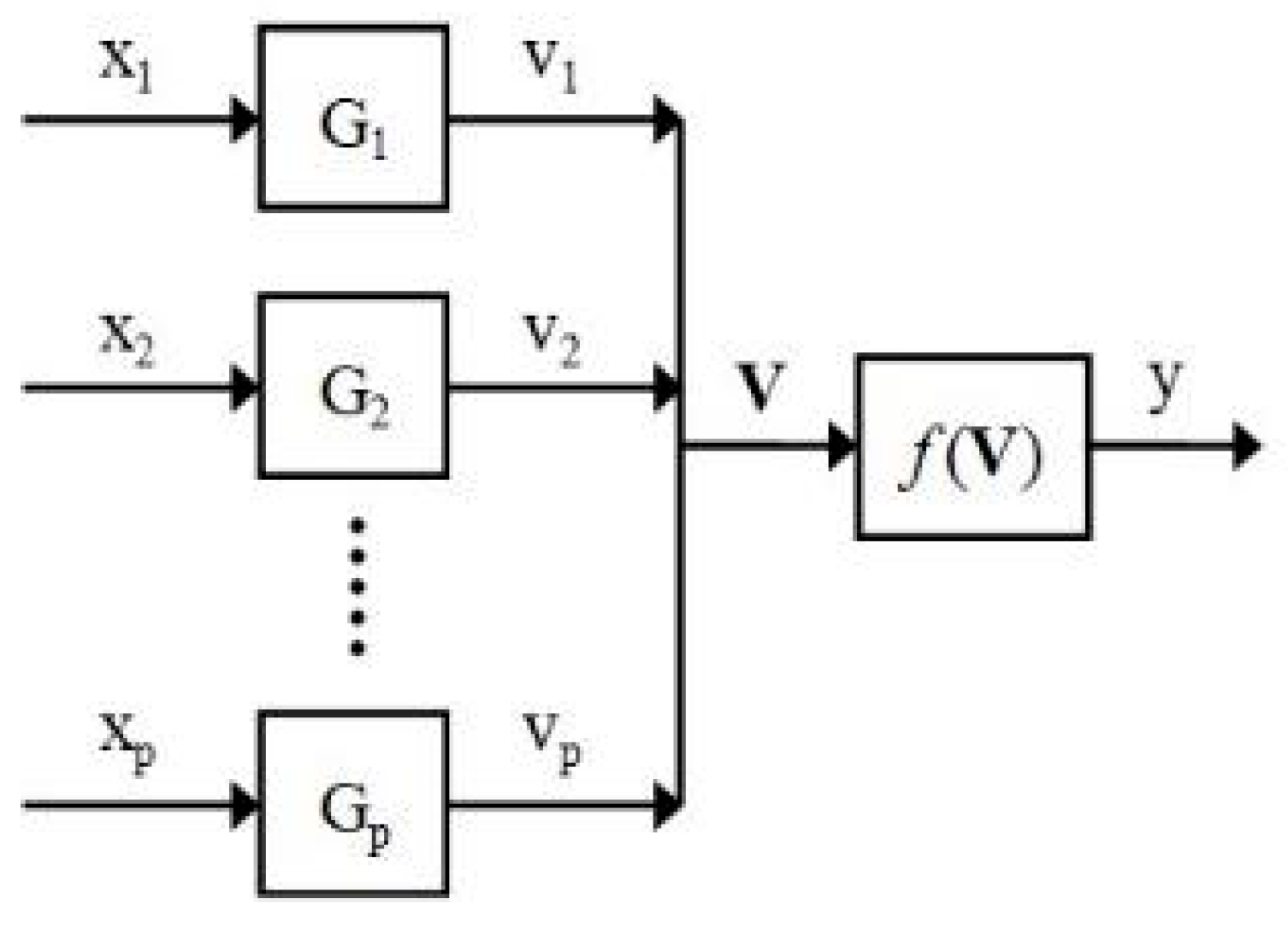
3.1. Weight Data Set Results
The second author provided a data set with the output (y) as the subject’s weight and several nutrient variables as the inputs. The data set covered a period of 9 years, during which time body weight was measured daily using a beam scale that was confirmed by a second measurement using a digital scale, and all food intake was weighed and recorded. The food intake was converted to individual nutrients by a nutritional database that includes information from the USDA Foods Database and many commercial sources, which was available through Calorie Count (sponsored by About.com) for the first 5.2 years and then through Nutritionix.com for the last 3.8 years of data collection. The data was compiled over consecutive 5-, 7-, or 10-day periods, with 28 occasional lapses of 5 to 28 days.
