A Brief Overview of Restricted Mean Survival Time Estimators and Associated Variances
Abstract
1. Introduction
2. Restricted Survival Times
3. Estimators of RMST and Associated Variance
3.1. Parametric Methods
- Likelihood and -method based approach.Estimation of requires knowledge of . Estimates for under censoring can be obtained with maximum likelihood. Given the censoring indicator and we can define the likelihood function for asFor most cases there are no closed-form solutions for and , however, numerical estimates are enough. Once estimates for are available, can be estimated either by using in closed form equations, or as plug-ins in numerical integration. The variance for by the -method is given bywhere is the element of the population parameter.
- M-estimation (or estimating equation)M-estimation seeks solution to the vector equation . Here, are independently and identically distributed restricted survival times, is a p-dimensional parameter and is a known -function that does not depend on i or n [17]. M-estimates are asymptotically normal with variance . is a sandwich variance estimator given bywhere,and
- The second cumulantThe variance equivalent to the second cumulant of a probability distribution of . If there is no censoring present and can be estimated asleading to .This estimator is not practical due to censoring. When we have censoring in the data Rosyton & Parmar [20] suggested multiplying with a positive scaling factor , so that if no censoring and otherwise. The scaling factor can be estimated with help of Monte Carlo simulation.Alternatively, the variance can be estimated with the Stute estimator [6]. Adopting the notation of Stute [21], (i.e., ) and and a transformation of X so that . Here, whereBoth approaches require assumptions about the distribution of the survival times and censoring times. These two estimators of variance will not be further evaluated, however they do represent an important aspect of .
3.2. Flexible Parametric Survival Methods
3.3. Non-Parametric Methods
3.3.1. The Kaplan–Meier-Method
3.3.2. Pseudo-Observations
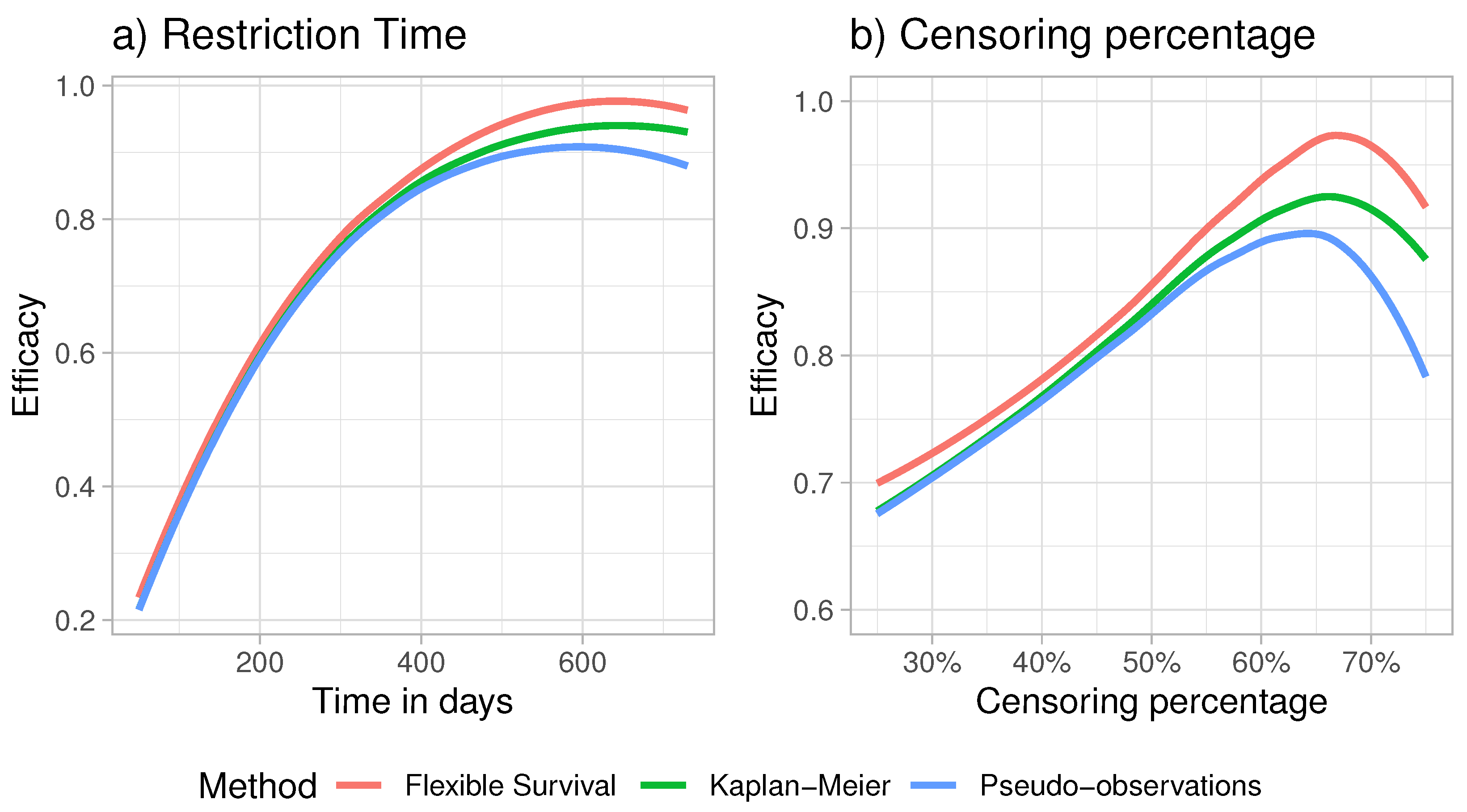
4. Simulation Studies
4.1. Relative Efficacy under Parametric Assumption
4.2. Estimation with Unknown Distribution Function
4.3. Parametric Estimation under Model Misspecification
5. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kaplan, E.L.; Meier, P. Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 1958, 53, 457–481. [Google Scholar] [CrossRef]
- Miller, R.G., Jr. What price kaplan-meier? Biometrics 1983, 1077–1081. [Google Scholar] [CrossRef]
- Jullum, M.; Hjort, N.L. What price semiparametric Cox regression? Lifetime Data Anal. 2019, 25, 406–438. [Google Scholar] [CrossRef] [PubMed]
- Meier, P.; Karrison, T.; Chappell, R.; Xie, H. The price of Kaplan–Meier. J. Am. Stat. Assoc. 2004, 99, 890–896. [Google Scholar] [CrossRef]
- Stute, W. The central limit theorem under random censorship. Ann. Stat. 1995, 23, 422–439. [Google Scholar] [CrossRef]
- Stute, W. Kaplan–meier integrals. Handbook Stat. 2003, 23, 87–104. [Google Scholar]
