Repeated-Measures Analysis in the Context of Heteroscedastic Error Terms with Factors Having Both Fixed and Random Levels
Abstract
:1. Introduction
2. Materials and Methods
2.1. An Illustrative Data Structure
2.2. Construction of a Linear Mixed-Effects Model in CRD
2.3. Linear Mixed-Effects Model in RMD
2.4. Model Assumptions
2.4.1. Sphericity (Circularity) Assumption
2.4.2. Compound Symmetry Assumption
2.5. Estimation Techniques
2.6. Methods of Inference
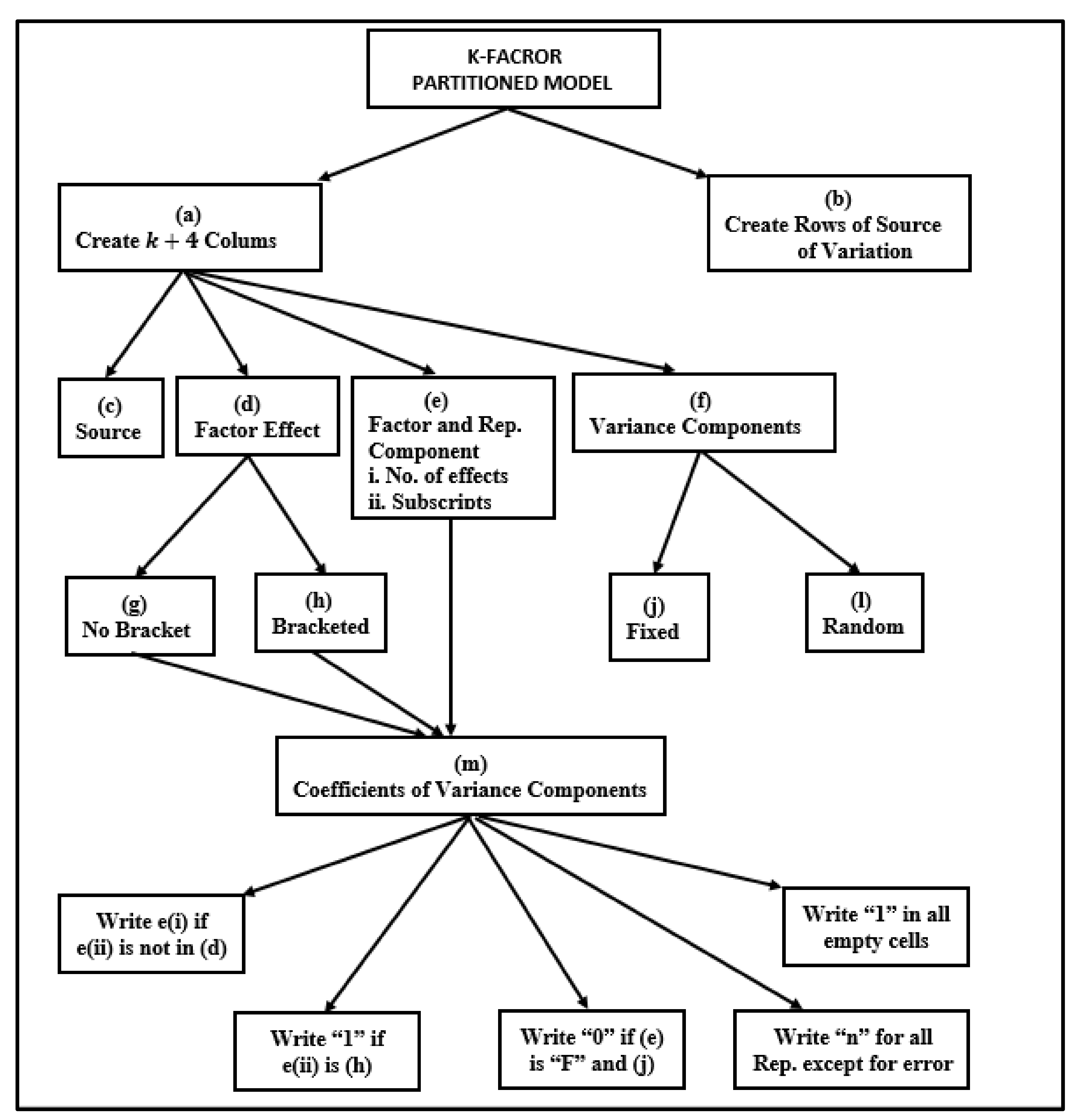
2.6.1. Process for Deriving Expected Mean Squares
- (a)
- Based on the model involved, construct a two-way table with column headings corresponding to the source of variation, effect labels, each of the subscripts included in the model, and row headings corresponding to each source of variation in the ANOVA table.