Given its limited size, our modeling work divided this data set into training and validation sets. Thus, our study does not include a test set. In addition, our objective is strictly modeling, rather than physiological assessments and interpretation. More specifically, our modeling objective is to obtain the highest rfit,val to illustrate the modeling effectiveness of TDR in real, free-living modeling of highly dynamic data sets. Moreover, the modeling goal here is to achieve an rfit,val > 0.90 for a relatively small set of input variables. The second author is interested in physiological understanding, and these results can provide a baseline fit for follow-up studies using other input combinations in this data set and other modeling ideas that she may have for this data set and other ones.
The full data set consisted of 14 nutrients: potassium, carbohydrates, calories, proteins, fats, sugar, saturated, cholesterol, fiber, vitamin A, vitamin C, calcium, iron, and salt. The sampling time, Δ
t, equals approximately one (1) week on average. The data covers a time range of 565Δ
t or about nine consecutive years. We found the following four inputs to best model the weight of this subject without any additional ones significantly impacting the fit: calories (
x1), fiber (
x2), proteins (
x3), and potassium (
x4). Single-input static (i.e., nondynamic) linear regression (S
LR) and
TDR results, with the same S
LR structures, are given in
Table 1 and
Table 2, respectively, and graphical results are given in
Figure 3 and
Figure 4, respectively. The first half of these figures are the training model results, and the second half are the validation model results.
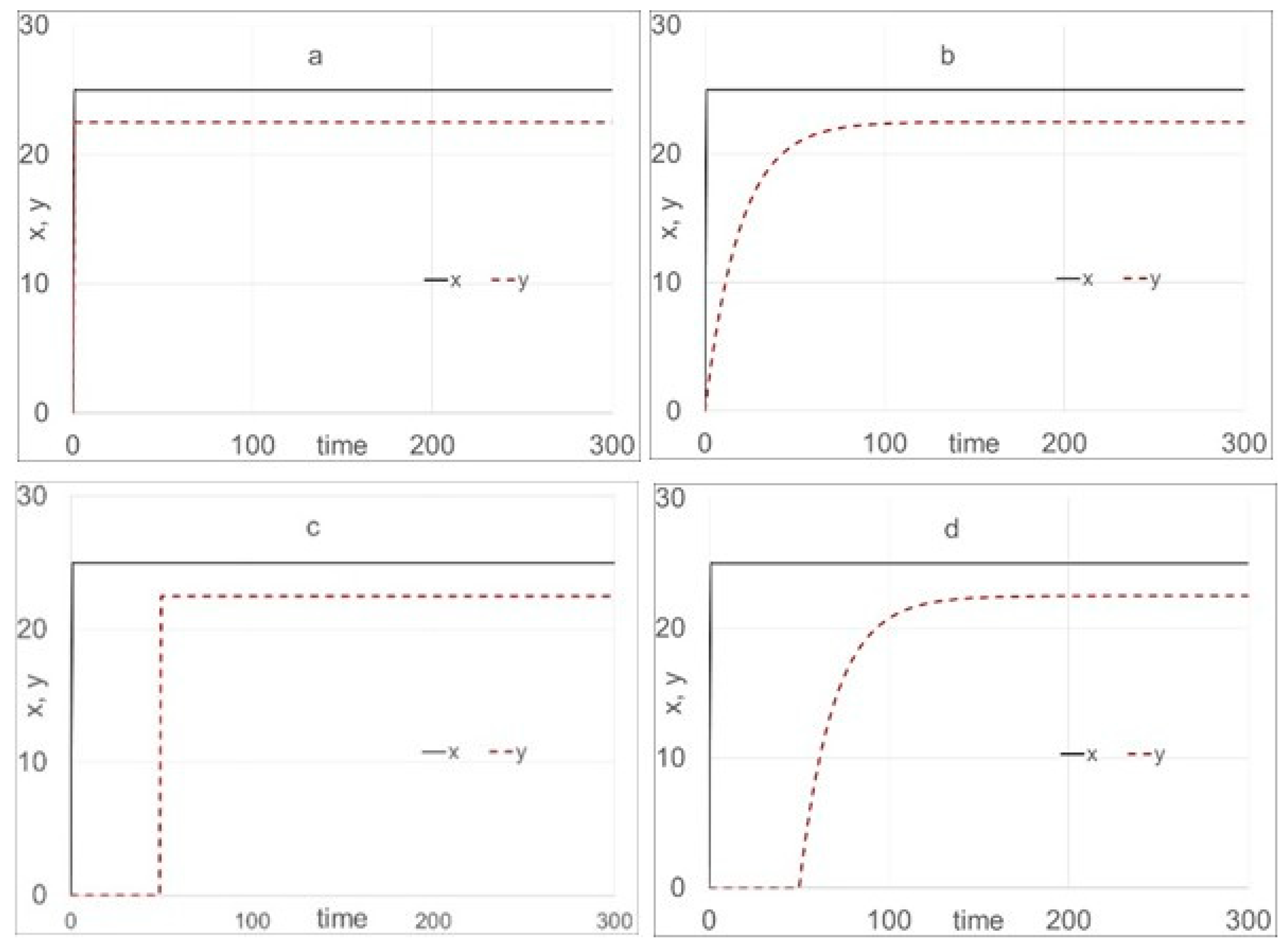
TDR modeling for this data set is based on Equations (9) to (11), i.e., the first-order, multiple-input, dynamic modeling structure. Note that for protein, r
fit,val values are for a single-input static model, and the dynamic models are 0.450 and 0.517, respectively, indicating that protein appears to be only slightly dynamic.
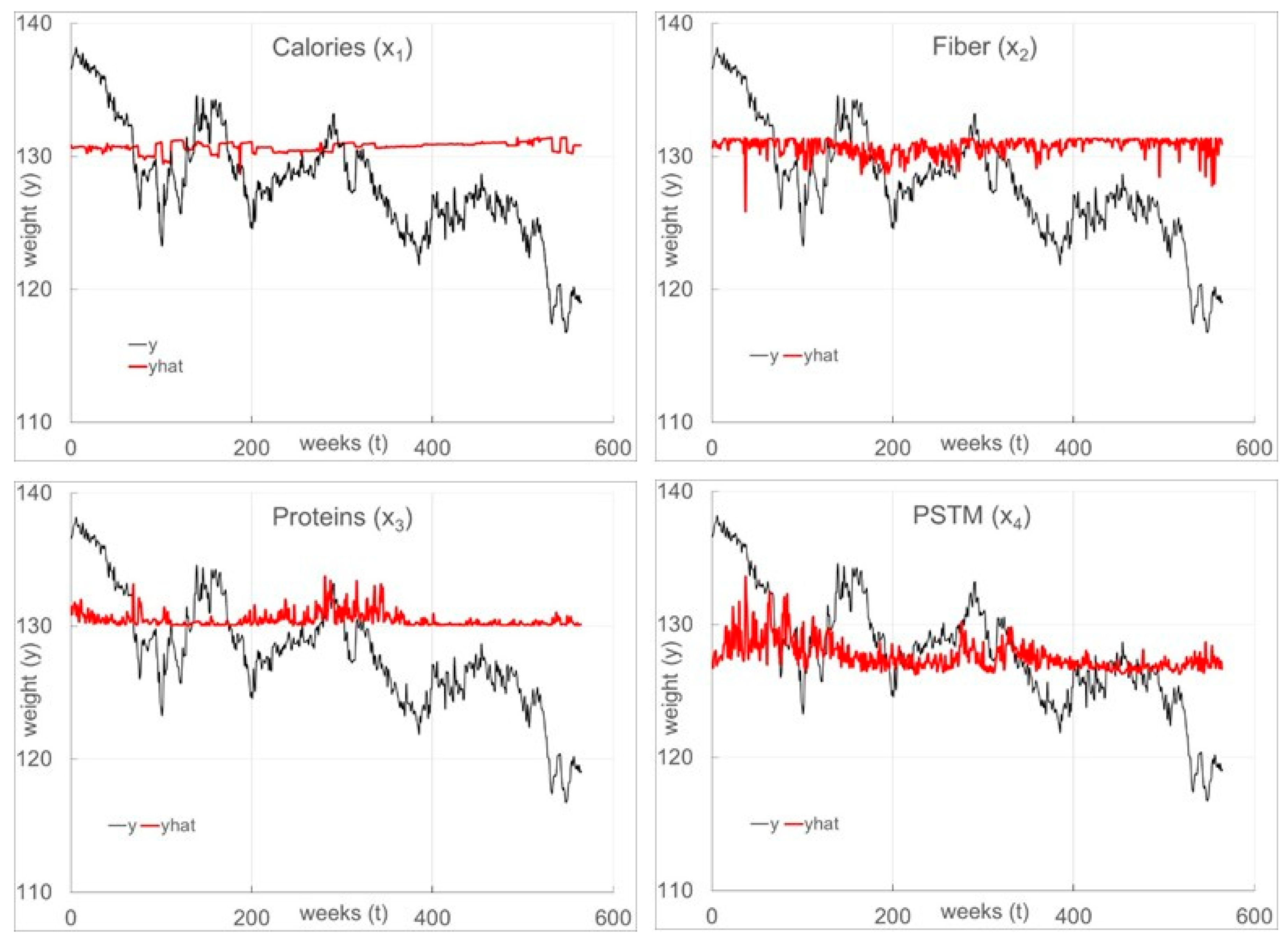
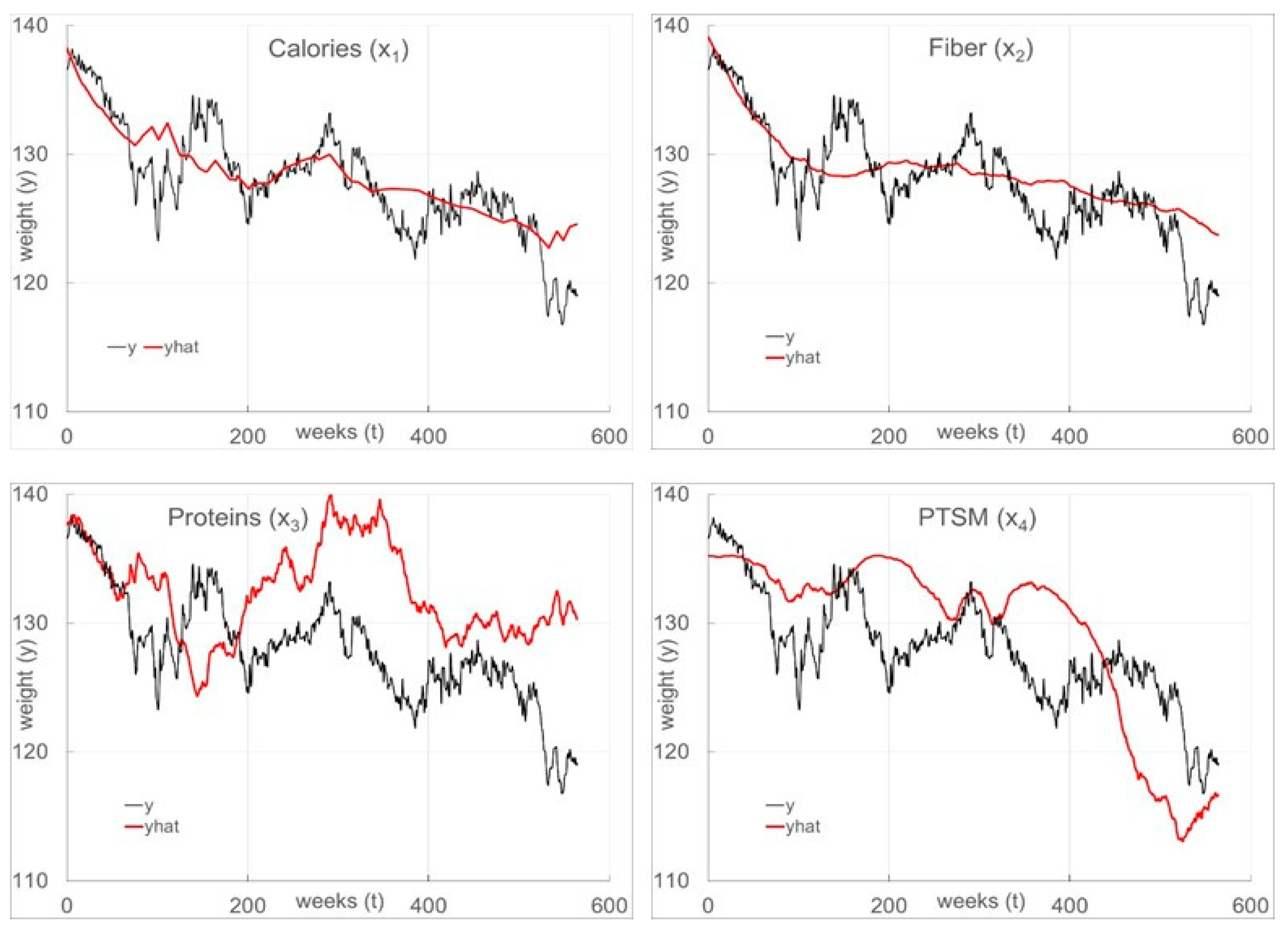
As shown in
Table 3 (
SLR) and
Table 4 (
TDR), with all four inputs in the model (the expectation functions are given in Equations (23) and (24), respectively), the structures consist of all eight (8) linear and quadratic terms. Note that
vi is determined from
xi based on Equation (11). As indicated in
Table 4, the range for
τ is 16 (protein) to 954 weeks. The plots showing the fits for both methods with all four inputs in the model are given in
Figure 5. The
TDR fit is excellent given the freely existing nature of this data set in contrast to the very poor
SLR fit.
3.2. Distillation Data Sets Results
The weight case example in the previous section is static modeling versus dynamic modeling when the process is dynamic. This section compares dynamic methods,
NARMAX versus
