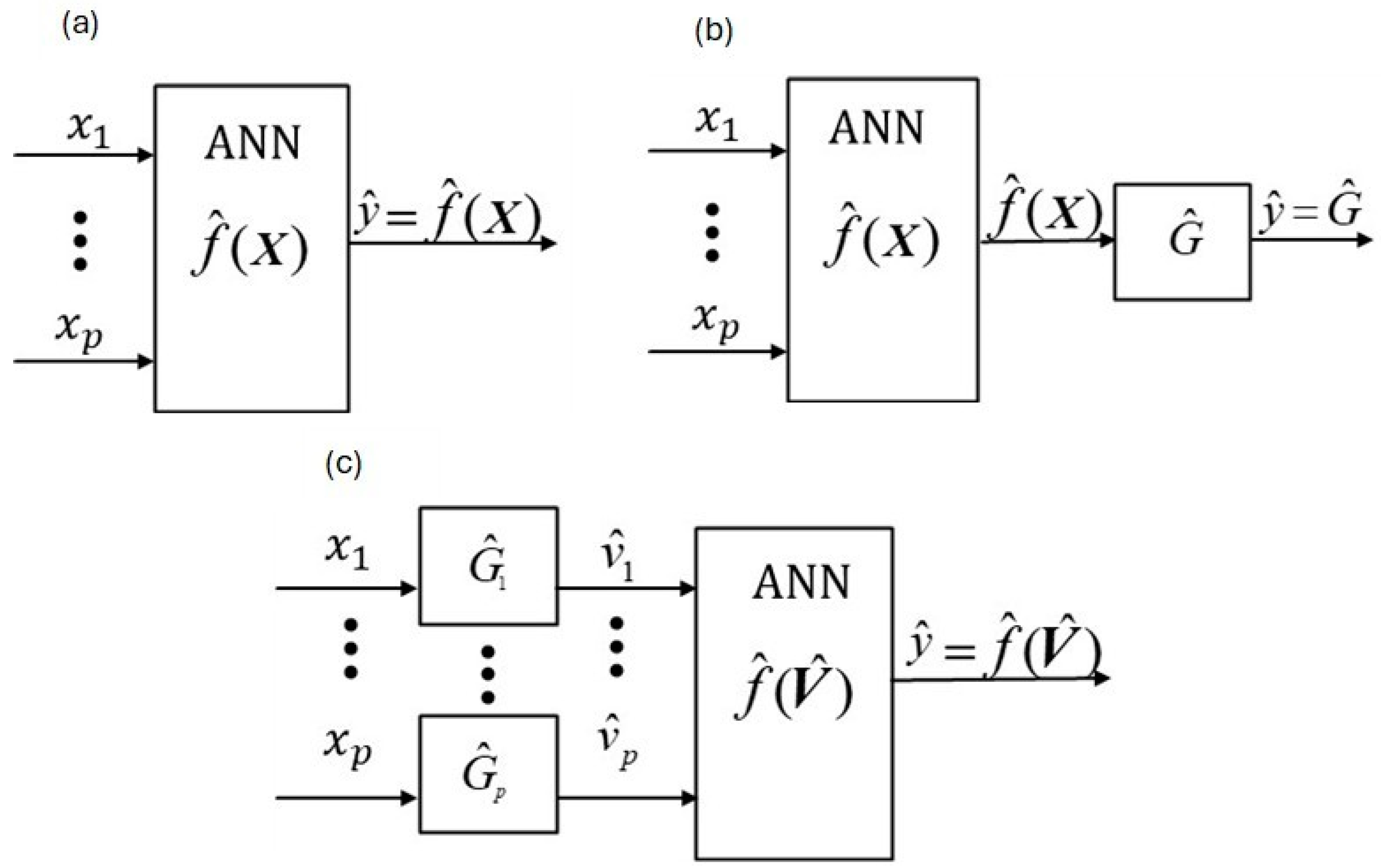
The classical
ANN approach is a one-box, empirical modeling methodology as illustrated in
Figure 1a. As shown,
p measured
xi inputs enter the
ANN function,
, which is an estimated (as denoted by “^”) empirical function with constant coefficients that are adjusted under some criterion, commonly least-squares estimation, to maximize agreement (i.e., fit) between its modeled output,
, and its measured output,
y (e.g.,
SGC), where
is a
pth dimensional vector of measured inputs. In
Figure 1a,
can be a static function, e.g., a nonlinear regression function, or a combined static and empirically dynamic function of lag variables, e.g., a Long- and Short-Term Memory (
LSTM) [
4,
5] function.
Phenomenologically dynamic and empirically static methodologies are illustrated in
Figure 1b (
PINN [
6]) and
Figure 1c (
W-PINN [
1]). Refs. [
6,
7] named their methodology “physically-informed-neural-network” (
PINN). The critical difference between
Figure 1a (classical
ANN) and
Figure 1b,c are the number of stages.