- Akritas, M.G. The central limit theorem under censoring. Bernoulli 2000, 6, 1109–1120. [Google Scholar] [CrossRef]
- Uno, H.; Wittes, J.; Fu, H.; Solomon, S.D.; Claggett, B.; Tian, L.; Cai, T.; Pfeffer, M.A.; Evans, S.R.; Wei, L.J. Alternatives to hazard ratios for comparing efficacy or safety of therapies in noninferiority studies. Ann. Intern. Med. 2015, 163, 127. [Google Scholar] [CrossRef]
- Uno, H.; Claggett, B.; Tian, L.; Inoue, E.; Gallo, P.; Miyata, T.; Schrag, D.; Takeuchi, M.; Uyama, Y.; Zhao, L.; et al. Moving beyond the hazard ratio in quantifying the between-group difference in survival analysis. J. Clin. Oncol. 2014, 32, 2380. [Google Scholar] [CrossRef]
- Hasegawa, T.; Misawa, S.; Nakagawa, S.; Tanaka, S.; Tanase, T.; Ugai, H.; Wakana, A.; Yodo, Y.; Tsuchiya, S.; Suganami, H.; et al. Restricted mean survival time as a summary measure of time-to-event outcome. Pharm. Stat. 2020. [Google Scholar] [CrossRef]
- Stensrud, M.J.; Aalen, J.M.; Aalen, O.O.; Valberg, M. Limitations of hazard ratios in clinical trials. Eur. Heart J. 2019. [Google Scholar] [CrossRef] [PubMed]
- Meier, P. Estimation of a distribution function from incomplete observations. J. Appl. Probab. 1975, 12, 67–87. [Google Scholar] [CrossRef]
- Andersen, P.K.; Klein, J.P.; Rosthøj, S. Generalised linear models for correlated pseudo-observations, with applications to multi-state models. Biometrika 2003, 90, 15–27. [Google Scholar] [CrossRef]
- Andersen, P.K.; Borgan, O.; Gill, R.D.; Keiding, N. Statistical Models Based on Counting Processes; Springer Science & Business Media: Berlin, Germany, 2012. [Google Scholar]
- Andersen, P.K.; Pohar Perme, M. Pseudo-observations in survival analysis. Stat. Methods Med. Res. 2010, 19, 71–99. [Google Scholar] [CrossRef]
- Royston, P.; Parmar, M.K. The use of restricted mean survival time to estimate the treatment effect in randomized clinical trials when the proportional hazards assumption is in doubt. Stat. Med. 2011, 30, 2409–2421. [Google Scholar] [CrossRef]
- Stefanski, L.A.; Boos, D.D. The calculus of M-estimation. Am. Stat. 2002, 56, 29–38. [Google Scholar] [CrossRef]
- Wang, J.L. Asymptotic Properties of M-estimators Based on Estimating Equations and Censored Data. Scand. Stat. Theory Appl. 1999, 26, 297–318. [Google Scholar] [CrossRef]
- Boos, D.D.; Stefanski, L. Likelihood Construction and Estimation. In Essential Statistical Inference; Springer: Berlin, Germany, 2013; pp. 27–124. [Google Scholar]
- Royston, P.; Parmar, M.K. Restricted mean survival time: An alternative to the hazard ratio for the design and analysis of randomized trials with a time-to-event outcome. BMC Med. Res. Methodol. 2013, 13, 152. [Google Scholar] [CrossRef]
- Stute, W. The jackknife estimate of variance of a Kaplan-Meier integral. Ann. Stat. 1996, 24, 2679–2704. [Google Scholar] [CrossRef]
- Royston, P.; Parmar, M.K. Flexible parametric proportional-hazards and proportional-odds models for censored survival data, with application to prognostic modelling and estimation of treatment effects. Stat. Med. 2002, 21, 2175–2197. [Google Scholar] [CrossRef]