- (b)
- Above each subscript, write the associated number of factor levels and insert on top either an “F” if the factor levels are fixed or an “R” if the factor levels are random.
- (c)
- Create an extra column on the extreme right for the variance components corresponding to the source of variation, and insert the appropriate random variance component () or fixed variance component () for each source of variation.
- (d)
- Compare the column subscript and the factor effect in each row, and write the number of levels corresponding to that subscript if the column subscript is not included in the factor effect label. Otherwise, leave it blank.
- (e)
- For rows that have effects that contain bracketed subscripts, write a “1” under the column if the subscript is included in the bracket.
- (f)
- For each row that has a fixed variance component (), put a zero in the cell headed by an “F” when the subscript is included in the effect label.
- (g)
- Enter a “1” in all remaining blank cells.
- (h)
- To obtain the expected mean squares for each effect, identify all the variance components associated with that effect label. Cover the column(s) headed by the effect subscript(s) in that effect, and obtain the coefficient of each of the identified components from the product of the entries in the column(s) headed by the uncovered subscript(s). Include the variance component with the coefficient of 1 in the list.
2.6.2. Hypothesis Testing for Fixed Effects
2.6.3. Hypothesis Testing for Random Effects
2.6.4. Combined Analysis
3. Results
3.1. Checking Model Assumptions
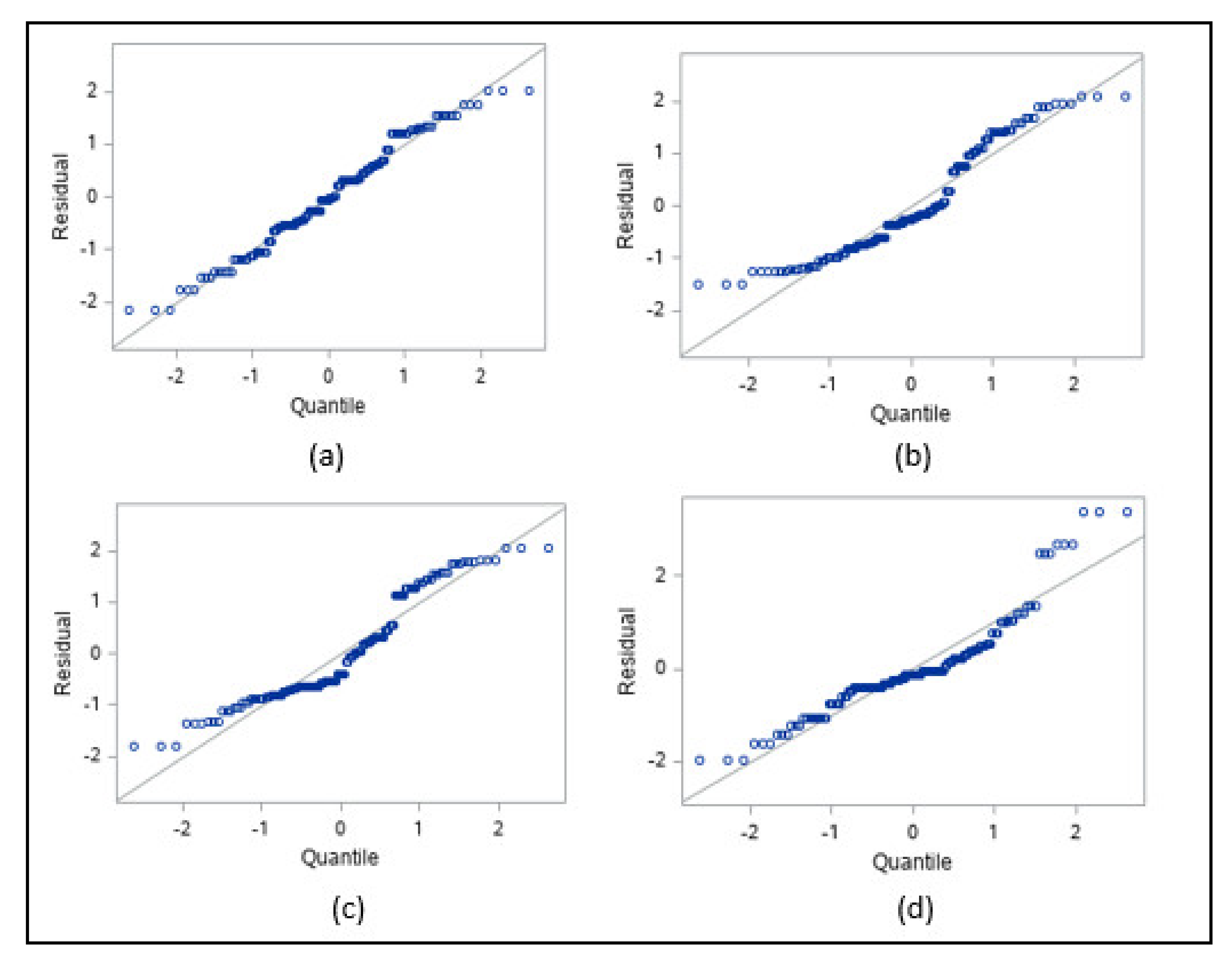
3.1.1. Normality and Outliers
3.1.2. Sphericity and/or Compound Symmetry
- FFF model: , , , significant;
- FRF:, , , significant;
- RFF: , , , significant;
- RRF: , , , significant.