Figure 1a has one stage for static and dynamic structures, and the two-stage methods in
Figure 1b,c have one stage for static structures and one stage for dynamic structures. The
PINN dynamic block is not restricted to linear dynamic structures as it is for
W-PINN. However, nonlinear dynamic behavior is modeled when the dynamic outputs of
W-PINN (i.e.,
’s) are passed through
. A critical advantage of
W-PINN over
PINN is that each input has its own dynamic model structure, as illustrated by comparing
Figure 1b,c.
The next section,
Section 2, describes the
W-PINN methodology fundamentally and mathematically. The theoretical structure of a general and complete second-order dynamic structure is first given in
Section 2.1 as a differential equation and then transformed to its discrete-time version using backward difference derivatives. It then gives the explicit equation for
, where “
t” is the sampling time.
Section 2.2 gives important Stage 1 details and results in assisting in the understanding of the Stage 2 methodology.
Section 2.3 describes the JMP and Python Stage 2 methodologies.
Section 2.4 gives the mathematical details of Model 1 (input only model), Model 2 (input-output), and Model 1–2, a combination of the strengths of Models 1 and 2.
Section 2.5 gives the forecast model structure, i.e., the structure for
. Finally,
Section 2.6 gives the information and equations for the summary statistics.
Section 3 gives a table with all the numerical results for the three modeling methods. It also gives Model 1, Model 2, and Model 1–2 graphical results for the best fitting Stage 2 subject, Subject 2. For all three models, their
is 0.93.
Section 4 gives a discussion of the results and
Section 5 comments on work in progress and speculates on other possible future directions.
2. Materials and Methods
This section describes the two-stage
W-PINN methodology in detail. It also gives critical Stage 1
SGC modeling particulars used by [
2] to obtain the
’s posted on the website of the last author (see
https://drollins9.wixsite.com/derrickrollins, accessed on 6 October 2025) With both the
’s and
’s posted on this website, modelers have the option of using these data sets to build both stages or just the second stage, the aim of this work.
With the sampling rate, Δ
t, equal to 5 min, our
SGC models are forecasting 12 steps (i.e., 60 min) into the future. The forecast nature of this work is an artifact of the data sets we are using and their application and, thus, not a necessity of the methodology. As described in [
2], 12Δ
t is the estimated observable time it took for the manipulated variable (
MV), exogenous insulin, to cause
SGC to start decreasing after a bolus increase (or insulin injection). The 60 min estimate was very consistent for the subjects in this clinical study, as noted in [
2].
The models that this work develops are for an unobservant, 60 min, forecast monitoring scenario. “Unobservant” is meant to convey the protocol that the person(s) determining insulin changes have no knowledge of the forecast (
) estimates. In addition, even though the authors of [
2] worked diligently to obtain models that minimize
pairwise correlation, we note that it is still significantly present. Thus, for this reason, this work is best understood as an unobservant monitoring application and not applicable to closed-loop control.
2.1. W-PINN
Our
W-PINN approach uses backward difference derivatives (
BDD) to discretize second-order-plus-dead-time-plus-lead (
SOPDTPL) (for details of this methodology see [
8]) theoretical dynamic systems, the only type used in this work, as given in Equation (1) below. The dynamic system does not have to be initially at a steady state for our
W-PINN modeling methodology since the initial conditions are also estimated. Note that, Equation (1) is the expression for each of the
p-inputs.
with
where
,
, and
for
i = 1, …,
p.
xi(
t) is the value of the
ith input variable at
t, and
vi(
t) is the value of the
ith output variable at
t, in the units of
xi;
y(
t) is the output variable in its units at
t,
means the expected value (i.e., true mean) of
y(
t); and
is the true output (gain) function of
V(
t), the vector of the
vi(
t)’s. The variables
is the deadtime of the MV,
is the damping coefficient,
is the primary time constant, and
is the lead time constant. When
is a nonlinear function of
V(
t), as in the
ANN (i.e.,
W-PINN) case, Equations (1) and (2), taken together, have a Wiener block-oriented structure [
8], as shown in
Figure 1c.
The lead term is the first term on the right side of the equal sign in Equation (1). This term tends to “speed up” the response and provides what the process modeling and control community has termed “numerator dynamics” [
8,
9,
10]. Ref. [
8] developed a second-order, multiple-input, single-output, discrete-time, nonlinear Wiener dynamic approach using
BDD based on Equation (1). More specifically, using
BDD approximation applied to a sampling interval of Δ
t, an approximate discrete-time form of Equation (1) is
with
where
to satisfy the unity gain constraint. From Equation (3) with
,
After obtaining
for each input
i, the modeled output value, at time
t, is determined by entering these results into
, a static
ANN in this application, i.e.,
2.2. Stage 1 Modeling Method
This subsection gives important Stage 1 details and results in assisting in the understanding of the Stage 2 methodology. While missing output (i.e.,
SGC) measurements are acceptable, missing input values are not for discrete-time modeling. Activity tracker data were the only missing input data. These missing values were estimated by averaging the two values on both sides of a gap and filling in the gap with this value. Some gaps were several hours long. Blocked cross-validation [
11,
12] was used to guard against overfitting, with the first week as the training (
Tr) data set, and the second week as the validation (
Val) data set.
