reflimLOD: A Modified reflimR Approach for Estimating Reference Limits with Tolerance for Values Below the Lower Limit of Detection (LOD)
Abstract
1. Introduction
2. Problem Formulation
- (i)
- Values from the healthy population follow a (truncated) normal distribution , i.e., X is a random variable following a normal distribution truncated at with expected value and variance after a suitable Box–Cox transformationwith parameter . For , the truncation is necessary because the Box–Cox transformation can only be applied to positive values. We assume that the truncation has a negligible effect, i.e., , where is the cumulative distribution function of the standard normal distribution. A Yeo–Johnson transformation [8] instead of the Box–Cox transformation could avoid the artificial truncation. However, it is common practice in laboratory medicine to stay with the Box–Cox transformation as analyte concentrations cannot be negative.
- (ii)
- There is a limit of detection , i.e., for values below the , we cannot measure their exact value. We only have the information that the corresponding values are smaller than the LOD. We assume that less than 50% of the values from the healthy population are below the .
- (iii)
- Apart from the values from the healthy population, there can be additional pathological values. We make no specific assumption on the distribution of the pathological values except that they tend to be higher than the values from the healthy population and that no values from diseased individuals are below the (see Section 6.2). We also assume that there are fewer pathological values than from the healthy population.
3. Related Work
4. A Modified Version of reflimR-Tolerating Values Below LOD
- (a)
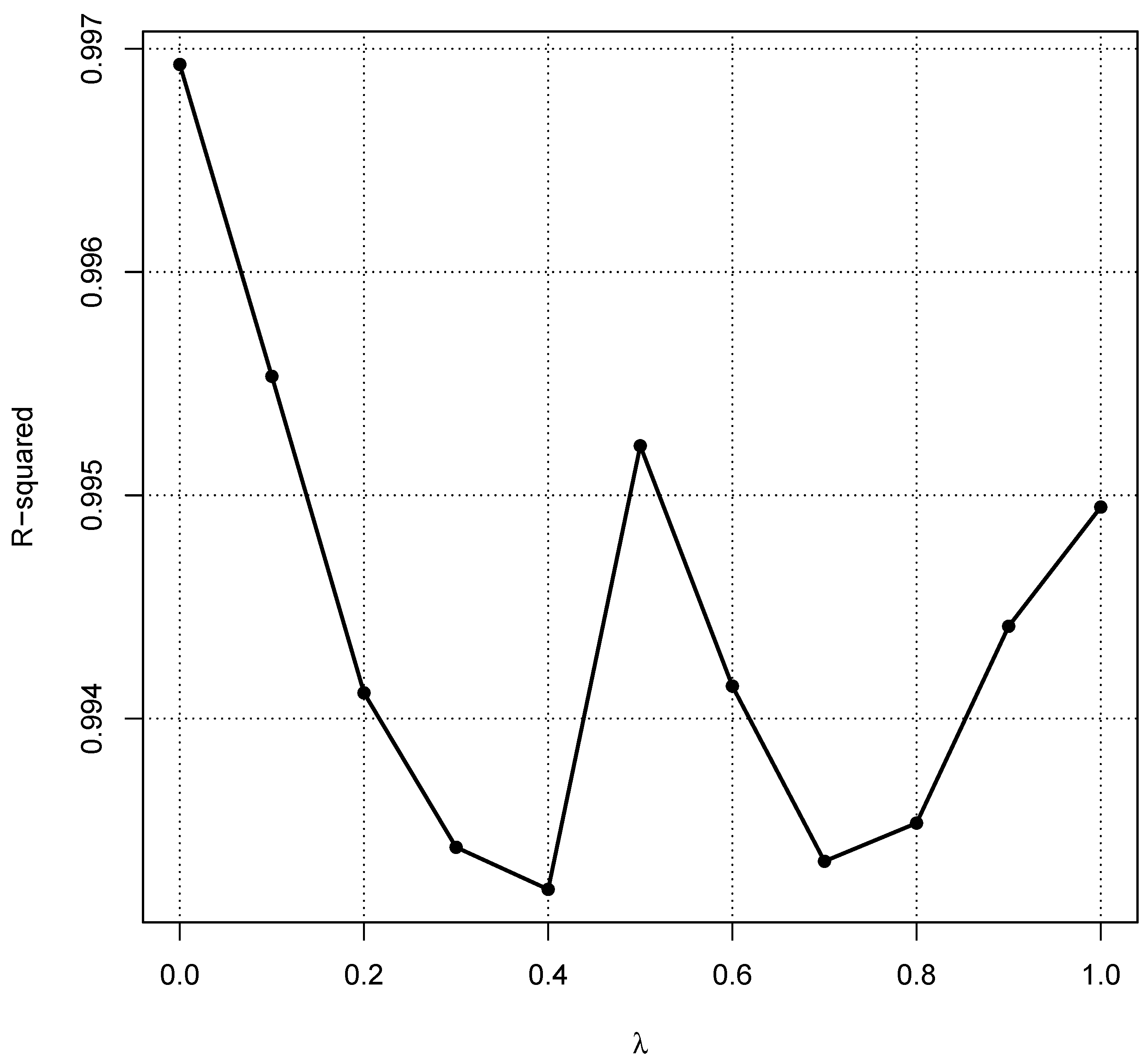
- In [14], it was suggested that the log-normal distribution should be the default assumption for the healthy population. LODreflimR either follows this recommendation and log-transforms the data or it provides estimated reference intervals for different values of the parameter for the Box–Cox transformation and uses modified quantile–quantile plots to decide which value of should be chosen. Details on the choice of will be provided in Section 4.4.
- (b)
- The truncation algorithm is modified. The truncation of the standard version of reflimR is based on the differences of the median to the first and third quartiles, i.e., the smaller difference is used to iteratively estimate the 2.5 and 97.5 percentiles of the healthy population. The LOD version uses only the difference between the third quartile and the median to iteratively estimate the 97.5 percentile of the healthy population. When there are more than 25% of the values below the LOD, the first quartile cannot be computed anyway. In contrast to the standard version of reflimR, only values beyond the estimated 97.5 percentile are discarded, i.e., all small values including those below the are kept. The underlying assumption is that there are no pathological small values. The modified truncation algorithm is described in Section 4.1.