3.2. Analysis of Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AIC | Akaike’s Information Criteria |
| ANOVA | Analysis of variance |
| AR(1) | First-order autoregressive |
| ARH(1) | Heterogeneous first-order autoregressive |
| CCl4 | Carbon tetrachloride |
| CHCl3 | Chloroform |
| CRD | Completely randomised design |
| CS | Compound symmetry |
| CSH | Heterogeneous compound symmetry |
| EU | Experimental unit |
| FFF | Fixed-Fixed-Fixed |
| FFR | Fixed-Fixed-Random |
| FRF | Fixed-Random-Fixed |
| FRR | Fixed-Random-Random |
| LHD | Lactic dehydrogenase |
| REML | Restricted maximum likelihood |
| RFF | Random-Fixed-Fixed |
| RFR | Random-Fixed-Random |
| RRF | Random-Random-Fixed |
| RRR | Random-Random-Random |
| SAS | Statistical Analysis Systems |
| VC | Variance components |
Appendix A
| SAS Code for Mixed-Model Analysis |
| /* Fitting FFR Model in Proc Mixed */ |
| FILENAME REFFILE ‘/home/u35581214/LDH Leakage Data.sav’; |
| PROC IMPORT DATAFILE = REFFILE |
| DBMS = SAV |
| OUT = LDH; |
| RUN; |
| /* View Repeated-Measures Data in Multivariate Form */ |
| Proc print data = LDH; |
| run; |
| /* Set Repeated-Measures Data to Univariate form */ |
| Data LDH_mult(keep = CCl4 CHCl3 Flask Time4 Time5 Time6) |
| LDH_univ(keep = CCl4 CHCl3 Flask Time Leakage); |
| set LDH; |
| output LDH_mult; |
| Leakage = Time4;Time = 1; output LDH_univ; |
| Leakage = Time5;Time = 2; output LDH_univ; |
| Leakage = Time6;Time = 3; output LDH_univ; |
| run; |
| /* View Data in Univariate and Multivariate Form */ |
| Proc print data = LDH_univ; |
| run; |
| Proc print data = LDH_mult; |
| run; |
| /* Subset or partition FRF from LHD univariate original Data */ |
| Data FRF; |
| set LDH_univ; |
| if (CCl4 = 2.5 AND CHCl3 = 0) then output; |
| if (CCl4 = 2.5 AND CHCl3 = 0) then output; |
| if (CCl4 = 2.5 AND CHCl3 = 0) then output; |
| if (CCl4 = 2.5 AND CHCl3 = 5) then output; |
| if (CCl4 = 2.5 AND CHCl3 = 5) then output; |
| if (CCl4 = 2.5 AND CHCl3 = 5) then output; |
| if (CCl4 = 5 AND CHCl3 = 0) then output; |
| if (CCl4 = 5 AND CHCl3 = 0) then output; |
| if (CCl4 = 5 AND CHCl3 = 0) then output; |
| if (CCl4 = 5 AND CHCl3 = 5) then output; |
| if (CCl4 = 5 AND CHCl3 = 5) then output; |
| if (CCl4 = 5 AND CHCl3 = 5) then output; |
| run; |
| Proc print data = FRF; |
| run; |
| /* Plot differences in leakages contributed by predictors */ |
| proc means noprint data = FRF nway; |
| var Leakage; |
| class CCl4 CHCl3 Flask Time; |
| output out = avgFRF mean = avgLeakage; |
| run; |
| proc print data = avgFRF; |
| run; |
| /* New data set called avg created*/ |
| /* Plot differences in leakage by predictor CHCl3 and Time */ |
| Proc gplot data = avgFRF; |
| plot avgLeakage*Time = CHCl3/haxis = 0 to 8 by 1 hminor = 0 vminor = 0; |
| symbol1 v = star c = blue i = join l = 1; |
| symbol2 v = plus c = red i = join l = 2; |
| title “Percentage leakage per time per CHCl3”; |
| run; Quit; |
| /* Partitioning FRF multivariate data for covariance analysis */ |
| Data FRF_mult; |
| set LDH_mult; |