TDR, using data generated by a real pilot distillation column obtained in [
29].
NARMAX is a specific class of
DR because of its empirical nature due to it using a lag variable
. As a lag-based approach,
NARMAX has all the limitations of
DR in relation to
TDR as mentioned above. In addition, since its structure is mathematically based and not physically based, obtaining good starting values can be significantly more challenging.
Ref. [
30] randomly obtained ten (10) freely existing pilot distillation column data sets over a three-year period, including nine (9) inputs and one output. One data set was used for training, one for validation, and eight for testing. These data sets were generated by undergraduate chemical engineering students for their unit operation lab course. For the training data set, Ref. [
30] could not find adequate starting values to obtain a
NARMAX lagged-based fit for any of the ten data sets using MATLAB
® (Version 8.3). However, Ref. [
29] derived the
NARMAX BDD transfer functions given by Equation (25) below. Note that Equations (18) (
Wiener) and (25) (
NARMAX) differ only in their denominators. More specifically,
Wiener BDD (Equation (18)) has a different denominator for each input, in contrast to
NARMAX BDD (Equation (25)), which has the same denominator for each input. Using Equation (25), Ref. [
29] successfully obtained training and validation
NARMAX-BDD-fitted models for the distillation column data sets using Excel
®. Note that while Equations (18) and (25) look similar, Equation (18) is a theoretically based modeling structure and is derived from first-principle modeling (e.g., a dynamic component mass balance) and Equation (25) is an empirically based modeling structure and cannot be derived from first-principle modeling but from a structure (e.g.,
NARMAX) using lagged variables.
After training/validating the model, the professor who oversees the lab arbitrarily (i.e., unbiasedly) obtained eight (8) test data sets over three years. The mean testing
rfit (i.e.,
rfit,ts) values for
Wiener TDR and
NARMAX BDD were 0.84 and 0.28, respectively (see
Table 5). Thus, in summary, for this distillation process, the
TDR (
WM) model was significantly better than the
NARMAX BDD (
NM) mode
l, and
NARMAX DR failed to model this process data.
3.3. Type 1 Diabetes Data Sets
Our third and final case presents the features and strengths of our TDR protocol for a forecast process control application. Our diabetes example serves this purpose and represents the first step in modeling this data set for a forecast control application. More specifically, the model structure for this case is a multiple-input, second-order, dynamic structure (see Equation (13)) with a first-order (i.e., linear) static structure. Thus, our objective is the completion of our first step, which is to obtain final or near-final estimates of the dynamic parameters using a first-order static function, the simplest one.