- (c)
- The output of the standard version of reflimR directly provides estimates for the 2.5 and 97.5 percentiles of the healthy population by the truncation limits. The version only provides an estimate for the upper truncation limit, i.e., an estimate for the 97.5 percentile but not for the 2.5 percentile. In Section 4.2, a very simple approach based on the together with the upper truncation limit is introduced to obtain an estimate for the reference interval. Section 4.3 describes as an alternative: a maximum likelihood approach using all values between the and the upper truncation limit.
4.1. Modified Truncation Algorithm
| Algorithm 1 The modified truncation algorithm. | ||
| 1: | procedure modTrunc(x,,) | ▹ Input x: measured values |
| 2: | ▹: No. of values | |
| 3: | ▹: Level of detection | |
| 4: | ▹ The value is appended | |
| 5: | ▹ times to x | |
| 6: | median(y) | ▹ Median of the measured values including s |
| 7: | quantile(y,0.75) | ▹ 75 percentile of the measured values |
| 8: | ▹ Including s | |
| 9: | ▹ Value for the upper truncation | |
| 10: | ▹ Remove all values from y. | |
| 11: | ▹ Equation (11) | |
| 12: | ▹ Equation (6) | |
| 13: | while do | ▹ Continue until no more values are truncated. |
| 14: | ||
| 15: | quantile() | |
| 16: | quantile() | |
| 17: | ||
| 18: | ▹ Remove all values from y. | |
| 19: | end while | |
| 20: | ▹ Remove the artificial values below . | |
| 21: | return | ▹ The non-truncated values and the upper truncation limit |
| 22: | end procedure | |
4.2. Quantile-Based Estimator: reflimLOD.Quant
- An estimate of the 97.5 percentile of the healthy population, beyond which all values are truncated.
- The measured values above the LOD and below .
- The value for the .
- The number of values below the .
4.3. Maximum Likelihood Estimator: reflimLOD.MLE
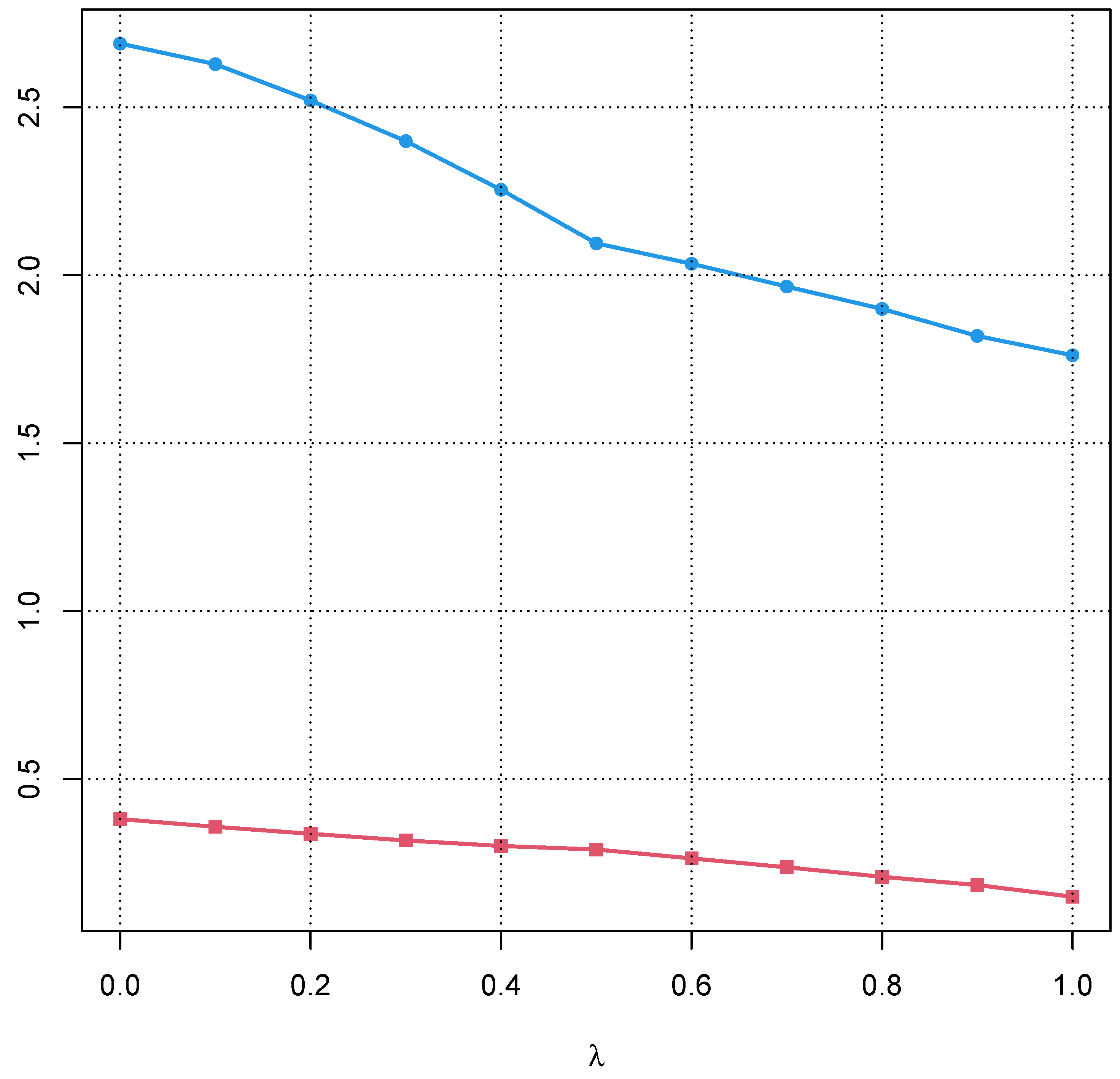
4.4. Choice of
- The healthy population follows a log-normal distribution with parameters and , i.e., the log-values follow a normal distribution with expected value and standard deviation .
- is chosen, so that almost 25% of the healthy population have values below the .
