Abstract
Soil anoxia is common in the annually thawed surface (‘active’) layer of permafrost soils, particularly when soils are saturated, and supports anaerobic microbial metabolism and methane (CH4) production. Rainfall contributes to soil saturation, but can also introduce oxygen, causing soil oxidation and altering anoxic conditions. We simulated a rainfall event in soil mesocosms from two dominant tundra types, tussock tundra and wet sedge tundra, to test the impacts of rainfall-induced soil oxidation on microbial communities and their metabolic capacity for anaerobic CH4 production and aerobic respiration following soil oxidation. In both types, rainfall increased total soil O2 concentration, but in tussock tundra there was a 2.5-fold greater increase in soil O2 compared to wet sedge tundra due to differences in soil drainage. Metagenomic and metatranscriptomic analyses found divergent microbial responses to rainfall between tundra types. Active microbial taxa in the tussock tundra community, including bacteria and fungi, responded to rainfall with a decline in gene expression for anaerobic metabolism and a concurrent increase in gene expression for cellular growth. In contrast, the wet sedge tundra community showed no significant changes in microbial gene expression from anaerobic metabolism, fermentation, or methanogenesis following rainfall, despite an initial increase in soil O2 concentration. These results suggest that rainfall induces soil oxidation and enhances aerobic microbial respiration in tussock tundra communities but may not accumulate or remain in wet sedge tundra soils long enough to induce a community-wide shift from anaerobic metabolism. Thus, rainfall may serve only to maintain saturated soil conditions that promote CH4 production in low-lying wet sedge tundra soils across the Arctic.
1. Introduction
Permafrost soils consist of permanently frozen ground (permafrost) overlain by a shallow, seasonally thawed active soil layer (the ‘active layer’). These soils store nearly half of all global belowground organic carbon (~1300 Pg) [1]. The decomposition and net storage of organic carbon is controlled in large part by redox conditions in the active layer that are regulated by the availability of oxygen [2]. Permafrost affects redox conditions by confining soil water to the active layer, resulting in soil saturation and thus periodic or persistent soil anoxia [3,4]. Anoxic conditions develop when oxygen is consumed through biochemical processes (e.g., aerobic respiration) or geochemical reactions (e.g., iron oxidation) faster than it is introduced to the soil by diffusion, plant roots, or infiltrating rainwater [5,6]. Permafrost soils support anaerobic microbial metabolism and serve as a major source of methane (CH4) when organic carbon is decomposed under anoxic conditions [7,8,9].
In the Arctic, permafrost soils serve as a net sink for atmospheric carbon [10,11], but the net carbon dioxide (CO2) balance of these soils with the atmosphere is changing with recent warming [12,13] that has thawed large regions of permafrost and deepened the active layer [14,15,16]. Warming has also increased rainfall intensity, frequency, and accumulation in active layer soils [17,18,19] that has led to deeper permafrost thaw [20] and greater CH4 release from thawing permafrost soils [21]. Thus, permafrost thaw and changes in drainage and precipitation will likely lead to greater fluctuations in water levels and redox conditions in the active layer [6], which may trigger increased microbial activity that leads to faster rates of organic carbon decomposition and the release of CO2 to the atmosphere [22,23,24,25]. Research has shown that soil draining and water-level variation leads to rapid aerobic decomposition of soil organic carbon to CO2 [26,27,28,29,30,31]. Previous research has also found that adding oxygen-rich simulated rainwater rapidly changed redox conditions in saturated active layer soils, which increased the abiotic degradation of organic carbon to CO2 [5]. What remains unknown is the impact of rainfall and altered redox conditions on the microbial communities in these active layer soils, and specifically their metabolic capacity for anaerobic CH4 production and aerobic respiration following soil oxidation.