This third and final case consists of modeling
SGC (
y) for eleven (11) Type 1 diabetes data sets. Each one consists of about two weeks of free-living data, with a sensor glucose sampling rate of five (5) minutes. This context is feedback forecast control, where the controlled variable (
CV) is
SGC and the manipulated variable (
MV) is exogenous (i.e., infused) insulin with a dead-time of
θMV. Each data set has twelve inputs consisting of three (3) nutrients, seven (7) activity tracker variables, insulin infusion with bolus and basal injection amounts combined as one (1) input, and the
time of day (i.e., a 24 h clock). The first author’s research group collected these data sets about 14 years ago when the Medtronic glucose sensor required replacement every three to four days and had a much longer warm-up period than current devices which are replaced weekly. Thus, the amount of missing
SGC data is much higher than current Medtronic sensors. Subject-reported food logs for approximating carbohydrate, fat, and protein amounts are used. We note that while the activity tracker was innovative for this study at the time, it did not have the most critical sensor, in our opinion, heart rate. Moreover, its technology is considerably less advanced than today’s wearable activity devices and is thus obsolete by today’s standards. For more information about this study, see [
18]. The eleven data sets are available publicly on the homepage of the first author.
Basically, forecasting is the use of current information to predict a future outcome. Weather forecasting seeks to predict future weather conditions but not change them. We call this “forecast monitoring” (
FM). “Automatic forecast control” (A
FC), our objective, seeks to accurately predict the future value of the controlled variable (
CV) and then manipulate conditions in the present to drive the
CV to its target (i.e., set point, SP) effectively after this future time distance is reached. Thus,
AFC models must be “cause-and-effect” and developed and evaluated in outpatient studies. An
FM methodology that models a correlation structure will fail in an outpatient, free-living,
AFC study if it is not sufficiently “cause-and-effect”. All the
SGC models that we have found in the literature model the correlation structure (e.g., see [
30,
31,
32,
33]).
Ref. [
18] modeled these data sets using the second-order dynamic structure given by Equation (13) with the second-order, multiple linear regression, static structure given by Equation (12). They named this approach the Wiener Modeling Method (
WMM). Ref. [
17] extended
WMM to a coupled modeling method (
CMM) with the addition of unmeasured endogenous insulin. The
WMM and
CMM modeling results are very similar. However, as pointed out in [
17], the critical benefits of
CMM over
WMM are the inference of the unmeasured endogenous insulin concentration and a sounder phenomenological understanding and behavior.
The first two data sets in this work (weight and distillation) have static response surfaces that are highly first-order (i.e., linear). However, for the diabetes data sets, the static response surfaces are highly nonlinear and highly interactive. These attributes make modeling these eleven data sets considerably more challenging.
The approach of [
18], used for comparison with the proposed approach, had the following conditions and attributes in addition to the ones mentioned above. First, fifteen-minute meal announcements for all three food variables were used. Secondly, no dead-time was used for exogenous insulin or any inputs other than the three food variables. Thirdly, all modeling was carried out on one Excel
® worksheet. Fourthly, the following two-step model identification strategy was used to maximize validation,
rfit. In the first step, all the inputs and their dynamic parameters were fitted simultaneously using a first-order static model. Its best fit served as the starting point for the second step, i.e., to find the best full (i.e., second-order) model fit (see Equation (12)).
The following two procedures were the same for [
18] and the proposed
TDR approach for feedback control. Firstly, missing activity data were estimated by averaging the two values on both sides of a gap and filling in the gap with this value. Some gaps were several hours long. Secondly, cross-validation was used, with the first week as the training (
Tr) data set and the second week as the validation (
Val) data set, to guard against overfitting the models.