- Values from a pathological population are added following a log-normal distribution with parameters and for the associated normal distribution.
- Altogether, 500 values are generated, 400 from the healthy population and 100 from the pathological population.
5. Analysis and Validation
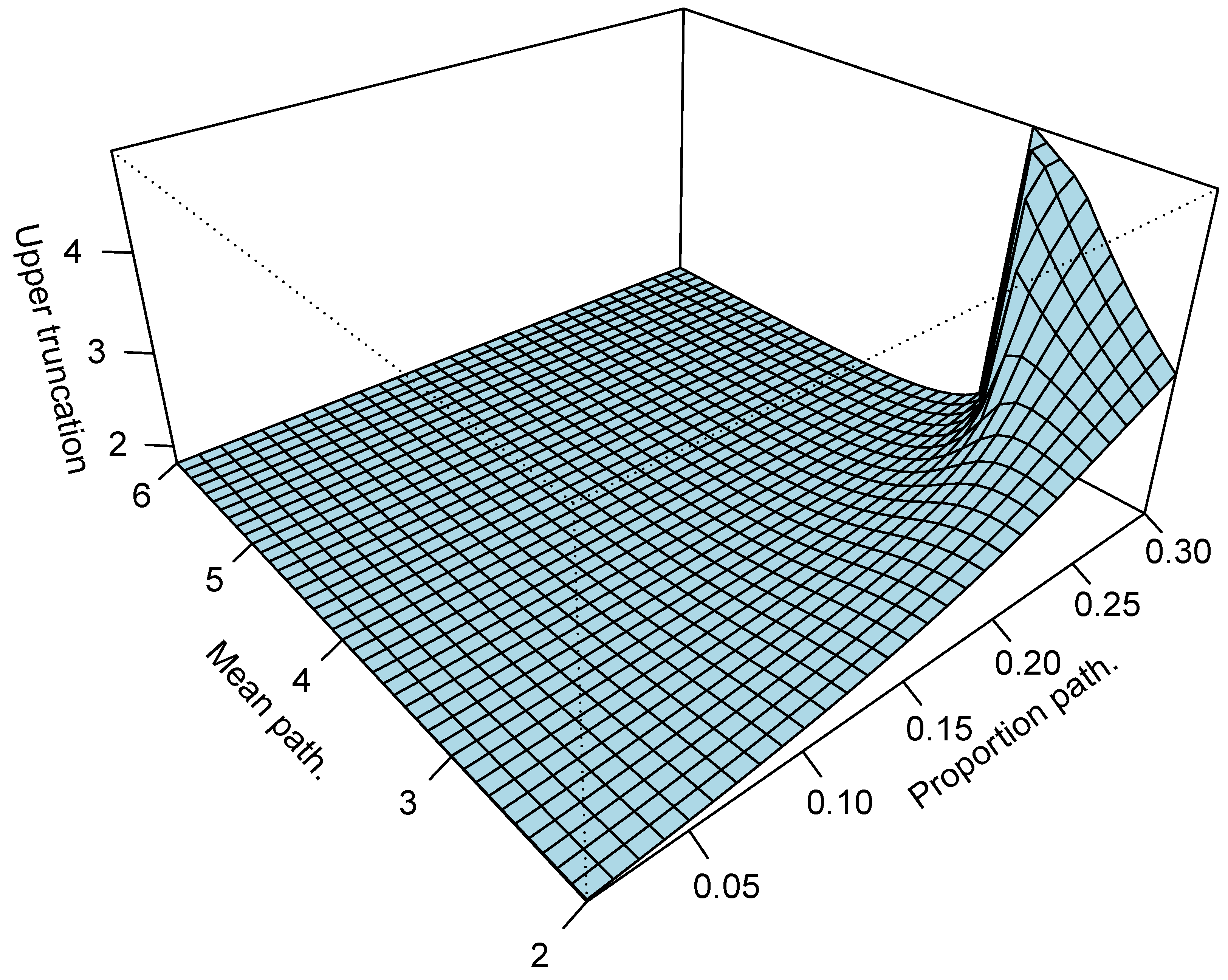
5.1. Bias of the Sample Interquartile Range
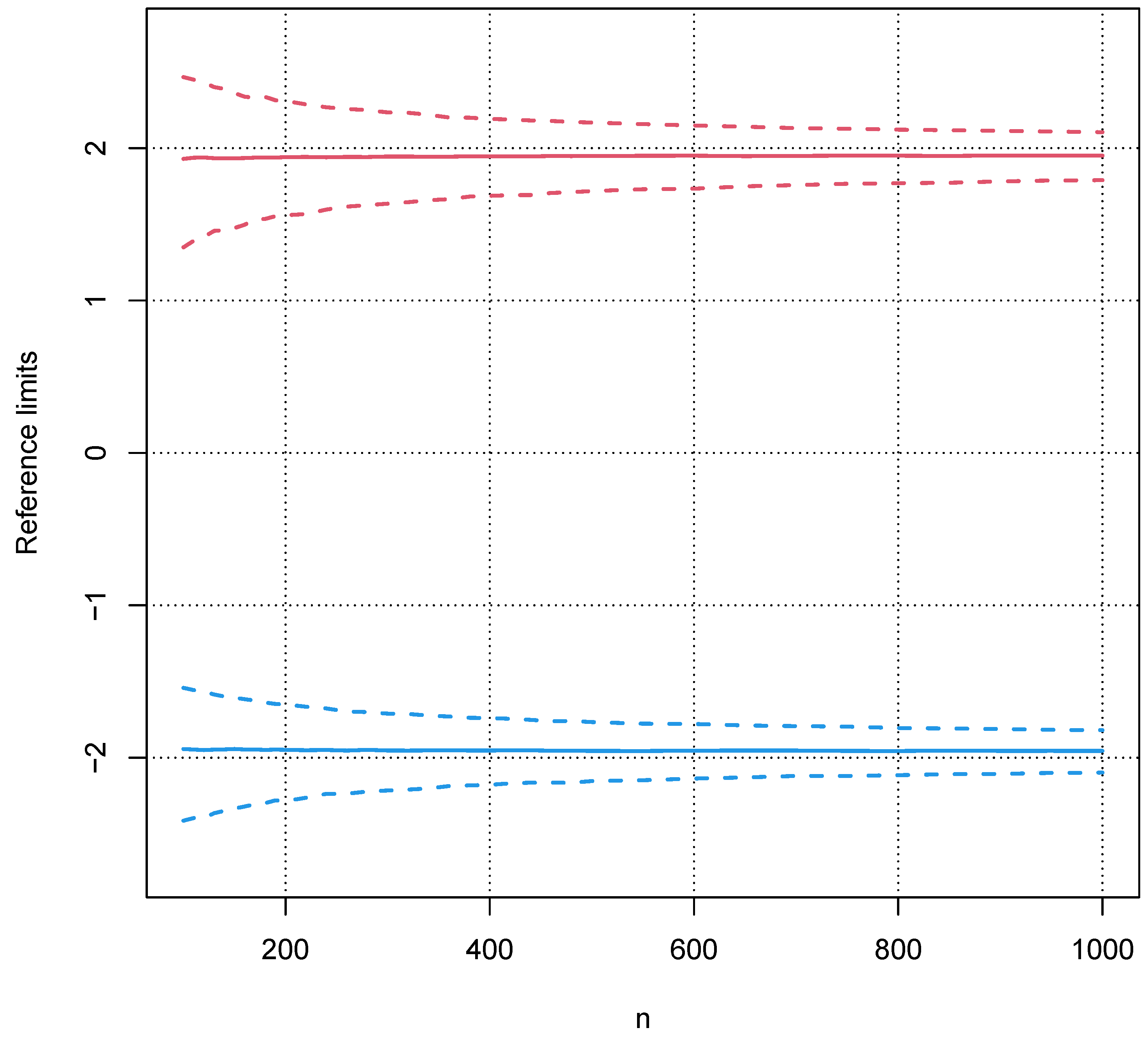
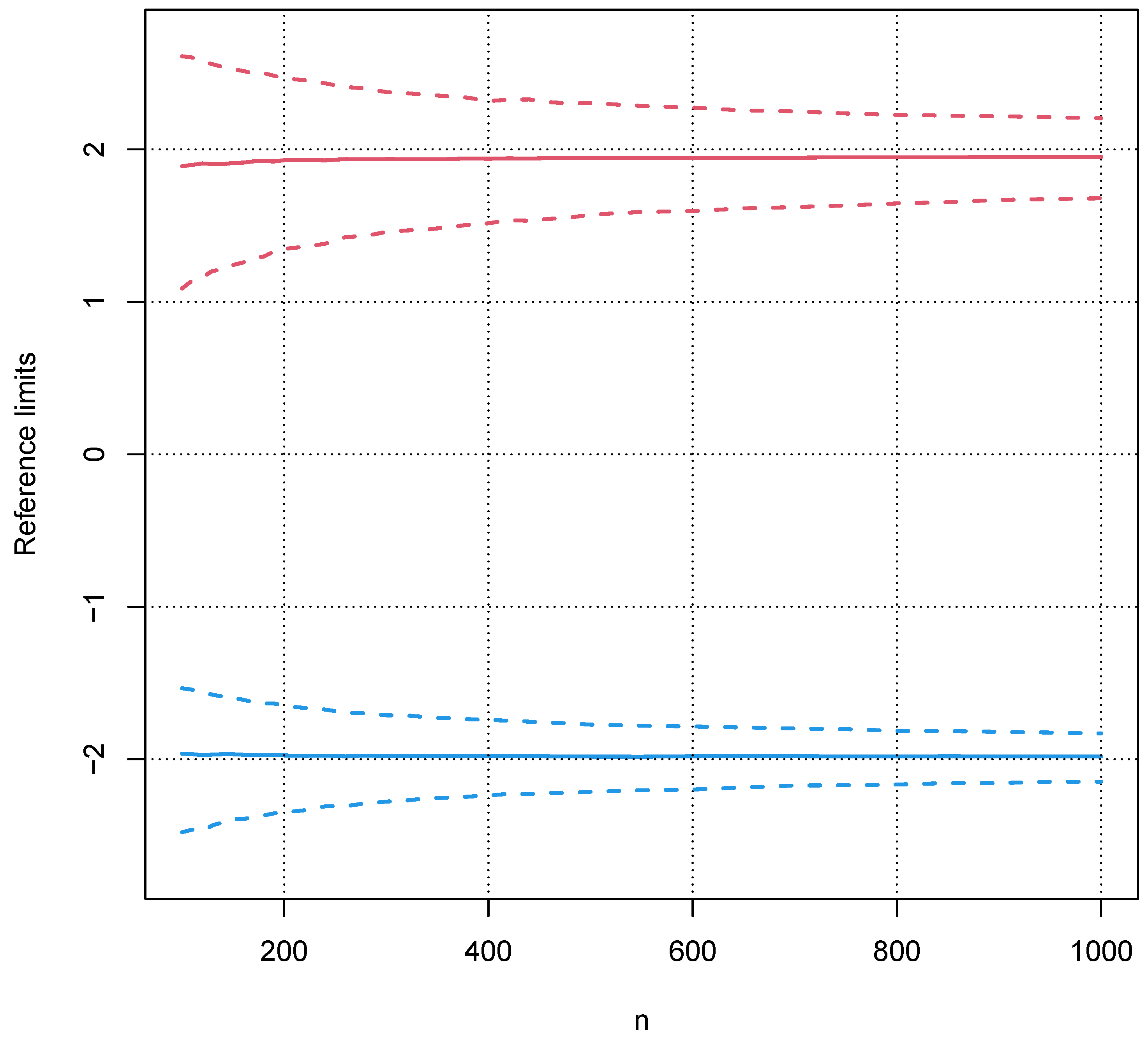
5.2. Influence of a Pathological Population on the Truncation Algorithm
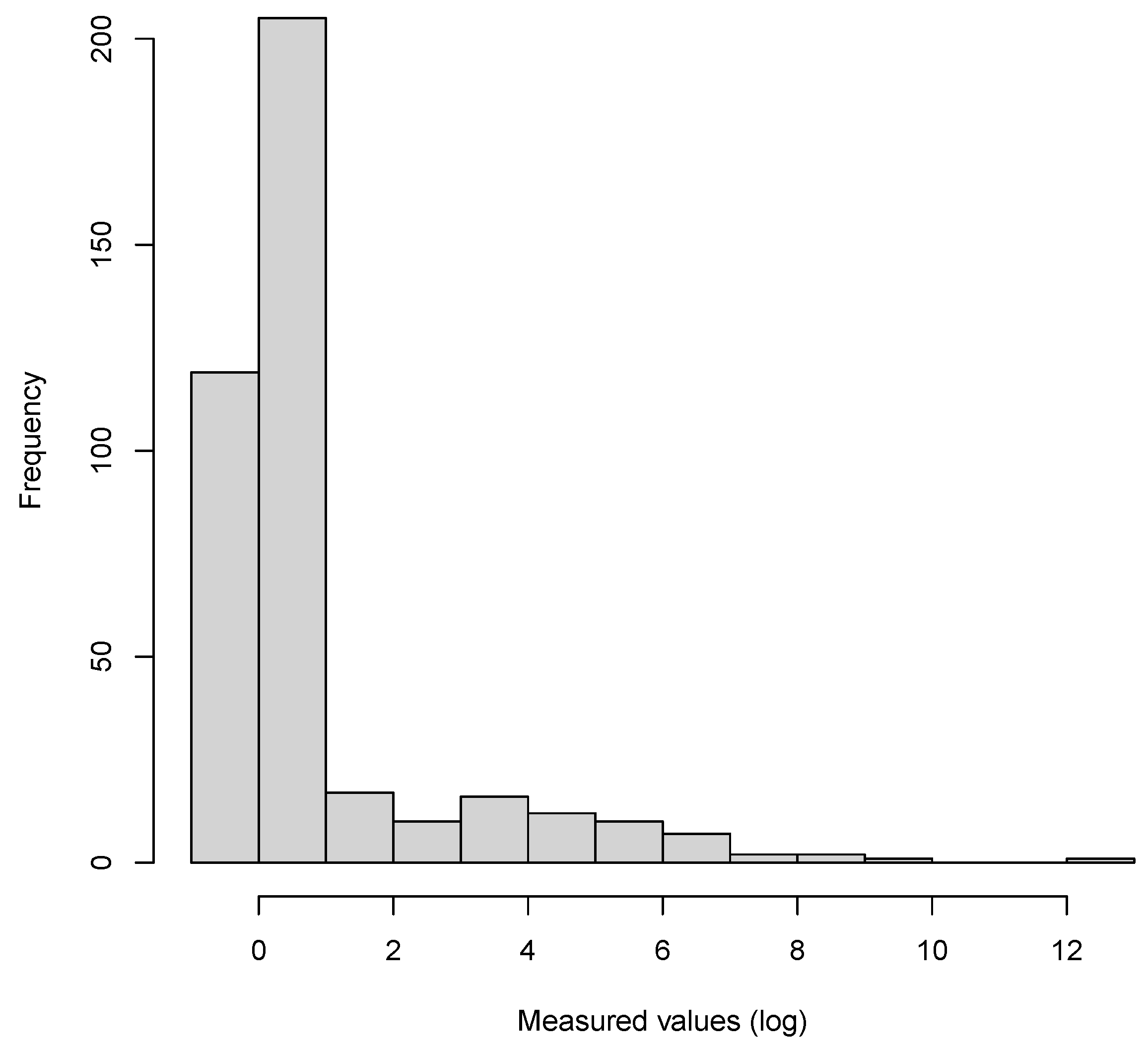
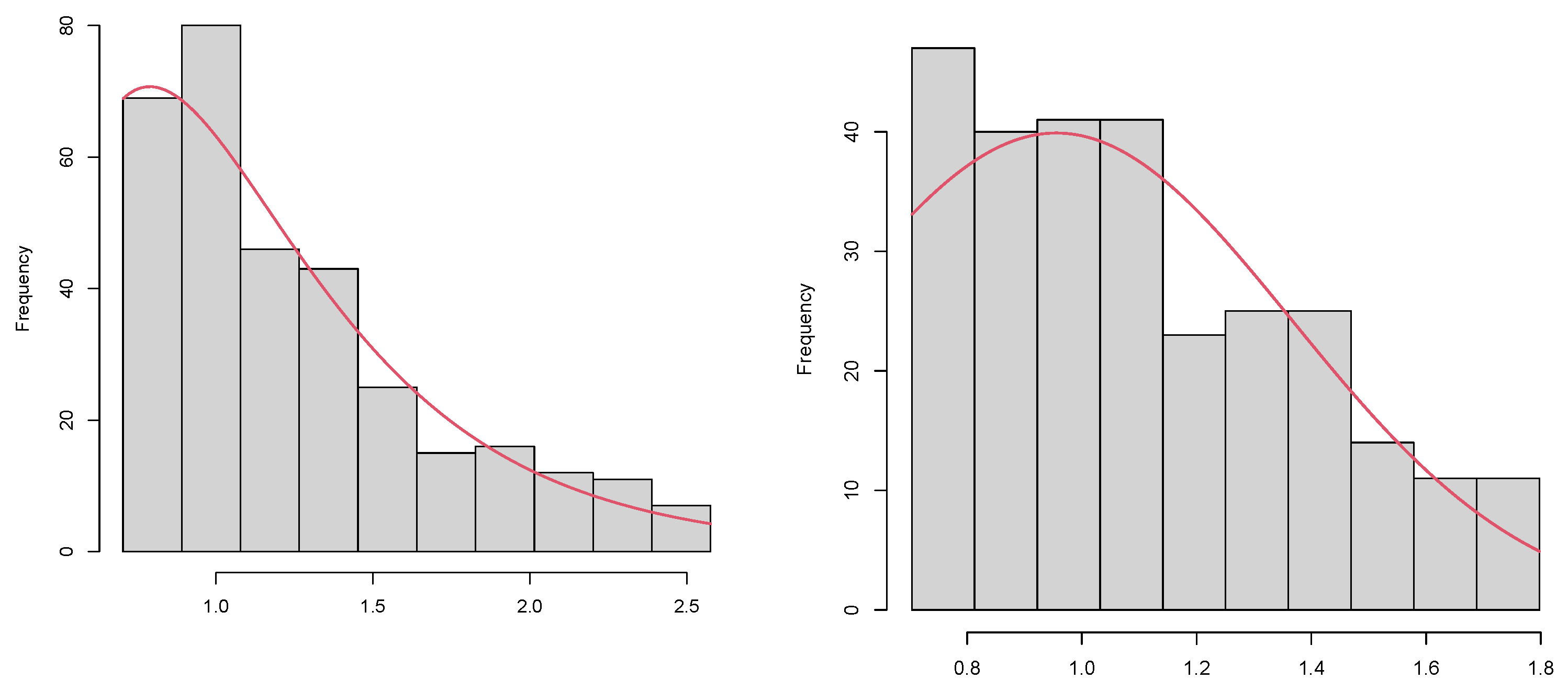
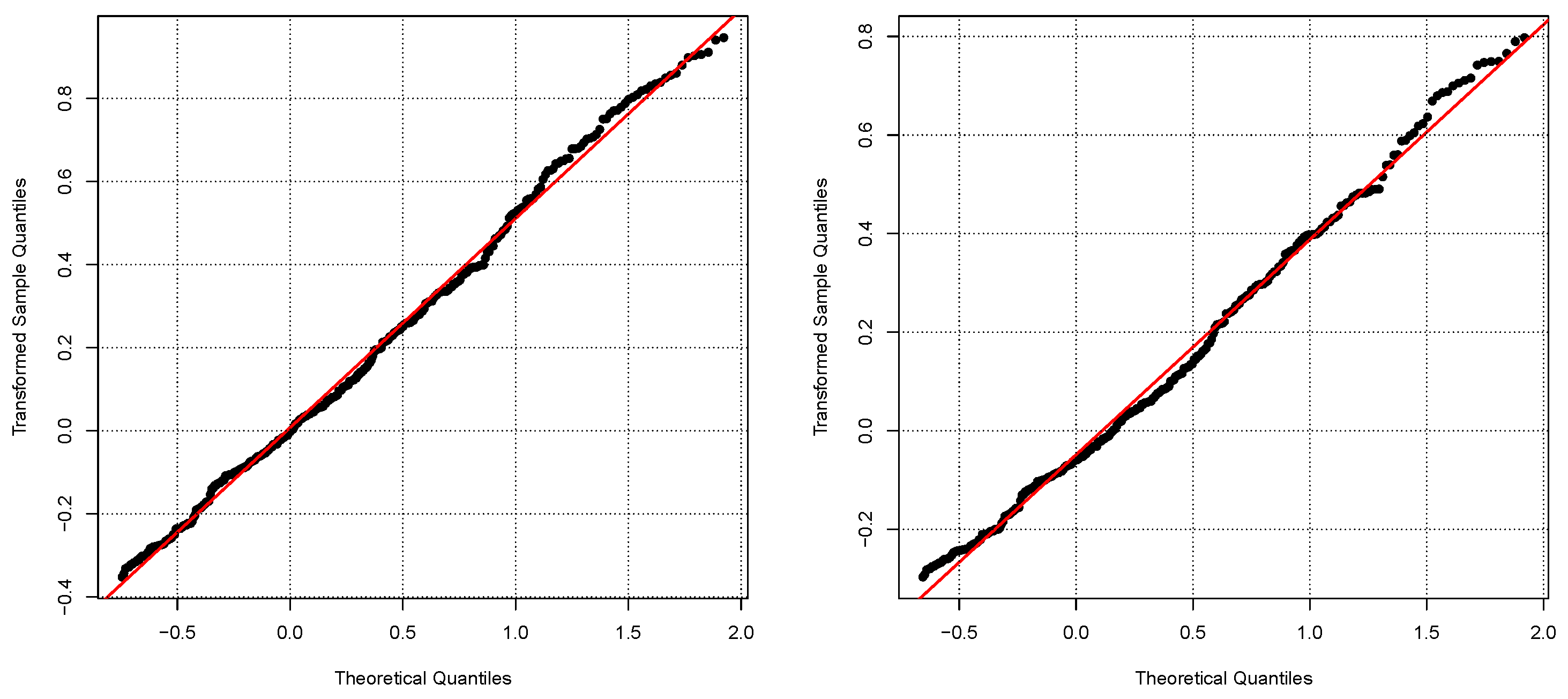
5.3. Validation with Real Data
6. Conclusions and Discussion
6.1. Conclusions
6.2. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Horowitz, G.; Altaie, S.; Boyd, J.C.; Ceriotti, F.; Garg, U.; Horn, P.; Pesce, A.; Sine, H.E.; Zakowski, J. Defining, Establishing, and Verifying Reference Intervals in the Clinical Laboratory; Tech Rep Document EP28-A3C; Clinical & Laboratory Standards Institute: Wayne, PA, USA, 2010. [Google Scholar]
- Jones, G.; Haeckel, R.; Loh, T.; Sikaris, K.; Streichert, T.; Katayev, A.; Barth, J.; Ozarda, Y. Indirect methods for reference interval determination: Review and recommendations. Clin. Chem. Lab. Med. 2018, 57, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, G.; Klawitter, S.; Trulson, I.; Adler, J.; Holdenrieder, S.; Klawonn, F. A novel tool for the rapid and transparent verification of reference intervals in clinical laboratories. J. Clin. Med. 2024, 13, 4397. [Google Scholar] [CrossRef]