Redox conditions regulate the microbial decomposition of soil organic carbon into CH4 or CO2 by controlling microbial gene expression in response to oxygen availability [31]. Under anoxic conditions, organic carbon is degraded stepwise through anaerobic pathways of fermentation and acetogenesis that supply alcohols and organic acids as substrates for methanogenesis [32]. In oxidized soils, aerobic heterotrophic microbes decompose biopolymers into simple sugars [33] or oxidize CH4 to CO2 as an energy source [34]. Thus, the microbial response to rainfall within anoxic active layer soils will likely be controlled by changes in soil redox conditions via an increase in dissolved oxygen (O2) concentration. However, the extent to which microbial communities can respond to rainfall-induced changes in soil redox conditions, or the duration of their response following rainfall, has yet to be determined.
Rainfall and soil oxidation may also stimulate microbial community responses that differ between distinct tundra types. The low arctic landscape is dominated by two tundra types, tussock tundra and wet sedge tundra, that differ in landscape position, dominant vegetation, soil properties, redox conditions, and microbial community composition [35,36,37,38,39,40]. Previous studies in the low arctic that use next-generation sequencing techniques to characterize the microbial taxa inhabiting permafrost soils have focused on polygonal tundra of the coastal plains [32,41] or boreal forests and shrub-dominated tundra in the discontinuous permafrost zone [42,43,44]. Current characterizations of the microbial communities within tussock tundra and wet sedge tundra are limited to phospholipid fatty acid (PLFA) analyses [38] and terminal restriction fragment length polymorphism (T-RFLP) analyses [45], which limits our ability to predict how these distinct microbial communities may respond metabolically to rainfall-induced changes in soil redox conditions. It is likely that the extent of the microbial response to rainfall and soil oxidation among these tundra types will be constrained by the microbial species composition and their functional genes, as well as the expression of their genes under changing soil redox conditions; we test that idea in this paper.
Here, we provide mechanistic explanations for the microbial community response to rainfall-induced soil oxidation using an integrated meta-omics approach coupled with soil oxygen modeling. Meta-omics studies reveal the composition and diversity of soil microbial communities, their functional potential, and their in situ gene expression during environmental disturbances without the need for cultivation [46]. As such, we determined the taxonomic composition and genomic potential of the soil microbial communities within tussock tundra and wet sedge tundra using 16S rRNA amplicons and metagenomics. Using metatranscriptomics we determined microbial gene expression within the communities of both tundra types under initial anoxic conditions and again following a simulated rainfall event to oxidize the soil. To determine the magnitude and temporal extent of rainfall-induced soil oxidation, we modeled both the accumulation of dissolved O2 added to the soil in rainfall and the consumption of dissolved O2 following rainfall as the saturated soils re-acclimated. Our modeled concentrations of dissolved O2 after rainfall were then coupled with the observed patterns in microbial gene expression in response to rainfall at 4-h and 24-h following the rainfall event. We also conducted a separate soil incubation experiment to measure rates of microbial respiration and CH4 production under anoxic and oxic conditions that mimic pre- and post-rainfall soil redox conditions. We hypothesized that an increase in soil oxygenation by rainfall would (1) decrease the microbial expression for anaerobic metabolism genes as well as the rates of CH4 production, while (2) increasing growth expression for aerobic microbial taxa and subsequent CO2 emissions in response to the rapid shift in soil redox conditions. We also hypothesized that the extent of rainfall-induced changes in microbial gene expression patterns depends on the genomic potential of tussock tundra or wet sedge tundra microbial communities to respond to changing soil redox conditions, as well as on the amount of dissolved O2 supplied to the soil.
2. Materials and Methods
2.1. Mesocosm Design
A total of 12 intact soil-plant cores (diameter 28 cm; length 31 ± 4 cm) were collected from two dominant tundra types, tussock tundra and wet sedge tundra, near Toolik Lake, Alaska (68°38′ N, 149°43′ W) between July and August 2017 and transferred to 20 L plastic buckets to establish the mesocosm experiment. Triplicate mesocosms from each tundra type were separated into two distinct study groups: (1) biotic response cores (N = 6) and (2) abiotic response cores (N = 6), where results from the abiotic response cores are previously described [5]. The full mesocosm experiment was split into two study groups to avoid the effects of destructive soil sampling necessary for the biotic response cores. The mesocosm experimental procedures for biotic and abiotic cores were identical and consisted of an acclimation period under static waterlogged conditions (i.e., no flowing water) for 4–7 days to generate reducing conditions observed in intact soils in the field [47,48], followed by a flushing period over 1–4 h to mimic the effects of brief and rapid changes in redox conditions induced by rainfall (Figure S1). The flushing period consisted of ten individual flushes of 2 L of DI water per flush for each replicate mesocosm. The total volume of water flushed was chosen to represent rainfall events up to and in excess of the natural rainfall pattern near Toolik Field Station, which received ~270 mm of rainfall between 1 June and 30 September 2017 (Environmental Data Center, Toolik Field Station). Additional details of the mesocosm design are provided in the Supplemental Materials.