As our knowledge has evolved, we have come to recognize critical and unique challenges in modeling SGC that this proposed forecast modeling TDR (f-TDR) approach overcomes. The first, and perhaps most critical, is the dead-time application. An accurate method to estimate the dead-time for exogenous insulin (θMV) and the appreciation of the criticality of θMV as the modeled dead-time for all inputs, except those with announcements, was not known or understood by the original modelers. Insulin injection decreases blood glucose concentration (BGC) but not until an unknown time after injection. For these eleven subjects, insulin injections are highly correlated with increasing and high SGC because these subjects tended to inject insulin when their BGC was high or from the knowledge that it would be increasing. However, any model with θMV too small (as in the WMM and CMM studies) will have a physiologically incorrect increase in SGC behavior with increasing insulin infusion (i.e., a positive correlation). It is critical to note that the WMM and CMM models had dead-time (i.e., θ > 0) for only the food variables, as mentioned above. Thus, this modeling condition alone makes the WMM and CMM approaches unacceptable for a control modeling application such as this one. It is critical to note that, in this context, θMV is not actually the exogenous insulin dead-time but what we call “the model effective dead-time,” which is the value that has the detectably correct phenomenological behavior and gives the best fit of the model to the data. Next, we explain how we estimate θMV.
The proposed f-TDR approach has the correct exogenous insulin (i.e., MV) cause-and-effect relationship for process control and significantly improves soundness, admissibility, and rfit, in comparison to the WMM and CMM studies, using the following novel modeling protocol. Firstly, combining bolus and basal insulin reduced the number of inputs by one. Secondly, with only the insulin input in the model, and starting with θMV equal to 60 min, we moved θMV one sample distance (i.e., 5 min) at a time in the direction of increasing rfit,val until rfit,val stopped increasing. Using this scheme, we estimated the model effective dead-time, θMV to be 60 min (12Δt), or close to this value, in all 11 modeling cases. Thirdly, for each input i, we used an f-TDR dead-time rule of except for the announcement inputs and time of day (since its value is known for any time in the future). This criterion is necessary because real-time model prediction is a forecasting application since an MV change does not detectably affect SGC until θMV time in the future. Fourthly, for each input, f-TDR obtains an SOPDTPL (i.e., Equation (13)) dynamic and first-order static fit (i.e., Equation (12) with one input) on its own (i.e., individual) Excel® worksheet. This simplest static structure is used because the focus is on obtaining good starting values for the dynamic parameters one input at a time and for each input separately.
As this process proceeds, the single-input fitted results are examined and evaluated based on fitting unique dynamic information in the data. To maximize this objective, we applied a strategy of using different starting values. Fifthly, each input’s fitted first-order static parameter and dynamic parameters are transferred to one Excel® worksheet as starting values for fitting the multiple-input model.
For each subject, starting with carbohydrates, then insulin, then the two other food inputs, then the tracker inputs, and, lastly, time of day, a multiple-input, second-order dynamic (i.e., Equation (13) for each input) and a first-order static (i.e., Equation (12) with αij = 0, ) fitted structure is obtained in two steps. The first step, one input at a time, varies its dynamic parameters, the static intercept, and its first-order static parameter to improve the validation fit for the f-TDR multiple-input static model. The input is retained in the model if, and only if, rfit,val improves. The second step seeks to further improve rfit,val by allowing all retained static parameters to change with all the dynamic parameters fixed.
Ref. [
18] modeling results using the (
WMM) full model static, multiple-input structure and our first-order static, multiple-input
f-TDR structure are given in
Table 6. Keep in mind, as explained above, that the W
MM approach of [
18] is not admissible in this forecast control application. As
Table 6 shows,
WMM, even with its full static and
versus
modeling advantages, the mean
f-TDR rfit,val is significantly higher, more specifically, 0.68 versus 0.62. In addition, the smallest
f-TDR rfit,val is 0.59 versus 0.43 for
WMM.
For one
f-TDR case, Subject 11, we fit a static linear regression model structure with significant second-order interactions and third-order main effects. It resulted in an
rfit,val of 0.79, significantly better than 0.74 but not substantially better. In addition, we performed it one term at a time to keep from overextending the optimization tool, and thus, the modeling exercise required substantial time, expertise, and effort. Stepwise linear regression algorithms [
34] cannot be used in this application because, to our knowledge, none exist for cross-validation. However, since artificial neural network (
ANN) algorithms are designed for nonlinear static modeling and inherently use cross-validation, our future work to improve the fit of these eleven (11) data sets will focus on new ways to apply
ANN algorithms and tools to extend this
f-TDR approach to nonlinear dynamic and nonlinear static Wiener
BDD modeling structures using a cross-validation tool-box. This work will use a new and novel
physically informed dynamic nonlinear modeling approach [
35,
36].