- Ichihara, K.; Boyd, J.C. IFCC Committee on Reference Intervals and Decision Limits (C-RIDL). An appraisal of statistical procedures used in derivation of reference intervals. Clin. Chem. Lab. Med. 2010, 48, 1537–1551. [Google Scholar] [CrossRef] [PubMed]
- Sikaris, K. Separating disease and health for indirect reference intervals. J. Lab. Med. 2021, 45, 55–68. [Google Scholar] [CrossRef]
- Ammer, T.; Schützenmeister, A.; Prokosch, H.U.; Rauh, M.; Rank, C.M.; Zierk, J. refineR: A Novel Algorithm for Reference Interval Estimation from Real-World Data. Sci. Rep. 2021, 11, 16023. [Google Scholar] [CrossRef] [PubMed]
- Haeckel, R.; Wosniok, W.; Torge, A.; Junker, R. Reference limits of high-sensitive cardiac troponin T indirectly estimated by a new approach applying data mining. A special example for measurands with a relatively high percentage of values at or below the detection limit. J. Lab. Med. 2021, 45, 87–94. [Google Scholar] [CrossRef]
- Yeo, I.-K.; Johnson, R.A. A new family of power transformations to improve normality or symmetry. Biometrika 2000, 87, 954–959. [Google Scholar] [CrossRef]
- Wosniok, W.; Haeckel, R. A new indirect estimation of reference intervals: Truncated minimum chi-square (TMC) approach. Clin. Chem. Lab. Med. 2019, 57, 1933–1947. [Google Scholar] [CrossRef]
- Concordet, D.; Geffré, A.; Braun, J.P.; Trumel, C. A new approach for the determination of reference intervals from hospital-based data. Clin. Chim. Acta 2009, 405, 43–48. [Google Scholar] [CrossRef]