2.2. Soil Properties
Soil properties for pH, conductivity (μS cm−1), temperature (°C), dissolved soil O2 (mg O2 L−1), and soil moisture (% water), the latter of which was calculated as the volumetric water content (VWC) of the soil, were measured directly at 10–20 cm depth in the biotic response cores at the end of the anoxic acclimation period prior to rainfall treatments. Soil properties for bulk density (g soil dry cm−3), porosity (% soil volume), organic matter content (%), and organic carbon content (% C) were determined from subsamples of the biotic response cores taken at the end of the experiment. All tussock tundra soil cores were composed of an upper organic soil layer (0–20 cm) and a lower mineral soil layer (20–30 cm) whereas all wet sedge tundra soil cores were composed entirely of organic soil (0–30 cm). Additional details are provided in the Supplemental Materials.
2.3. Soil Sample Collection
For all biotic response cores, an initial soil sampling event was conducted at the end of the anoxic acclimation period, representing sampling time point “T0”. Mesocosm replicates were then flushed to simulate rainfall as described above (Figure S1). Additional soil sampling events were conducted at 4-h (“T4”) and 24-h (“T24”) following simulated rainfall. At each sampling time point, duplicate soil cores (diameter 2.54 cm, length 30 cm) were extracted from each mesocosm replicate and homogenized by depth in 10-cm increments. The 10–20 cm soil increment, composed of organic soil in all mesocosms, was chosen for all downstream microbial analyses and preserved in RNAlater Stabilization Reagent (Qiagen, Hilden, Germany) in sterile tubes at 4 °C for 18 h and then stored at −80 °C until thawed for extraction (N = 18).
2.4. Modeling Soil O2 Accumulation and Consumption Rates
A mass balance model was developed to estimate the rates of soil O2 accumulation introduced through rainfall and soil O2 consumption following rainfall, as well as the final concentrations of soil O2 at each sampling time point in the mesocosm experiment (Figure S2). Measurements derived from both the abiotic response cores [5] and the biotic response cores (this study) in the mesocosm experiment were incorporated into the mass balance model to estimate the rates of rainfall O2 (Frain) and soil O2 consumption (Sall) (Figure S2). The rates of soil O2 diffusion (Fatm) were considered to be zero during this relatively short experiment, and there was no groundwater O2 (Fground) or soil O2 leaching (Fleach) (Figure S2). The initial concentration of soil O2 (Mt; mg O2 L−1) in the biotic response cores was measured at the end of the anoxic acclimation period (T0) prior to simulated rainfall (details in Supplemental Materials). No additional measurements for soil O2 concentration were taken in the biotic response cores, and soil measurements for tussock tundra and wet sedge tundra reported for the abiotic response cores [5] were used to model rates of soil O2 accumulation during the DI water flush (Frain; mg O2 L−1 h−1) and calculate the final soil O2 concentration at the end of the DI water flush (Mf; mg O2 L−1) in the biotic response cores (details in Supplemental Materials). Likewise, additional soil measurements for tussock tundra and wet sedge tundra reported for the abiotic response cores [5] were used to model the rates of soil O2 consumption during the re-acclimation phase following simulated rainfall (Sall; mg O2 L−1 h−1) and calculate the final soil O2 concentration at 4-h (Mf+4; T4) and 24-h (Mf+24; T24) following the DI water flush (details in Supplemental Materials).