In Stage 1, all inputs were first modeled separately on their own Excel worksheet with a first-order linear regression static function. The tool that one chooses to use for this step, as well as all the steps, is a matter of preference. However, we encourage modelers to break the modeling process down for this, and all large/complex data sets, as we have for this case. Note that for the other two modeling cases in [
2], this decomposition procedure was not used.
For each case, insulin was modeled first. The estimated deadtime, i.e.,
, was set at 60 min and was varied one Δ
t forwards and backwards at a time to find the value that gave the best fit. For all the inputs, the estimate of
θMV,
, was determined to be 60 min, i.e., 12 Δ
t. The food variables were the only ones with announcements, and the carbohydrate input was the only one found to have a deadtime less than
(see [
8] for details on how to incorporate announced inputs into this approach). The “time of day” input has no
deadtime, and the deadtime for all other inputs was
, except for fats that had deatimes that were much larger than
, as determined by model estimation. After estimating the dynamic coefficients for each input (i.e., Equations (4)–(6)), these values were copied to an Excel worksheet as the dynamic structure starting values for fitting the SOPDTPL dynamic, and first-order static, multiple-input model (i.e., Equation (10)). With
as a first-order multiple linear regression static function,
, for Stage 2 was determined.
2.3. Stage 2 Model Development Modeling Methods
The objective of this work is the development, evaluation, and comparison of two, Stage 2, W-PINN modeling approaches using from Stage 1 to obtain for each of the eleven data sets for the two approaches. The first approach used the JMP ANN toolbox to approximately find the smallest SSE (i.e., SSR) by fitting many cases and selecting the best one. All analyses were conducted using JMP Pro Version 16 (SAS Institute Inc, Cary, NC, USA) for neural network construction, preprocessing, and model optimization. More specifically, JMP was used to create and optimize a three-layer ANN structure. The ANN model began as a fully connected single-layer perceptron, functioning as a decision-making node. This architecture consisted of three layers: the input layer, a hidden transfer layer, and the output layer. Each node in the transfer layer received weighted inputs from the input layer, and the final predictions were made based on the output layer’s activations. The transfer functions used within the model were a combination of linear and Gaussian transformations, and boosting techniques were applied to enhance the model’s performance. To ensure the inputs were appropriately scaled and transformed, continuous covariates were preprocessed by fitting them to a Johnson Su distribution. Using maximum likelihood estimation, this preprocessing step helped transform the data closer to normality, thereby mitigating the effects of skewed distributions and outliers. The general fitting approach aimed to minimize the negative log-likelihood of the observed data, augmented by a penalty function to regulate the model complexity. Specifically, a sum-of-squares penalty, applied to a scaled and centered subset of the parameters, was used to address the overfitting problem that often occurs with ANN models. This penalty was based on the magnitude of the squared residuals (i.e., ), helping to stabilize the parameter estimates and improve the model’s optimization. Cross-validation was performed using the holdout method to assess the model’s ability to generalize to new data. The training set consisted of the initial data range, while the validation set represented future observations, ensuring the model’s predictive capacity for unseen data was tested.
The second approach is our newly developed, novel,
ANN structure that is coded in the Python language, with the scipy.optimize library with a Tan(h) activation function running the dual annealing optimization solver. As
Section 3 will show, both (JMP and Python) nonlinear regression, static stage-two,
ANN approaches significantly improved fit in comparison with the linear regression, static stage-two, first-order approach used by [
2].