- Scott, D.W. Averaged shifted histogram. WIREs Comput. Stat. 2010, 2, 160–164. [Google Scholar] [CrossRef]
- Klawonn, F.; Riekeberg, N.; Hoffmann, G. Importance and uncertainty of λ-estimation for Box-Cox transformations to compute and verify reference intervals in laboratory medicine. Stats 2024, 7, 172–184. [Google Scholar] [CrossRef]
- Bowley, A.L. Elements of Statistics; P.S. King & Son: London, UK, 1901. [Google Scholar]
- Haeckel, R.; Wosniok, W. Observed, unknown distributions of clinical chemical quantities should be considered to be log-normal: A proposal. Clin. Chem. Lab. Med. 2010, 48, 1393–1396. [Google Scholar] [CrossRef] [PubMed]
- Nelder, J.A.; Mead, R. A simplex algorithm for function minimization. Comput. J. 1965, 7, 308–313. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Whaley, D.L. The Interquartile Range: Theory and Estimation. Electronic Theses and Dissertations. Master’s Thesis, East Tennessee State University, Johnson City, TN, USA, 2005. Paper 1030. Available online: https://dc.etsu.edu/etd/1030 (accessed on 7 July 2024).
- Zierk, J.; Arzideh, F.; Kapsner, L.A.; Prokosch, H.-U.; Metzler, M.; Rauh, M. Reference interval estimation from mixed distributions using truncation points and the Kolmogorov-Smirnov distance (kosmic). Sci. Rep. 2020, 10, 1704. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, G.; Allmeier, N.; Kuti, M.; Holdenrieder, S.; Trulson, I. How Gaussian mixture modelling can help to verify reference intervals from laboratory data with a high proportion of pathological values. J. Lab. Med. 2024, 48, 251–258. [Google Scholar] [CrossRef]