- Irwin, J. The standard error of an estimate of expectation of life, with special reference to expectation of tumourless life in experiments with mice. Epidemiol. Infect. 1949, 47, 188–189. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.P.; Moeschberger, M.L. Survival Analysis: Techniques for Censored and Truncated Data; Springer Science & Business Media: Berlin, Germany, 2006. [Google Scholar]
- Eaton, A.; Therneau, T.; Le-Rademacher, J. Designing clinical trials with (restricted) mean survival time endpoint: Practical considerations. Clin. Trials. 2020, 1740774520905563. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Nadarajah, S. Reliability for lifetime distributions. Math. Comput. Model. 2003, 37, 683–688. [Google Scholar] [CrossRef]
- Jullum, M.; Hjort, N.L. Parametric or nonparametric: The FIC approach. Stat. Sin. 2017, 951–981. [Google Scholar] [CrossRef]
- Lawrence, J.; Qiu, J.; Bai, S.; Hung, H.J. Difference in Restricted Mean Survival Time: Small Sample Distribution and Asymptotic Relative Efficiency. Stat. Biopharm. Res. 2019, 11, 61–66. [Google Scholar] [CrossRef]
- Schall, R. The empirical coverage of confidence intervals: Point estimates and confidence intervals for confidence levels. Biom J. 2012, 54, 537–551. [Google Scholar] [CrossRef] [PubMed]
- Stute, W.; Wang, J.L. The jackknife estimate of a Kaplan—Meier integral. Biometrika 1994, 81, 602–606. [Google Scholar]
- Azarang, L.; de Uña-Álvarez, J.; Stute, W. The Jackknife estimate of covariance of two Kaplan–Meier integrals with covariables. Stats 2015, 49, 1005–1025. [Google Scholar] [CrossRef]
- Akritas, M.G. Bootstrapping the Kaplan—Meier Estimator. J. Am. Stat. Assoc. 1986, 81, 1032–1038. [Google Scholar]
- Burr, D. A comparison of certain bootstrap confidence intervals in the Cox model. J. Am. Stat. Assoc. 1994, 89, 1290–1302. [Google Scholar] [CrossRef]
- Deheuvels, P.; Mason, D.M.; Shorack, G.R. Some results on the influence of extremes on the bootstrap. Ann. l’I.H.P. Prob. Stat. 1993, 29, 83–103. [Google Scholar]
- Andrews, D.W. Inconsistency of the bootstrap when a parameter is on the boundary of the parameter space. Econometrica 2000, 68, 399–405. [Google Scholar] [CrossRef]
- Tian, L.; Jin, H.; Uno, H.; Lu, Y.; Huang, B.; Anderson, K.M.; Wei, L. On the empirical choice of the time window for restricted mean survival time. Biometrics 2020. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Kuan, P.F. Comparison of the restricted mean survival time with the hazard ratio in superiority trials with a time-to-event end point. Pharm. Stat. 2018, 17, 202–213. [Google Scholar] [CrossRef]
- Jackson, C. flexsurv: A Platform for Parametric Survival Modeling in R. J. Stat. Softw. 2016, 70, 1–33. [Google Scholar] [CrossRef]
- Lambert, P.C.; Royston, P. Further development of flexible parametric models for survival analysis. Stata J. 2009, 9, 265–290. [Google Scholar] [CrossRef]
- Luo, X.; Huang, B.; Quan, H. Design and monitoring of survival trials based on restricted mean survival times. Clin. Trials 2019, 16, 616–625. [Google Scholar] [CrossRef]
- Parner, E.T.; Andersen, P.K. Regression analysis of censored data using pseudo-observations. Stata J. 2010, 10, 408–422. [Google Scholar] [CrossRef]