2.5. Nucleic Acid Extraction and Sequencing
Microbial RNA and DNA were extracted from soil samples collected at the 10–20 cm depth in each tundra mesocosm at each sampling time point (N = 18) in the biotic response cores using the RNeasy PowerSoil Total RNA Isolation kit (Qiagen) and RNeasy PowerSoil DNA Elution Accessory kit (Qiagen), respectively, following the manufacturer protocol. PCR amplicon sequencing of the 16S ribosomal RNA genes was conducted on the extracted DNA following the Earth Microbiome Project protocol (https://www.earthmicrobiome.org/protocols-and-standards/16s/; accessed on 10 March 2021) (N = 18; details in Supplemental Materials). Samples for 16S amplicons were sequenced at the University of Michigan Microbiome Core with Illumina MiSeq 2 × 150 bp paired-end reads. Metagenome sequences were generated from extracted DNA (N = 15; details in Supplemental Materials) and sequenced at the University of Michigan Advanced Genomics Core with Illumina HiSeq (4000) 150 bp paired-end reads. Metatranscriptome reads were generated from total RNA extractions (N = 12; details in Supplemental Materials) and sequenced at the University of Michigan Advanced Genomics Core with Illumina HiSeq (4000) 150 bp paired-end reads.
2.6. 16S rRNA Gene Analysis
The 16S amplicon sequences were paired, quality filtered, dereplicated, and aligned to the SILVA reference database [49] (v.132) with chimeras removed using the VSEARCH algorithm [50], all using MOTHUR [51]. The remaining quality-controlled (QC) reads were classified using a Bayesian classifier with the RDP training set [52] (set 16). Reads were then clustered at 97% similarity to form operational taxonomic units (OTUs) and rarefied to 9565 sequences per sample (average 14,562 QC sequences per sample prior to rarefying; Table S1) with raw counts transformed into relative abundance (average 95% community coverage for rarefied samples; Table S1). The consensus taxonomy for each OTU was assigned using the RDP classifier trained to the SILVA database (v.132). OTUs classified as Eukaryote, Chloroplast, Mitochondria, and Unknown were removed, with raw counts for remaining OTUs and their taxonomic classification provided in an OTU table (Dataset S1).
2.7. Metagenome Assembly
DNA sequences were trimmed and quality-filtered with the BBDuk algorithm from BBMap [53] with dereplication. Quality-controlled metagenome reads from each sample (average 36.8 × 106 QC reads per sample; Table S2) were normalized to 20× kmer coverage with kmers below 5× coverage removed using BBNorm from BBmap [53] prior to co-assembling using MEGAHIT [54]. The co-assembled contigs were indexed using Bowtie2 [55] and short reads from each sample were individually mapped to the indexed contigs using BBMap [53]. A contigs database was created from the co-assembly using Anvi’o [56] with the abundance information for all contigs merged into an Anvi’o profile. Coding sequences (CDS) within the contigs database were predicted with PRODIGAL [57] while HMMER [58] was used to search for and tabulate the occurrence of single-copy housekeeping genes for bacteria and archaea from two collections [59,60]. CDS were also annotated to the Kyoto Encyclopedia of Genes and Genomes database (KEGG) [61] as well as taxonomically annotated using the online GhostKOALA program [62] and imported into the contigs database. Counts per annotation were normalized to genes per million (GPM) [63] to reduce biases associated with library size and CDS length, and to express all counts as a portion of one million (Dataset S2). The mapping information for each sample mapped to the contigs database was merged into a single Anvi’o profile prior to binning contigs into metagenome assembled genomes (MAGs) using CONCOCT [64] and MetaBAT [65]. Redundant MAGs derived from the independent binning algorithms were dereplicated using DASTools [66] to generate the final MAGs dataset, where MAGs with >70% completeness and <10% redundancy were retained. Retained MAGs were queried against the Genome Taxonomy Database (GTDB) [67] using the contigs and profile databases within Anvi’o to generate a consensus taxonomy for each MAG and its relative abundance across all samples.