- Haeckel, R.; Wosniok, W.; Arzideh, F. Equivalence limits of reference intervals for partitioning of population data. Relevant differences of reference limits. LaboratoriumsMedizin 2016, 40, 199–205. [Google Scholar] [CrossRef]
- Hoffmann, G.; Klawonn, F.; Lichtinghagen, R.; Orth, M. The zlog value as a basis for the standardization of laboratory results. J. Lab. Med. 2017, 41, 20170135. [Google Scholar] [CrossRef]
- Anker, S.; Morgenstern, J.; Adler, J.; Brune, M.; Brings, S.; Fleming, T.; Kliemank, E.; Zorn, M.; Fischer, A.; Szendroedi, J.; et al. Verification of sex- and age-specific reference intervals for 13 serum steroids determined by mass spectrometry: Evaluation of an indirect statistical approach. Clin. Chem. Lab. Med. 2023, 61, 452–463. [Google Scholar] [CrossRef]
- Ozarda, Y.; Sikaris, K.; Streichert, T.; Macri, J. Distinguishing reference intervals and clinical decision limits—A review by the IFCC committee on reference intervals and decision limist. Crit. Rev. Clin. Lab. Sci. 2018, 55, 420–431. [Google Scholar] [CrossRef]
- Ceriotti, F.; Henny, J. Are my laboratory results normal? Considerations to be made concerning reference intervals and decision limits. EJIFCC 2008, 19, 106–114. [Google Scholar]