- Overgaard, M.; Andersen, P.K.; Parner, E.T. Regression analysis of censored data using pseudo-observations: An update. Stata J. 2015, 15, 809–821. [Google Scholar] [CrossRef]
- Klein, J.P.; Gerster, M.; Andersen, P.K.; Tarima, S.; Perme, M.P. SAS and R functions to compute pseudo-values for censored data regression. Comput. Methods Programs Biomed. 2008, 89, 289–300. [Google Scholar] [CrossRef]

| -Method | M-Estimator | Kaplan–Meier | Flexible Parametric | Pseudo-Obs | |
|---|---|---|---|---|---|
| n = 50, CP = 50 % | |||||
| 230.1 | 230.1 | 230.7 | 231.2 | 230.7 | |
| 368.0 | 362.1 | 437.5 | 429.3 | 446.9 | |
| eff | 0.98 | - | 0.82 | 0.84 | 0.81 |
| Coverage | 0.94 | 0.94 | 0.93 | 0.93 | 0.94 |
| n = 50, CP = 75 % | |||||
| 230.2 | 230.2 | 230.2 | 228.0 | 228.4 | |
| 724.6 | 712.9 | 792.1 | 794.6 | 1068.7 | |
| eff | 0.98 | - | 0.90 | 0.90 | 0.66 |
| Coverage | 0.93 | 0.92 | 0.89 | 0.89 | 0.90 |
| n = 100, CP = 50 % | |||||
| 230.9 | 230.9 | 231.4 | 231.6 | 231.4 | |
| 185.2 | 182.2 | 221.2 | 217.0 | 223.4 | |
| eff | 0.98 | - | 0.83 | 0.84 | 0.81 |
| Coverage | 0.96 | 0.95 | 0.95 | 0.95 | 0.95 |
| n = 100, CP = 75 % | |||||
| 229.9 | 229.9 | 229.4 | 229.1 | 229.1 | |
| 367.0 | 365.1 | 421.8 | 406.3 | 485.6 | |
| eff | 0.99 | - | 0.86 | 0.89 | 0.75 |
| Coverage | 0.95 | 0.94 | 0.92 | 0.94 | 0.94 |
| Kaplan–Meier | Flexible Parametric | Pseudo-Obs | |
|---|---|---|---|
| Log-logistic, n = 50 | |||
| 183.4 | 183.9 | 183.4 | |
| 402.6 | 397.6 | 413.9 | |
| Coverage | 0.93 | 0.94 | 0.93 |
| Log-logistic, n = 100 | |||
| 182.2 | 182.7 | 182.2 | |
| 201.7 | 199.4 | 204.2 | |
| Coverage | 0.94 | 0.94 | 0.94 |
| Exp-mix, n = 50 | |||
| 224.5 | 224.6 | 224.5 | |
| 444.4 | 435.4 | 454.3 | |
| Coverage | 0.95 | 0.95 | 0.95 |
| Exp-mix, n = 100 | |||
| 225.5 | 225.7 | 225.5 | |
| 224.8 | 220.5 | 227.1 | |
| Coverage | 0.96 | 0.95 | 0.96 |
| Bias | MSE | |||
|---|---|---|---|---|
| = 50 | ||||
| Parametric(-met) | 45.11 | 0.35 | −1.48 | 2.54 |
| Parametric (M-est) | 45.11 | 0.39 | −1.48 | 2.58 |
| Kaplan–Meier | 46.63 | 0.91 | 0.04 | 0.91 |
| = 365 | ||||
| Parametric (-met) | 188.24 | 191.42 | 6.04 | 227.96 |
| Parametric (M-est) | 188.24 | 213.10 | 6.04 | 249.63 |
| Kaplan–Meier | 182.54 | 203.06 | 0.35 | 203.18 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nemes, S.; Bülow, E.; Gustavsson, A. A Brief Overview of Restricted Mean Survival Time Estimators and Associated Variances. Stats 2020, 3, 107-119. https://doi.org/10.3390/stats3020010
Nemes S, Bülow E, Gustavsson A. A Brief Overview of Restricted Mean Survival Time Estimators and Associated Variances. Stats. 2020; 3(2):107-119. https://doi.org/10.3390/stats3020010
Chicago/Turabian StyleNemes, Szilárd, Erik Bülow, and Andreas Gustavsson. 2020. "A Brief Overview of Restricted Mean Survival Time Estimators and Associated Variances" Stats 3, no. 2: 107-119. https://doi.org/10.3390/stats3020010
APA StyleNemes, S., Bülow, E., & Gustavsson, A. (2020). A Brief Overview of Restricted Mean Survival Time Estimators and Associated Variances. Stats, 3(2), 107-119. https://doi.org/10.3390/stats3020010