Our
W-PINN JMP approach uses a classical
ANN p-input variable layer (see
Figure 1) where
enters node
i,
i = 1, …,
p, as shown in
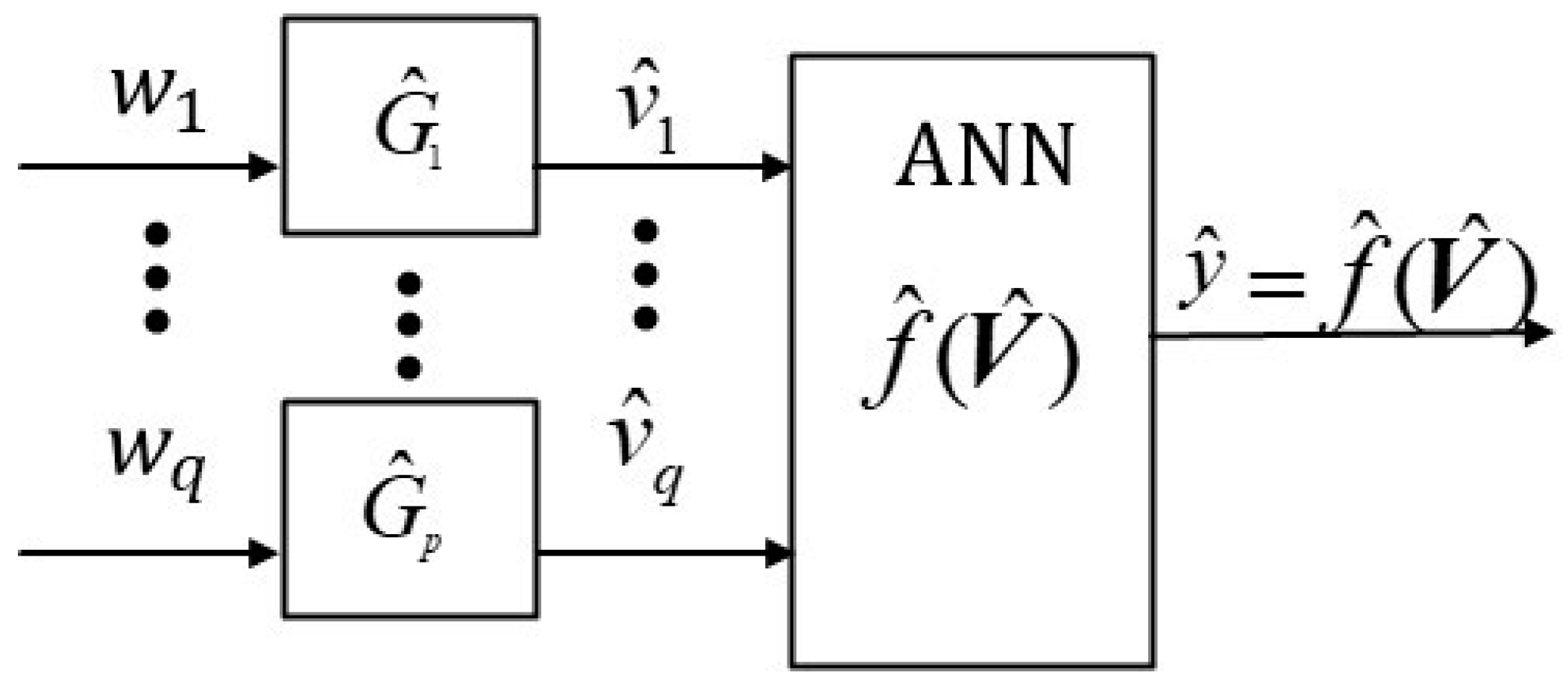
Figure 1c. In contrast, our proposed
W-PINN Python approach uses a q-input factor layer where
(see
Figure 2). For example, it uses terms like quadratic factors (e.g.,
) and interaction factors (e.g.,
) as inputs to the input layer, where
represents input factor
i (e.g.,
). Moreover, our proposed
W-PINN Python methodology uses
q input factors, and not
p input variables, as shown in
Figure 2 below.
2.4. Three Input Models
We developed three types of input model structures for this application. The first one we call the “input only model” or “Model 1.” All the inputs in this structure have a deadtime except for announcement inputs that can have deadtimes less than like carbohydrates, equal to like proteins, greater than like fats, and zero like time of day, as mentioned above.
The second one we call the “input-output model” or “Model 2.” It combines the input-only structure of Model 1 (i.e., Equation (10)) with a model of weighted residuals (i.e., bias correction, see [
8]), a minimum of
distance in the past (note that, this is model building and not model forecasting), as shown in Equation (11) below (see [
8] for the derivation).
Equation (11) has no value if any residual is not determinable due to missing output measurements. Thus, unlike Model 1, which has estimates for all t since it uses only input data, Model 2 will not have an estimate when an output value is missing.
The final model, Model 1–2, is a combination of the strengths of Models 1 and 2. More specifically, for Model 1–2,
2.5. Forecast Structures
Equation (10), , is the fitted structure for Model 1, i.e., estimate of the output, , at the current time, t. There are no missing input values in Equation (10). This is why missing armband data had to be estimated. In addition, non-announcement input values to obtain Equation (10) must be at least a distance of in the past. This requirement is because the model developed input lag must be the same as the forecast input lag, i.e., at t, for forecasting a distance into the future.
After obtaining
, its transformation into the
kΔ
t forecast form, i.e., the online version, is given by Equation (13) below:
where from Equation (7), with
t =
t +
kΔ
t,
Note that, if k = 12 and Δt = 5 min, Equation (14) is forecasting 60 min into the future. Thus, all the non-announcement inputs must have, i.e., use, a model building and forecast prediction deadtime of at least 60 min.
2.6. Statistical Analyses
Formal statistics inference tools/methodologies such as confidence intervals, hypothesis testing, etc., are not applicable to dynamic modeling because response data are time-correlated, an inherent nature of time delay and time lag (i.e., dynamic) behavior [
12]. Thus, it is not possible to randomize the occurrence (i.e., time order) of trials. Consequently, only informal inference, i.e., direct comparison of the values of statistical features (numerical and/or visual), is applicable to dynamic modeling inference. Moreover, this work uses the following statistics.