- Virani, S.S.; Aspry, K.; Dixon, D.L.; Ferdinand, K.C.; Heidenreich, P.A.; Jackson, E.J.; Jacobson, T.A.; McAlister, J.L.; Neff, D.R.; Gulati, M.; et al. The importance of low-density lipoprotein cholesterol measurement and control as performance measures: A joint Clinical Perspective from the National Lipid Association and the American Society for Preventive Cardiology. J. Clin. Lipidol. 2023, 17, 208–218. [Google Scholar] [CrossRef]
- Rebelos, E.; Tentolouris, N.; Jude, E. The role of vitamin D in health and disease: A narrative review on the mechanisms linking vitamin D with disease and the effects of supplementation. Drugs 2023, 83, 665–685. [Google Scholar] [CrossRef]
- Lazar, D.R.; Lazar, F.-L.; Homorodean, C.; Cainap, C.; Focsan, M.; Cainap, S.; Olinic, D.M. High-sensitivity troponin: A review on characteristics, assessment, and clinical implications. Dis. Markers 2022, 2022, 9713326. [Google Scholar] [CrossRef]
- Helfenstein Fonseca, F.A.; de Oliveira Izar, M.C. High-sensitivity C-reactive protein and cardiovascular disease across countries and ethnics. Clinics 2016, 71, 235–242. [Google Scholar] [CrossRef]
- Al-Mekhlafi, A.; Klawitter, S.; Klawonn, F. Standardization with zlog values improves exploratory data analysis and machine learning for laboratory data. J. Lab. Med. 2024, 48, 215–222. [Google Scholar] [CrossRef]
- Cheng, Y.; Regnier, M. Cardiac troponin structure-function and the influence of hypertrophic cardiomyopathy associated mutations on modulation of contractility. Arch. Biochem. Biophys. 2017, 601, 11–21. [Google Scholar] [CrossRef]
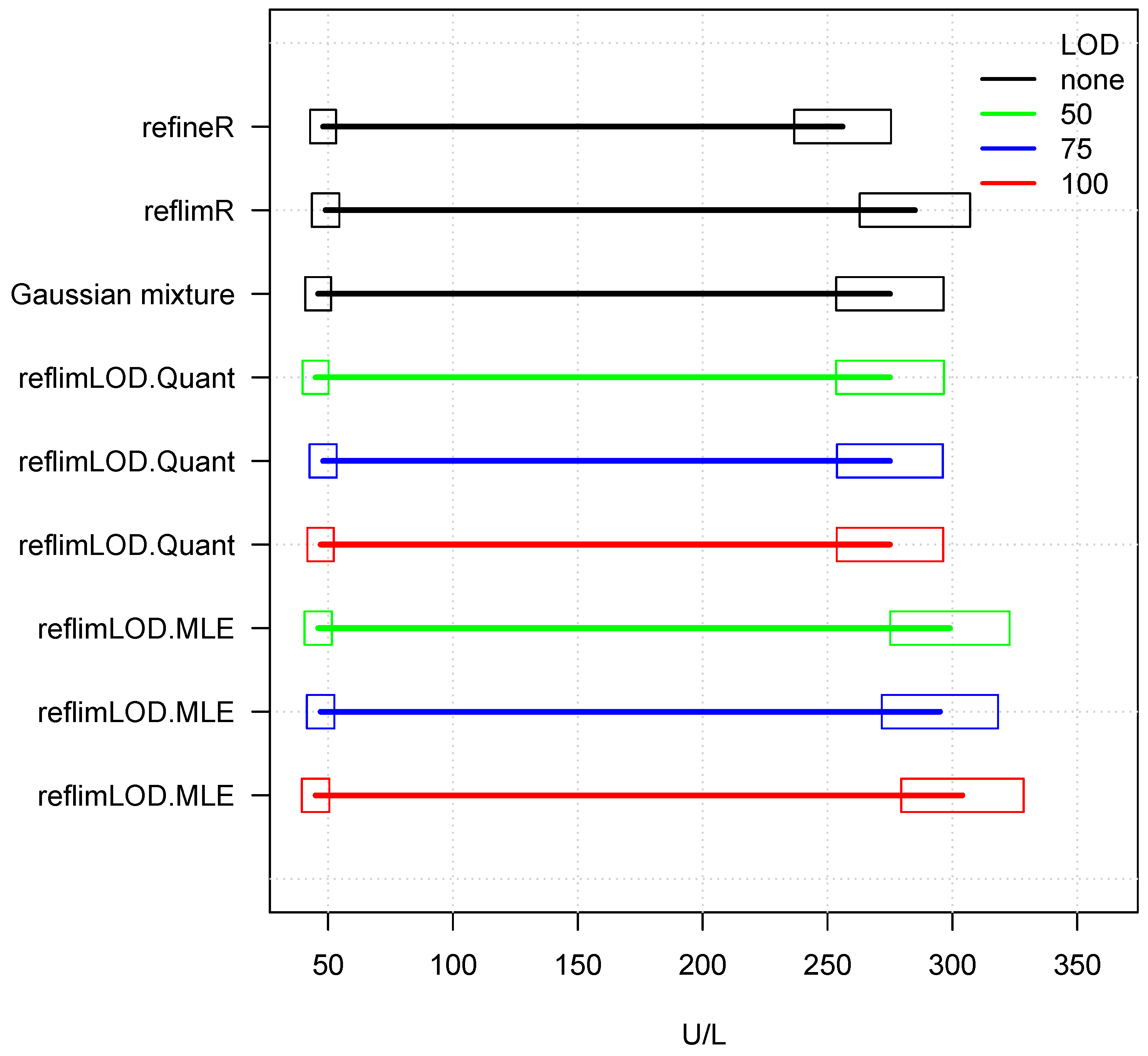
| Method | Lower Limit | Upper Limit | No. of Values | |