| if (CCl4 = 2.5 AND CHCl3 = 0) then output; |
| if (CCl4 = 2.5 AND CHCl3 = 0) then output; |
| if (CCl4 = 2.5 AND CHCl3 = 0) then output; |
| if (CCl4 = 2.5 AND CHCl3 = 5) then output; |
| if (CCl4 = 2.5 AND CHCl3 = 5) then output; |
| if (CCl4 = 2.5 AND CHCl3 = 5) then output; |
| if (CCl4 = 5 AND CHCl3 = 0) then output; |
| if (CCl4 = 5 AND CHCl3 = 0) then output; |
| if (CCl4 = 5 AND CHCl3 = 0) then output; |
| if (CCl4 = 5 AND CHCl3 = 5) then output; |
| if (CCl4 = 5 AND CHCl3 = 5) then output; |
| if (CCl4 = 5 AND CHCl3 = 5) then output; |
| run; |
| Proc print data = FRF_mult; |
| run; |
| /* Sphericity Test using PROC MIXED */ |
| /* Sphericity test H0: Sphericity holds */ |
| proc mixed data = FRF method = reml cl ic covtest; |
| class CCl4 CHCl3 Time Flask; |
| model Leakage = CCl4|CHCl3; |
| random CHCl3 CCl4*CHCl3 CHCl3*Time CCl4*CHCl3*Time /s; |
| repeated / subject = Flask(CCl4*CHCl3) type = un; |
| run; |
| proc mixed data = FRF method = reml cl ic covtest; |
| class CCl4 CHCl3 Time Flask; |
| model Leakage = CCl4|CHCl3; |
| random CHCl3 CCl4*CHCl3 CHCl3*Time CCl4*CHCl3*Time /s; |
| repeated / subject = Flask(CCl4*CHCl3) type = HF; |
| run; |
| /* Normality Q–Q plots */ |
| ods graphics on; |
| proc mixed data = FRF plots = influenceestplot; |
| class CCl4 CHCl3 Time Flask; |
| model Leakage = CCl4 Time CCl4*Time/residual; |
| random CHCl3 CCl4*CHCl3 CHCl3*Time CCl4*CHCl3*Time; |
| repeated/subject = Flask(CCl4*CHCl3) type = cs r; |
| run; |
| ods graphics off; |
| /* Checking Covariance Structure */ |
| proc corr data = FRF_mult cov; |
| var Time4 Time5 Time6; |
| run; |
| /* Fit the model by PROC MIXED and compare covariance structures */ |
| proc mixed data = FRF method = reml cl ic covtest; |
| class CCl4 CHCl3 Time Flask; |
| model Leakage = CCl4 Time CCl4*Time/s; |
| random CHCl3 CCl4*CHCl3 CHCl3*Time CCl4*CHCl3*Time; |
| repeated/subject = Flask(CCl4*CHCl3) type = cs r; |
| run; |
| proc mixed data = FRF method = reml cl ic covtest; |
| class CCl4 CHCl3 Time Flask; |
| model Leakage = CCl4 Time CCl4*Time; |
| random CHCl3 CCl4*CHCl3 CHCl3*Time CCl4*CHCl3*Time/s; |
| repeated/subject = Flask(CCl4*CHCl3) type = arh(1) r; |
| lsmeans CCl4/pdiff cl adjust = tukey; |
| run; |
| proc mixed data = FRF method = reml cl ic covtest; |
| class CCl4 CHCl3 Time Flask; |
| model Leakage = CCl4 Time CCl4*Time; |
| random CHCl3 CCl4*CHCl3 CHCl3*Time CCl4*CHCl3*Time/s; |
| repeated/subject = Flask(CCl4*CHCl3) type = ar(1) r; |
| lsmeans CCl4/pdiff cl adjust = tukey; |
| run; |
| /* Scrapping Data for the Combined Model FB */ |
| Data FB_univ; |
| set LDH_univ; |
| if (CHCl3 = 10) then output; |
| if (CHCl3 = 25) then output; |
| run; |
| /* Scrapping Data for the FA x FB Combined Model */ |
| Data FAFB_univ; |
| set LDH_univ; |
| if (CCl4 = 2.5 and CHCl3 = 10) then output; |
| if (CCl4 = 2.5 and CHCl3 = 25) then output; |
| if (CCl4 = 5 and CHCl3 = 10) then output; |
| if (CCl4 = 5 and CHCl3 = 25) then output; |
| run; |
| Proc print data = FB_univ; |
| run; |
| /* Fitting the combined model FB (for narrow inferential scope) */ |