2.8. Metatranscriptome Assembly
RNA sequences were trimmed and quality-filtered with the BBDuk algorithm from BBMap [53] without dereplication to determine the abundance of transcribed genes. Quality-controlled metatranscriptome reads (average 26.3 × 106 QC reads per sample; Table S3) were mapped to KEGG-annotated coding sequences (CDS) indexed from the metagenome assembly using BBMap to generate pile-up files (average read depth per gene), while SAMtools was used to extract counts and CDS lengths from the BBMap output [53]. On average, 65% of the metatranscriptome CDS were expressed (>1 count per read) with 40% of expressed CDS assigned KEGG functional annotations, of which 99% were also assigned taxonomic annotations. Counts per annotation were normalized to transcripts per million (TPM) [63] to reduce biases associated with library size and CDS length, and to express all counts as a portion of one million (Dataset S3). This workflow assembled metatranscriptomic reads into functionally and taxonomically annotated KEGG orthologs (KOs) by mapping reads to whole genome sequences derived from the co-extracted DNA-based metagenomes.
2.9. Taxonomic Composition and Diversity
The taxonomic composition and diversity of the total microbial communities within tussock tundra and wet sedge tundra soils were determined from the 16S rRNA amplicons and metagenomes. Within the 16S rRNA amplicons, taxonomic composition was based on the classification of OTUs clustered at 97% similarity (Dataset S1). Within the metagenomes, the taxonomic composition and diversity metrics were based on the relative abundance of GPM-normalized genes annotated as KEGG tier III “Ribosomes” (03010 Ribosome PATH: ko03010) further separated at the KEGG tier IV level into annotations for “small subunit ribosomal protein” (ssu) analogous to bacterial and archaeal 16S ssu rRNA and fungal 18S ssu rRNA (Dataset S4). Similarly, the taxonomic composition and diversity of the active microbial communities within tussock tundra and wet sedge tundra soils were determined from the TPM-normalized metatranscriptomes annotated as the KEGG tier IV ribosomal category “ssu” (Dataset S4). Pairwise comparisons between microbial taxa within total or active tundra communities were calculated using paired t-tests or between sampling time points within tundra communities using two-way ANOVA based on differences in relative abundance. Shannon-Wiener diversity index was calculated with raw taxonomic gene counts and assessed via two-way ANOVA to determine the effects of tundra type on total and active taxonomic alpha diversity. Beta diversity treatment effects were estimated using permutational multivariate analysis of variance (PERMANOVA) based on Bray-Curtis dissimilarity distance between taxonomic gene counts using the vegan package [68] in the R software environment [69], and PCoA plots were generated using the ggplot2 package [70]. Throughout this paper, all statistical analyses were considered significant at p < 0.05.
2.10. Genomic Potential and Functional Gene Expression
The genomic potential and functional gene expression within the total and active communities in tussock tundra and wet sedge tundra soils were determined from the GPM-normalized metagenomes and TPM-normalized metatranscriptomes, respectively, based on the relative abundance of genes annotated to KEGG tiers II-IV. Pairwise comparisons among functional genes between tundra communities were calculated using paired t-tests or between sampling time points within tundra communities using two-way ANOVA based on differences in relative abundance and visualized with Z-score transformations using pheatmap [71] in R. Diversity analyses of the functional genes were performed using the vegan package [68] in R. Shannon-Wiener diversity index was calculated with raw functional gene counts and assessed via two-way ANOVA with post hoc Tukey honest significant differences tests to determine the effect of tundra type on functional alpha diversity. Beta diversity treatment effects were estimated using PERMANOVA based on Bray-Curtis dissimilarity distance between GPM- or TPM-normalized functional gene counts with the R-vegan function adonis.
2.11. Differential Gene Expression
Differential gene expression (DGE) analysis was applied to the TPM-normalized metatranscriptomes, where DGE between sampling time points within each tundra type was determined using the glmFit function within edgeR [72] based on KO values, after setting calcnormfactors to “none” to avoid default TMM normalization by edgeR (Dataset S5). The generalized linear models were fit to a design matrix based on the experimental design to test for differential expression between sampling time points while adjusting for differences between mesocosm replicates at each sampling time point. This was considered an additive linear model with mesocosm replicates at each sampling time point as the blocking factor. DGE for TPM-normalized KOs were considered differentially expressed if the false discovery rate (FDR) [73] value of p was < 0.05.