|---|---|---|---|---|
| refineR | 48 | 256 | 0 | 0 |
| reflimR | 49 | 285 | 0 | 0 |
| Gaussian mixture | 46 | 275 | 0 | 0 |
| reflimLOD.Quant | 45 | 275 | 50 | 33 |
| reflimLOD.Quant | 48 | 275 | 75 | 132 |
| reflimLOD.Quant | 47 | 275 | 100 | 296 |
| reflimLOD.MLE | 46 | 299 | 50 | 33 |
| reflimLOD.MLE | 47 | 295 | 75 | 132 |
| reflimLOD.MLE | 45 | 304 | 100 | 296 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klawonn, F.; Hoffmann, G.; Holdenrieder, S.; Trulson, I. reflimLOD: A Modified reflimR Approach for Estimating Reference Limits with Tolerance for Values Below the Lower Limit of Detection (LOD). Stats 2024, 7, 1296-1314. https://doi.org/10.3390/stats7040075
Klawonn F, Hoffmann G, Holdenrieder S, Trulson I. reflimLOD: A Modified reflimR Approach for Estimating Reference Limits with Tolerance for Values Below the Lower Limit of Detection (LOD). Stats. 2024; 7(4):1296-1314. https://doi.org/10.3390/stats7040075
Chicago/Turabian StyleKlawonn, Frank, Georg Hoffmann, Stefan Holdenrieder, and Inga Trulson. 2024. "reflimLOD: A Modified reflimR Approach for Estimating Reference Limits with Tolerance for Values Below the Lower Limit of Detection (LOD)" Stats 7, no. 4: 1296-1314. https://doi.org/10.3390/stats7040075
APA StyleKlawonn, F., Hoffmann, G., Holdenrieder, S., & Trulson, I. (2024). reflimLOD: A Modified reflimR Approach for Estimating Reference Limits with Tolerance for Values Below the Lower Limit of Detection (LOD). Stats, 7(4), 1296-1314. https://doi.org/10.3390/stats7040075