| proc mixed data = FB_univ method = reml cl ic covtest; |
| class CCl4 CHCl3 Time Flask; |
| model Leakage = CCl4 CHCl3 CCl4*CHCl3 Time CCl4*Time CHCl3*Time CCl4*CHCl3*Time/s; |
| repeated/subject = Flask(CCl4*CHCl3) type = arh(1) r; |
| run; |
| /* Fitting the combined model FB (for broad inferential scope) */ |
| proc mixed data = FB_univ method = reml cl ic covtest; |
| class CCl4 CHCl3 Time Flask; |
| model Leakage = CHCl3 Time CHCl3*Time/s; |
| random CCl4 CCl4*CHCl3 CCl4*Time CCl4*CHCl3*Time; |
| repeated/subject = Flask(CCl4*CHCl3) type = ar(1) r; |
| run; |
References
- Fitzmaurice, G.M.; Davidian, M.; Verbeke, G.; Molenberghs, G. (Eds.) Longitudinal Data Analysis; Chapman and Hall/CRC Handbooks of Modern Statistical Methods; Chapman & Hall/CRC: Boca Raton, FL, USA, 2009. [Google Scholar]
- Pan, J.; Shang, J.A. Simultaneous Variable Selection Methodology for Linear Mixed Models. J. Stat. Comput. Simul. 2018, 88, 3323–3337. [Google Scholar] [CrossRef]
- Demidenko, E. Mixed Models: Theory and Applications with R, 2nd ed.; Wiley Series in Probability and Statistics; Wiley: Hoboken, NJ, USA, 2013. [Google Scholar]
- Verbeke, G.; Molenberghs, G. Linear Mixed Models for Longitudinal Data; Springer Series in Statistics; Springer: New York, NJ, USA; Berlin, Heidelberg, 2000. [Google Scholar]
- Verbeke, G. The Linear Mixed Model. A Critical Investigation in the Context of Longitudinal Data Analysis. Ph.D. Dissertation, Catholic University of Leuven, Faculty of Sciences, Department of Mathematics,, Leuven, Belgium, 1995. [Google Scholar]
- Verbeke, G.; Lesaffre, E. The Effect of Misspecifying the Random-Effects Distribution in Linear Mixed Models for Longitudinal Data. Comput. Stat. Data Anal. 1997, 23, 541–556. [Google Scholar] [CrossRef]
- Davis, C.S. Statistical Methods for the Analysis of Repeated Measurements; Springer Texts in Statistics; Springer: New York, NY, USA, 2002. [Google Scholar]
- Muller, K.E.; Edwards, L.J.; Simpson, S.L.; Taylor, D.J. Statistical Tests with Accurate Size and Power for Balanced Linear Mixed Models. Statist. Med. 2007, 26, 3639–3660. [Google Scholar] [CrossRef] [PubMed]
- Hickey, G.L.; Mokhles, M.M.; Chambers, D.J.; Kolamunnage-Dona, R. Statistical Primer: Performing Repeated-Measures Analysis. Interact. Cardiovasc. Thorac. Surg. 2018, 26, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Matuschek, H.; Kliegl, R.; Vasishth, S.; Baayen, H.; Bates, D. Balancing Type I Error and Power in Linear Mixed Models. J. Mem. Lang. 2017, 94, 305–315. [Google Scholar] [CrossRef]
- Fitzmaurice, G.M.; Laird, N.M.; Ware, J.H. Applied Longitudinal Analysis, 2nd ed.; Wiley Series in Probability and Statistics; Wiley: Hoboken, NJ, USA, 2011. [Google Scholar]
- Chaka, L.; Njuho, P. Construction of a Linear Mixed Model with Each Factor Having Both Fixed and Random Levels: A Case of Split-Split-Plot Structure in a RCBD. Int. J. Agric. Stat. Sci. 2021, 17, 501–518. Available online: https://connectjournals.com/03899.2021.17.501 (accessed on 20 January 2022).