2.12. Oxygen-Regulated Gene Expression
Results from the DGE analysis (log fold-change) were evaluated for the differential expression of oxygen-regulated functional KOs (i.e., genes) between sampling time points within each tundra type. Genes identified as oxygen-regulated infers that their expression can be either positively or negatively affected by O2 availability and the differential expression of these genes is an indication that simulated rainfall changed soil redox conditions through an increase in dissolved O2 concentration. Select marker genes that are considered to be oxygen-regulated were chosen from previous studies [74] and includes genes from obligate anaerobic pathways of carbon fixation, fermentation, denitrification, and methanogenesis, as well as genes from obligate aerobic pathways of carbon fixation and methane oxidation. Additional oxygen-regulated genes from KEGG Metabolism categories were chosen from the reference database in Wu and Moore [75], which identified KO groups of which gene distributions were statistically different for aerobic vs. anaerobic environmental conditions. In the reference study, complete prokaryotic genomes in NCBI’s Microbial Genome Project Database were used to statistically predict correlations between functional KO groups and oxygen requirements (see [75] for details).
2.13. Iron Redox Cycling Gene Expression
The microbial pathways for iron oxidation and dissimilatory iron reduction were quantified as additional proxies for obligate aerobic or anaerobic gene expression, respectively. Established gene annotation pipelines such as GhostKOALA [62], used in this study, generally do not incorporate hidden Markov models (HMMs) capable of annotated iron redox cycling genes [76]. We re-annotated the metatranscriptome contigs using FeGenie [76], which provides a comprehensive database of HMMs based on genes related to iron oxidation or reduction in bacteria and archaea. Counts within each iron gene category were summarized as their relative abundance against the sum of all identified iron genes with only those genes identified for iron oxidation or reduction presented in this study. Pairwise comparisons with two-way ANOVA were conducted across sampling time points within the iron oxidation or reduction gene categories to determine if rainfall-induced soil oxidation changed obligate redox-dependent gene expression.
2.14. Microbial Respiration and Methane Production
The rates of microbial respiration (CO2) and net methane (CH4) production were measured from a separate soil incubation experiment using a subset of each homogenized soil sample originally collected from the 10–20 cm sampling depth in each tussock tundra and wet sedge tundra mesocosm in the biotic response cores at each sampling time point (N = 18). Each soil sample (average of 15 ± 5 g per sample) was placed in an individual 250 mL glass jar sealed with an airtight lid containing a septum for gas sampling. Jars containing soil collected from the T0 sampling time point were purged with N2 to reproduce anoxic field conditions. Jars containing soil from the T4 and T24 sampling time points were purged with compressed air to reproduce oxic field conditions. Jars were incubated for 48 h in the dark at field temperature (5 °C). Following the 48-h incubation, 20 mL of headspace gas was collected using an evacuated syringe and immediately analyzed for CO2 and CH4 concentrations via gas chromatography on a Shimadzu GC-14A with TCD and FID detectors (Shimadzu, Kyoto, Japan). Gas concentrations were converted to rates of gas production per hour relative to the dry mass of soil. Pairwise comparisons with two-way ANOVA were conducted between sampling time points within each tundra type for the rates of CO2 and CH4 production.
3. Results
3.1. Soil Properties
We measured soil properties within tussock tundra and wet sedge tundra mesocosms in the biotic response cores at the end of the anoxic acclimation period (T0) prior to simulated rainfall. Soil pH and conductivity were lower in tussock tundra soil compared to wet sedge tundra soil (paired t-test; p = 0.03, 0.05, respectively; Table 1), with measured values consistent with previous studies conducted in younger landscape tundra soils in the Toolik Lake region [5,47,48]. Soil temperature, moisture, and dissolved O2 concentrations were similar between both tundra soils prior to simulated rainfall (paired t-test; p > 0.1 for all; Table 1). Organic matter content and its corresponding organic carbon content were lower in tussock tundra soil compared to the wet sedge tundra soil (paired t-test; p = 0.04, 0.04; Table 1). Bulk density was greater in tussock tundra soil compared to wet sedge tundra soil (paired t-test; p = 0.02; Table 1), with lower porosity than in wet sedge tundra soil (paired t-test; p = 0.02; Table 1).