The first, and most important, modeling statistic is
(which is bounded between −1 and 1), the fitted correlation of the measured
SGC,
, and the fitted
SGC,
, as given in Equation (15) below.
where
n is the number of samples in the set and the bar above a statistic means that it is its sample mean value. The equations to determine
AAD and
AD are, respectively,
The equation for
SSE (i.e.,
SSR, the sum of squared residuals), the more common name and used by JMP, is
The loss function
shown in Equation (19), used by the Python program, is the
SSE and an
ANN parameter penalty
σ is shown in Equation (20).
where
3. Results
Training and validation Stage 1 and Stage 2 summary statistics for the eleven subjects are given in
Table 1. All the results are
unless indicated otherwise. Recalling that each subject has a fixed
that was determined in Stage 1 using the first-order static structure as given in Equation (21) below:
As shown in
Table 1, Stage 1, Model 1,
results varied from 0.59 to 0.77, with a mean of 0.68. Moreover, Stage 2, Model 1,
results improved significantly over the Stage 1 results for both
ANN approaches. As shown, JMP Stage 2, Model 1,
results varied from 0.60 to 0.85, with a mean of 0.74. However, Python Stage 2, Model 1,
results are significantly better than JMP, varying from 0.72 to 0.93, with a mean of 0.82. As a result, Model 2 training and validation results and Models 1–2 validation results are given in
Table 1 for Python only. From Model 1 to Model 2, the Python mean
increased from 0.82 to 0.87, the minimum from 0.72 to 0.80, and the maximum of 0.93 did not change. In summary, Python Stage 2 results improved considerably over Stage 1 results and are significantly better than JMP Stage 2 results.
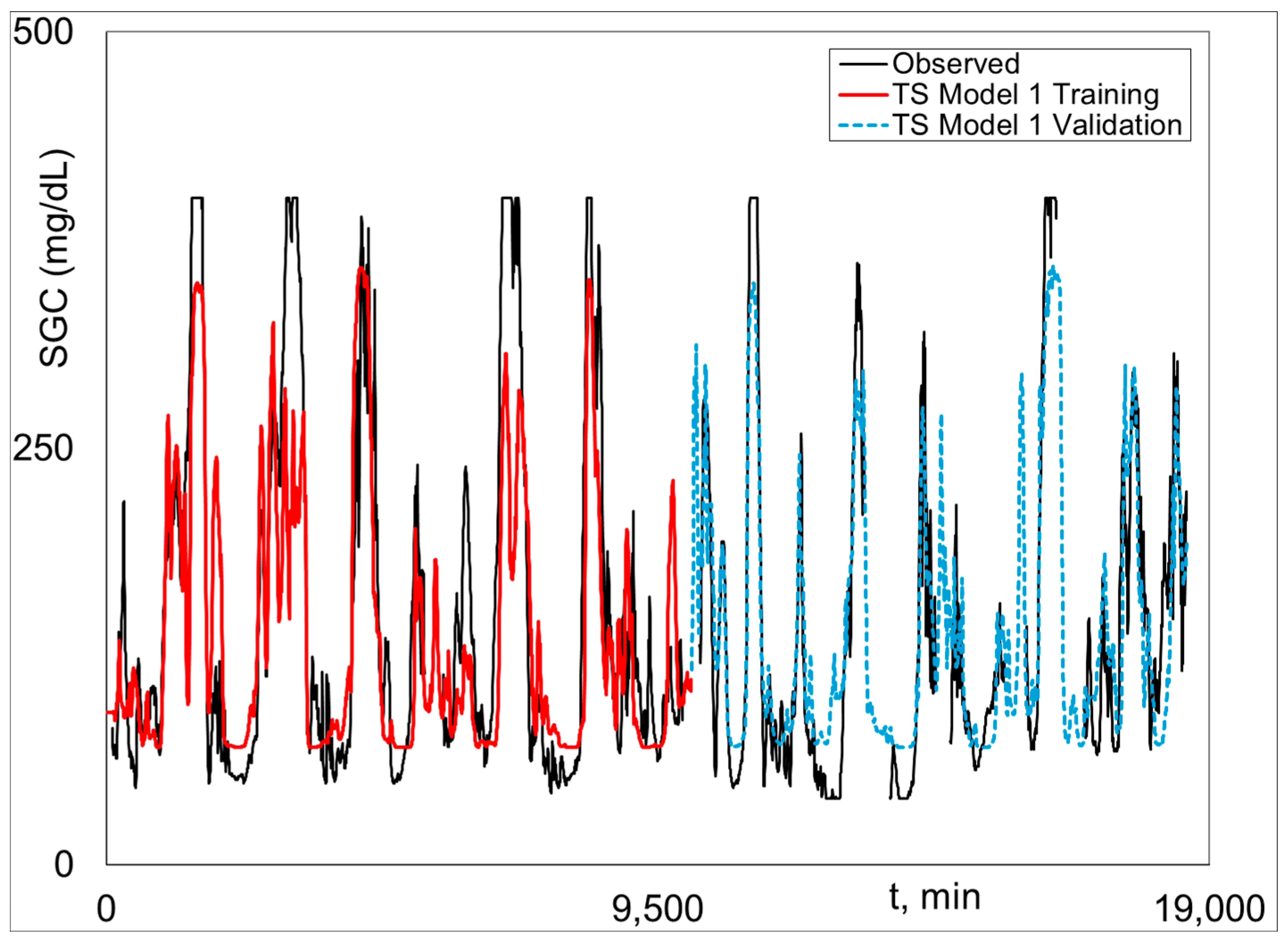
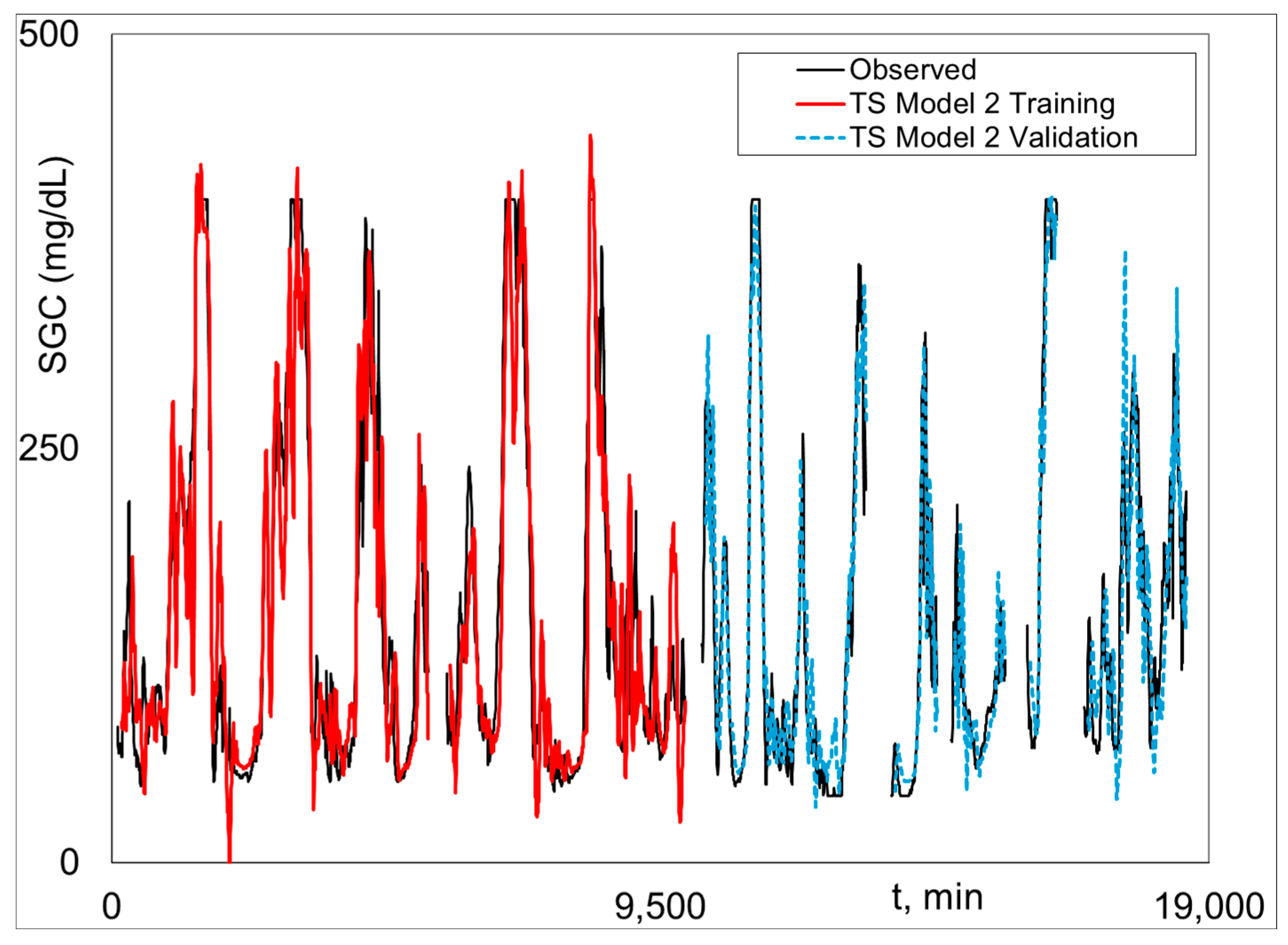
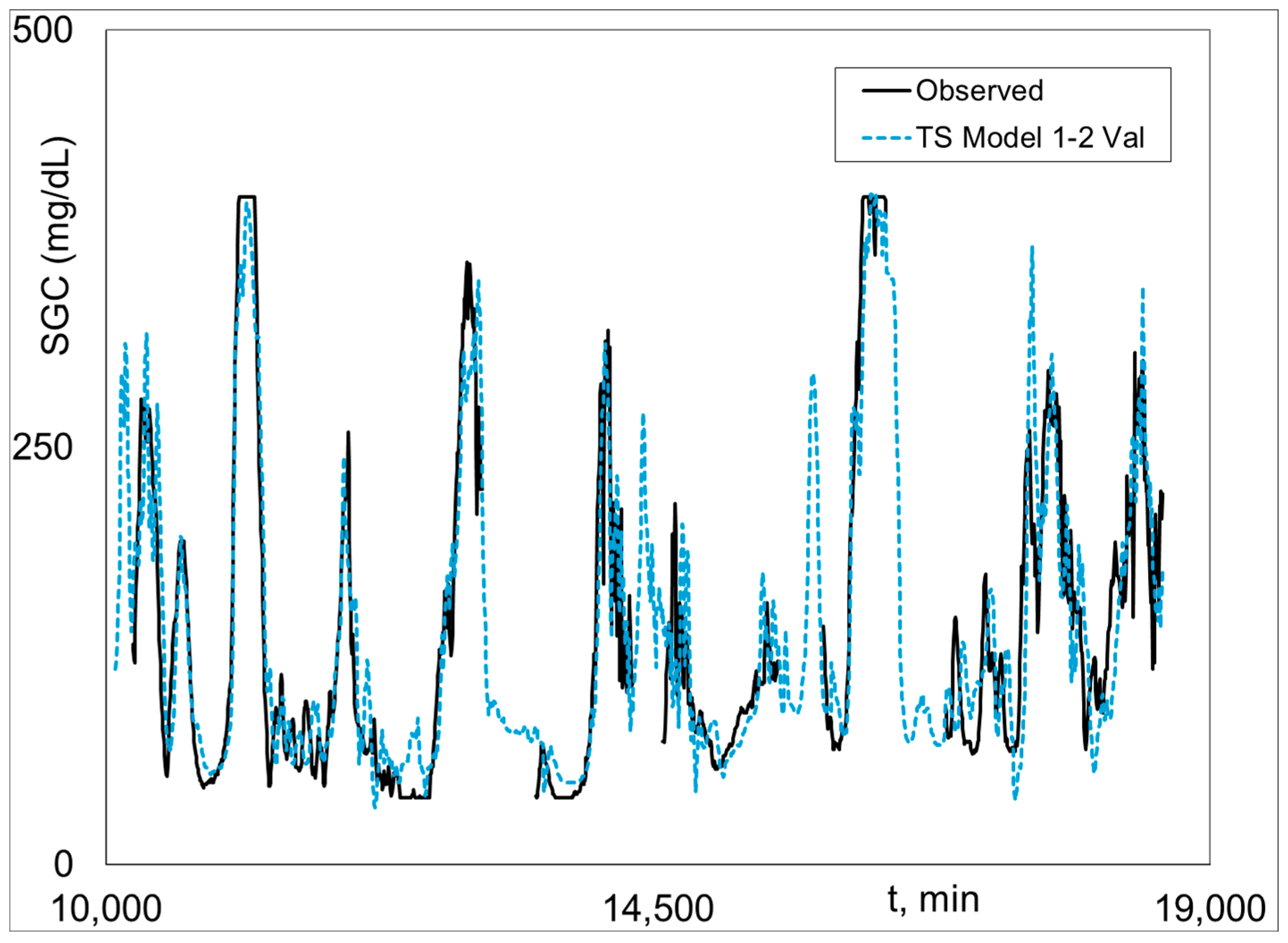
Graphical Python Stage 2 fitted and measured
SGC results for Subject 2 (the best case) are given (i.e., plotted) in
Figure 3,
Figure 4 and
Figure 5.
Figure 3 shows Model 1 training and validation.
Figure 4 is Model 2 training and validation.
Figure 5 presents the combined validation results, where Model 1 is plotted when there is no output data, and Model 2 is plotted when there is output data, i.e., the Model 1–2 validation plot. The Model 1–2 plots are associated with the results in the last three columns in
Table 1.
Figure 4 shows excellent fit of Model 3 and the highly realistic behavior of Model 1 when Model 2 results are not possible because of missing
SGC data (see Equation (11)).
The Python Stage 2, Model 1 contains fewer than 65 trainable parameters, performs 60 min-ahead inference in 0.9–1.4 ms on an Intel Core i7-11850H. This can be extrapolated to an estimated <20 ms on an ARM Cortex-M7 microcontroller, making it highly suitable for real-time embedded deployment. The linear dynamic stage is analytically stable with all poles inside the unit circle, the shallow tan(h) network is Lipschitz continuous, and the optimization consistently converges across all 11 subjects [
13]. Compared with an equivalent single-stage
LSTM (~65 k parameters), the proposed method could train 4–6× faster, requires orders of magnitude less memory, and achieves superior validation performance, confirming excellent computational efficiency and numerical stability for practical diabetes monitoring and control applications.