- Crowder, M.J.; Hand, D.J. Analysis of Repeated Measures, 1st ed.; Monographs on Statistics and Applied Probability; Chapman and Hall: London, UK; New York, NY, USA, 1990. [Google Scholar]
- Barnett, A.G.; Koper, N.; Dobson, A.J.; Schmiegelow, F.; Manseau, M. Using Information Criteria to Select the Correct Variance-Covariance Structure for Longitudinal Data in Ecology: Selecting the Correct Variance-Covariance. Methods Ecol. Evol. 2010, 1, 15–24. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Goonewardene, L.A. The Use of MIXED Models in the Analysis of Animal Experiments with Repeated Measures Data. Can. J. Anim. Sci. 2004, 84, 1–11. [Google Scholar] [CrossRef]
- Patterson, H.D.; Thompson, R. Recovery of Inter-Block Information When Block Sizes Are Unequal. Biometrika 1971, 58, 545–554. [Google Scholar] [CrossRef]
- Diffey, S.M.; Smith, A.B.; Welsh, A.H.; Cullis, B.R. A New REML (Parameter Expanded) EM Algorithm for Linear Mixed Models. Aust. N. Z. J. Stat. 2017, 59, 433–448. [Google Scholar] [CrossRef] [Green Version]
- Dempster, A.P.; Laird, N.M.; Rubin, D.B. Maximum Likelihood from Incomplete Data via the EM Algorithm. J. R. Stat. Society. Ser. B (Methodol.) 1977, 39, 1–22. [Google Scholar]
- Gennings, C.; Chinchilli, V.M.; Carter, W.H. Response Surface Analysis with Correlated Data: A Nonlinear Model Approach. J. Am. Stat. Assoc. 1989, 84, 805–809. [Google Scholar] [CrossRef]
- Njuho, P.M.; Milliken, G.A. Analysis of Linear Models with One Factor Having Both Fixed and Random Levels. Commun. Stat. Theory Methods 2005, 34, 1979–1989. [Google Scholar] [CrossRef]
- Njuho, P.M.; Milliken, G.A. Analysis of Linear Models with Two Factors Having Both Fixed and Random Levels. Commun. Stat. Theory Methods 2009, 38, 2348–2365. [Google Scholar] [CrossRef]
- Kotchaporn, S.; Araveeporn, A. Modifications of Levene’s and O’Brien’s Tests for Testing the Homogeneity of Variance Based on Median and Trimmed Mean. Thail. Stat. 2018, 16, 106–128. [Google Scholar]
- Sullivan, L.M. Repeated Measures. Circulation 2008, 117, 1238–1243. [Google Scholar] [CrossRef] [Green Version]
- Armstrong, R.A. Recommendations for Analysis of Repeated-Measures Designs: Testing and Correcting for Sphericity and Use of MANOVA and Mixed Model Analysis. Ophthalmic Physiol. Opt. 2017, 37, 585–593. [Google Scholar] [CrossRef] [Green Version]
- Freund, R.J.; Wilson, W.J.; Mohr, D.L. Design of Experiments. In Statistical Methods; Elsevier: Amsterdam, The Netherlands, 2010; pp. 521–576. [Google Scholar] [CrossRef]
- Geisser, S.; Greenhouse, S.W. An Extension of Box’s Results on the Use of the F Distribution in Multivariate Analysis. Ann. Math. Statist. 1958, 29, 885–891. [Google Scholar] [CrossRef]
- Huynh, H.; Feldt, L.S. Estimation of the Box Correction for Degrees of Freedom from Sample Data in Randomized Block and Split-Plot Designs. J. Educ. Stat. 1976, 1, 69. [Google Scholar] [CrossRef]
- Box, G.E.P. Some Theorems on Quadratic Forms Applied in the Study of Analysis of Variance Problems, I. Effect of Inequality of Variance in the One-Way Classification. Ann. Math. Statist. 1954, 25, 290–302. [Google Scholar] [CrossRef]
- Verma, J.P. Repeated Measures Design for Empirical Researchers; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015. [Google Scholar]
- Conover, W.J.; Guerrero-Serrano, A.J.; Tercero-Gómez, V.G. An Update on ‘a Comparative Study of Tests for Homogeneity of Variance’. J. Stat. Comput. Simul. 2018, 88, 1454–1469. [Google Scholar] [CrossRef]
- Ott, L.; Longnecker, M. An Introduction to Statistical Methods & Data Analysis, 7th ed.; Cengage Learning: Boston, MA, USA, 2016. [Google Scholar]
- Mauchly, J.W. Significance Test for Sphericity of a Normal N-Variate Distribution. Ann. Math. Statist. 1940, 11, 204–209. [Google Scholar] [CrossRef]
- Statistical Methods and Data Analytics. UCLA: Statistical Consulting Group. Available online: https://stats.oarc.ucla.edu/sas/seminars/sas-repeatedmeasures/ (accessed on 6 February 2022).