Table 1.
The chemical and physical soil properties in tussock tundra and wet sedge tundra mesocosms in the biotic response cores were measured at the end of the anoxic acclimation period (T0), and bulk soil properties were measured at the end of the experiment (mean ± SD; N = 3 per tundra ecosystem). Asterisks indicate significant differences in mean pairwise similarity between tussock and wet sedge tundra soils (paired t-test; * p < 0.05; NS = no statistically significant difference).
3.2. Soil O2 Accumulation and Consumption Rates and Final Concentrations
We added 20 L of DI water to all tundra mesocosms in the biotic response cores during the simulated rainfall event (Table 2). However, the duration of the DI water flush differed between tundra type due to differences in soil drainage rates, with the DI water flush taking an average of 3.77 h to completely drain through the tussock tundra soil and 1.13 h to completely drain through the higher-porosity wet sedge tundra soil (paired t-test; p = 0.02; Table 2). Our modeled rates of soil O2 accumulation (Frain; mg O2 L−1 h−1) at the 15-cm sampling depth during the DI water flush were similar between the tussock tundra and wet sedge tundra soils (paired t-test; p = 0.7; Table 2). Yet, due to differences in the duration of the DI water flush among tundra soils, the soil O2 concentration (Mf; mg O2 L−1) at the 15-cm sampling depth at the end of the DI water flush for tussock tundra increased 8.5-fold while increasing only 4-fold in wet sedge tundra (Table 2). Overall, the soil O2 concentration at the end of the DI water flush was 2.5-fold greater in the tussock tundra soil than in wet sedge tundra soil (paired t-test; p = 0.02; Table 2). Our modeled rates of soil O2 consumption (Sall; mg O2 L−1 h−1) during the re-acclimation phase following the DI water flush were greater in the tussock tundra soil compared to the wet sedge tundra soil (paired t-test; p = 0.03; Table 2).

Table 2.
Soil modeling flush variables, rates of soil O2 accumulation, and concentrations of soil O2 after simulated rainfall are for the biotic response cores. The soil O2 consumption rates are modeled from soil properties reported for the abiotic response cores [5] and are representative rates within tussock tundra and wet sedge tundra soils under static, waterlogged field conditions. Asterisks indicate significant differences in mean pairwise similarity among tundra soils (paired t-test; * p < 0.05; NS = no statistically significant difference).
3.3. Taxonomic Composition and Diversity of the Total and Active Microbial Communities
Using 16S rRNA amplicons and metagenomes annotated as KEGG tier III “Ribosomes”, we found that the total microbial communities from both tundra types were dominated by Bacteria (>98% relative abundance; Figure 1A,B), but there were significant differences in the relative abundance of bacterial phyla between tundra communities (paired t-test; p < 0.05 for all; Table 3). Specifically, bacterial taxa in the tussock tundra total community were dominated by Acidobacteria, Actinobacteria, Alphaproteobacteria, Gammaproteobacteria, Planctomycetes, and Verrucomicrobia (>85% of total community; Figure 1A; Table 3). Bacterial taxa in the wet sedge tundra total community were dominated by the same phyla as the tussock tundra community, but with elevated contributions of Betaproteobacteria, Deltaproteobacteria, Bacteriodetes, Chloroflexi, and Firmicutes (Figure 1B; Table 3). Archaeal taxa were composed primarily of methanogenic Euryarchaeota (Table 3), which were present only in the wet sedge tundra total community and included dominant taxa such as Methanobacterium, Methanocella, Methanomicrobia, Methanoregula, Methanosarcina, and Methanothrix (Datasets S1 and S4). Fungal taxa were composed primarily of Ascomycetes (Table 3), which were present almost exclusive to the tussock tundra community and included dominant taxa Aspergillus, Coccidioides, Glarea, Histoplasma, Marssonina, Phialocephala, and Sclerotinia (Dataset S4).