In this study, two physically based virtual forecasting sensor approaches were developed for obtaining the value of the response variable,
SGC, a
θMV time distance in the future, for a two-stage forecast modeling application. The first stage, a physically (i.e., theoretical) based dynamic modeling approach [
2] estimates the physically interpretable dynamic parameters from the measured inputs (
xi’s) with multiple physical constraints to obtain dynamic outputs (
vi’s). The
vi’s are the inputs to the second stage, a static
ANN structure. For the first method, this structure was determined by using the
ANN toolbox in JMP. For the second method, this structure was coded using Python. Both methods resulted in large average improvements over the Stage 1 results using a first-order linear regression static structure (see
Table 1). In addition, a critical advantage of these two approaches is that the modeling is much easier and much less time-consuming than the second order multiple linear regression (
MLR) approach. We do note, however, that the static behavior of these data sets is highly nonlinear. Thus, we strongly recommend
ANN over
MLR for the static model structure, i.e.,
Note that
ANN modeling is just a particular class of nonlinear regression. We also note that the
MLR model was applied to Subject 11, the highest
(0.79) in [
3], to compare the performance with the
ANN models. The
had a modest improvement from Stage 1 alone, going from 0.74 to 0.79, but much less than the 0.85 obtained by
P-ANN.
5. Conclusions and Future Work
The W-PINN approach is particularly powerful because each input xi is dynamically transformed to its vi counterpart and is the input to a static ANN. The proposed two-stage W-PINN approach greatly improved the SGC model fit for eleven historical diabetes data sets. A one-stage W-PINN approach, in its evaluation stage, is the next step in this research.
During his time as a professor, the corresponding author gained valuable insight into the limitations of empirical modeling through a real-world industrial application. A BS Chemical Engineering student, also pursuing an MS in Statistics, undertook a summer project at a leading Midwest chemical company, which was approved for her MS thesis. The project focused on developing a multivariate Statistical Process Control (SPC) monitoring methodology for a process line. Data were collected, and an SPC chart was developed, resulting in an excellent model fit. However, when the process exceeded control limits, adjustments to the manipulated variable based on this model failed to restore control. A subsequent attempt with new data and a revised control chart, despite another excellent fit, similarly failed to correct deviations when applied in a feedback control scenario. This experience highlighted that the control chart, designed for monitoring, was unsuitable for feedback control due to its reliance on empirical correlation rather than cause-and-effect relationships. Empirical SGC modeling, which uses free-living data and non-physiological structures, faces similar limitations, as it cannot adequately capture cause-and-effect dynamics critical for model-based control applications like automatic forecast control. In contrast, physically informed modeling, which integrates physiological information and structure with free-living data, offers inherent intelligence and a robust structure for potentially developing effective models for control applications. Our W-PINN methodology proposed in this manuscript is approaching the goal for the diabetes data set, but more research and creative screening of inputs are needed to fully realize the goal for SGC closed-loop feedback control. In addition, this advancement relies on a dual hormone scenario, one to decrease SGC, i.e., insulin, and one to as effectively and safely increase SGC, possibly glucagon. Nonetheless, our proposed two-stage methodology has promise, it seems, for physically based, highly nonlinear static systems or processes.
Type 1 and 2 diabetes
SGC modeling for monitoring can be effective (i.e., informative) using empirical or physically informed dynamic modeling approaches [
14,
15,
16,
17,
18]. Closed-loop Type 1
SGC automatic control is inherently forecast automatic control because a change in
MV, injected insulin, will take a time of
θMV to start lowering
SGC. For automatic closed-loop control, empirical dynamic modeling approaches are not likely to succeed because they lack a cause-and-effect relationship, unlike
PINN approaches. Insulin is the process variable that is changed to keep
SGC close to its set point, i.e., it is the manipulated variable (
MV). For a control system to do this well in a forecast feedback control scheme, the controlled variable,
, must be accurately estimated. An empirical method could possibly control
online accurately if the correlation structure remains the same as it was when the model was developed. However, it is not possible for the correlation structure of an empirical forecast modeling approach in this context to remain intact, i.e., fixed, in online forecast feedback control because the correlation structure changes each time the controller signal to the manipulated variable is transmitted. Thus, it is prudent to restrict free-living empirical modeling to monitoring open-loop processes but not to make decisions on how much to change a manipulated variable to make changes in the control variable.
There are several challenges in the data sets used in this work. First, they are nearly a decade and a half old, and SGC technology has advanced considerably, particularly in terms of missing and lost data. Secondly, wearable technology has advanced considerably in reliability, measured sensor technology such as heart rate, as well as in data management. In addition, there are advancements in ways to obtain accurate consumption of food nutrients. Thus, one (distant) future goal is to evaluate W-PINN using data generated by current technology.