- Moskowitz, D.S.; Hershberger, S.L.; American Psychological Association (Eds.) Modeling Intraindividual Variability with Repeated Measures Data: Methods and Applications; Multivariate Applications Book Series; L. Erlbaum Associates: Mahwah, NJ, USA, 2002. [Google Scholar]
- Hocking, R.R. The Analysis of Linear Models; Brooks/Cole Pub. Co: Monterey, CA, USA, 1985. [Google Scholar]
- Harville, D.A. Maximum Likelihood Approaches to Variance Component Estimation and to Related Problems. J. Am. Stat. Assoc. 1977, 72, 320–338. [Google Scholar] [CrossRef]
- SAS Institute Inc. SAS/STAT® 14.3 User’s Guide; SAS Institute Inc.: Cary, NC, USA, 2017. [Google Scholar]
- Milliken, G.A.; Johnson, D.E. Analysis of Messy Data. 3: Analysis of Covariance; Chapman & Hall/CRC: Boca Raton, FL, USA, 2002. [Google Scholar]
- Akaike, H. A New Look at the Statistical Model Identification. IEEE Trans. Automat. Contr. 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Kuehl, R.O. Design of Experiments: Statistical Principles of Research Design and Analysis, 2nd ed.; Brooks/Cole, Cengage Learning: Belmont, CA, USA, 2000. [Google Scholar]
- Wolfinger, R.D.; Chang, M. Comparing the SAS GLM and Mixed Procedures for Repeated Measures. In Proceedings of the Twentieth Annual SAS Users Groups Conference; SAS Institute Inc: Cary, NC, USA, 1995. [Google Scholar]
- Hamer, R.M. Mixed-Up Mixed Models: Things That Look Like They Should Work but Don’t, and Things That Look Like They Shouldn’t Work but Do. In Proceedings of the Twenty-Fifth Annual SAS® Users Group International Conference, Indianapolis, Indiana, 9–12 April 2000. [Google Scholar]
- McLean, R.A.; Sanders, W.L.; Stroup, W.W. A Unified Approach to Mixed Linear Models. Am. Stat. 1991, 45, 54–64. [Google Scholar] [CrossRef]
| Source of Variation | Factor Effect | F | R | F | Rep | Components of Variance |
|---|---|---|---|---|---|---|
| A | 0 | b | t | N | ||
| B | a | 1 | t | N | ||
| AB | 1 | 1 | t | N | ||
| EU | 1 | 1 | t | 1 | ||
| P | a | b | 0 | N | ||
| PA | 0 | b | 0 | N | ||
| PB | a | 1 | 1 | N | ||
| PAB | 1 | 1 | 1 | N | ||
| Error | 1 | 1 | 1 | 1 |
| Source of Variation | Sum of Squares | Degrees of Freedom | E(MS) |
|---|---|---|---|
| Factors | Time Period | |||||
|---|---|---|---|---|---|---|
| CCl4 | CHCl3 | Flask | 1 | 2 | … | t |
| 1 | 1 | 1 | ||||
| 2 | 1 | |||||
| 1 | ||||||
| 2 | 1 | 1 | ||||
| 2 | 1 | |||||
| 1 | ||||||
| 1 | 1 | |||||
| 2 | 1 | |||||
| 1 | ||||||
| Model FFF | Model FRF | ||||
|---|---|---|---|---|---|
| Covariance Structure | Number of Parameters | AIC | Covariance Structure | Number of Parameters | AIC |
| CS | 2 | −282.6 | CS | 4 | −361.3 |
| AR(1) | 2 | −443.1 | AR(1) | 3 | −492.4 |
| ARH(1) | 10 | −525.6 | ARH(1) | 13 | −506.3 |
| CSH | 10 | −356.7 | |||
| Model RFF | Model RRF | ||||
| Covariance Structure | Number of Parameters | AIC | Covariance Structure | Number of Parameters | AIC |
| CS | 4 | −450.1 | CS | 4 | −665.8 |
| AR(1) | 4 | −522.4 | AR(1) | 4 | −782.4 |
| ARH(1) | 13 | −509.3 | ARH(1) | 13 | −785.3 |
| Model | Effect | Numerator Degrees of Freedom | Denominator Degrees of Freedom | F | p-Value |
|---|---|---|---|---|---|
| FFF | CCl4 | 1 | 12 | 0.24 | 0.6363 |
| CHCl3 | 1 | 12 | 17.33 | 0.0013 ** | |
| CHCl3 | 2 | 12 | 1.35 | 0.2678 | |
| Time | 2 | 24 | 95.99 | <0.0001 ** | |
| CCl4 | 2 | 24 | 2.66 | 0.0908 | |
| CHCl3 | 2 | 24 | 20.91 | <0.0001 ** | |
| CHCl3 | 2 | 24 | 1.26 | 0.3023 | |
| FRF | CHCl3 | 1 | 1 | 0.5 | 0.6079 |
| Time | 2 | 2 | 2.17 | 0.3156 | |
| CCl4 | 2 | 2 | 2.83 | 0.2610 | |
| RFF | CHCl3 | 1 | 1 | 7.07 | 0.2290 |
| Time | 2 | 2 | 5.42 | 0.1557 | |
| CHCl3 | 2 | 2 | 3.26 | 0.2348 | |
| RRF | Time | 2 | 2 | 1.01 | 0.4978 |
| Model | Covariance Parameter | Estimate | Standard Error | Proportion of Variation Accounted for |
|---|---|---|---|---|
| FRF | CHCl3 | 0 | 0 | 0 |
| CHCl3 | 0.04249 | 0.04470 | 22.5 | |
| CHCl3 | 0 | 0 | 0 | |
| CHCl3 | 0.000621 | 0.000759 | 0.3 | |
| RFF | CCl4 | 0.000607 | 0.003056 | 0.05 |
| CHCl3 | 0 | 0 | 0 | |
| CCl4 | 0.000149 | 0.000809 | 0.02 | |
| CHCl3 | 0.000806 | 0.000984 | 0.08 | |
| RRF | CCl4 | 0.0011 | 0.00401 | 2.6 |
| CHCl3 | 0.00306 | 0.00677 | 7.1 | |
| CHCl3 | 0.00232 | 0.00396 | 5.4 | |
| CCl4 | 0.000002 | 0.00002 | 0.0 | |
| CHCl3 | 0.000065 | 0.000096 | 0.1 | |
| CHCl3 | 0.000013 | 0.000025 | 0.0 |
| Type III Tests of Fixed Effects in Combined Models | ||||||
|---|---|---|---|---|---|---|
| Model | AIC [CS] | Effect | Num DF | Den DF | F | Pr > F |
| FA | −127.6 [ARH(1)] | A | 1 | 24 | 3.36 | 0.0794 |
| B | 3 | 24 | 5.81 | 0.0039 ** | ||
| 3 | 24 | 9.10 | 0.0003 ** | |||
| C | 2 | 48 | 12.33 | 0.0001 ** | ||
| 2 | 48 | 2.52 | 0.0908 | |||
| 6 | 48 | 2.33 | 0.0468 ** | |||
| 6 | 48 | 0.76 | 0.6027 | |||
| FB | −127.6 [AR(1)] | A | 3 | 24 | 0.53 | 0.6631 |
| B | 1 | 24 | 16.3 | 0.0005 ** | ||
| 3 | 24 | 0.22 | 0.8801 | |||
| C | 2 | 48 | 24.82 | <0.0001 ** | ||
| 6 | 48 | 1.09 | 0.3795 | |||
| 2 | 48 | 7.82 | 0.0012 ** | |||
| 6 | 48 | 0.50 | 0.8064 | |||
| FB | −69.1 [AR(1)] | A | 1 | 12 | 0.14 | 0.7106 |
| B | 1 | 12 | 10.63 | 0.0068 ** | ||
| 1 | 12 | 0.83 | 0.3807 | |||
| C | 2 | 24 | 19.75 | <0.0001 ** | ||
| 2 | 24 | 0.58 | 0.5667 | |||
| 2 | 24 | 4.65 | 0.0196 ** | |||
| 2 | 24 | 0.26 | 0.7725 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaka, L.; Njuho, P. Repeated-Measures Analysis in the Context of Heteroscedastic Error Terms with Factors Having Both Fixed and Random Levels. Stats 2022, 5, 458-476. https://doi.org/10.3390/stats5020027
Chaka L, Njuho P. Repeated-Measures Analysis in the Context of Heteroscedastic Error Terms with Factors Having Both Fixed and Random Levels. Stats. 2022; 5(2):458-476. https://doi.org/10.3390/stats5020027
Chicago/Turabian StyleChaka, Lyson, and Peter Njuho. 2022. "Repeated-Measures Analysis in the Context of Heteroscedastic Error Terms with Factors Having Both Fixed and Random Levels" Stats 5, no. 2: 458-476. https://doi.org/10.3390/stats5020027
APA StyleChaka, L., & Njuho, P. (2022). Repeated-Measures Analysis in the Context of Heteroscedastic Error Terms with Factors Having Both Fixed and Random Levels. Stats, 5(2), 458-476. https://doi.org/10.3390/stats5020027