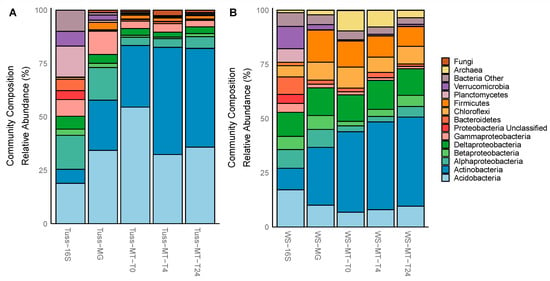
Figure 1.
Microbial community composition within (A) tussock tundra and (B) wet sedge tundra soils based on the mean relative abundance of 16S rRNA amplicons, GPM-normalized metagenome (MG), and TPM-normalized metatranscriptome (MT) genes belonging to Ribosome KEGG tier III pathway.

Table 3.
Relative abundance calculated for tussock tundra and wet sedge tundra communities averaged across all sampling time points based on KEGG tier III “Ribosome” category from the GPM-normalized metagenome (MG) and TPM-normalized metatranscriptome (MT) datasets (mean ± SD). Asterisks indicate the significance of differences in mean pairwise similarity among tundra communities (paired t-test; *** p < 0.001, ** p < 0.01, * p < 0.05; NS = no statistically significant difference).
As expected, the taxonomic composition of the total microbial communities did not differ between sampling time points (24-h) within tussock tundra or wet sedge tundra soils in any dataset (ANOVA; p > 0.05 for all). The total community from wet sedge tundra had significantly higher taxonomic alpha diversity of ribosome genes than the tussock tundra community (paired t-test; p < 0.01; Table S4). Beta diversity results indicated significant differences in taxonomic composition between tundra communities (p < 0.01), but no difference between sampling time points (p = 0.7) or the interaction of tundra community by sampling time points (PERMANOVA; p = 0.6; Table S5). The majority of our high-quality metagenome-assembled genomes (MAGs; >70% complete, <10% redundancy) were classified to the dominant bacterial phyla Acidobacteria, Actinobacteria, Chloroflexi, and multiple classes of Proteobacteria (Table S6). The majority of MAGs recovered were found predominantly in the wet sedge tundra community compared to the tussock tundra community based on their relative abundance (Table S6). However, MAGs with greater relative abundance in the tussock tundra community also had large genomes (Table S6).
We also determined differences in the relative abundance of active microbial taxa between tussock tundra and wet sedge tundra communities using the TPM-normalized metatranscriptomes annotated as KEGG tier III “Ribosomes.” Active taxa within each tundra community were an expressed subset of the total community identified in the metagenome (Figure 1A,B) with no significant differences in the relative abundance of active microbial taxa between sampling time points for tussock tundra or wet sedge tundra soils (ANOVA; p > 0.05). Within tussock tundra, Actinobacteria and Acidobacteria accounted for 83% of the active community (Table 3). Methanogenic Euryarchaeota were also detected in the tussock tundra active community, including taxa from Methanosarcina and Methanothrix (Dataset S4), although at significantly lower abundance than in the wet sedge tundra active community (paired t-test; p < 0.05 for all; Table S7). Within wet sedge tundra, Actinobacteria and Acidobacteria accounted for only 48% of the active community with high relative abundance of Deltaproteobacteria (13%), Firmicutes (10%), Chloroflexi (8%), and archaeal Euryarchaeota (7%) (Table 3). Deltaproteobacteria were dominated by Geobacter spp., representing 3% of the active community relative abundance (Table S7). The high relative abundance of active Euryarchaeota is due mainly to an increase in Methanosarcina and Methanothrix spp. (4%, 1%, respectively; Table S7).
3.4. Genomic Potential and Functional Gene Diversity of the Microbial Communities
The genomic potential of tussock tundra and wet sedge tundra microbial communities indicated that the KEGG tier II category “Metabolism” had the greatest relative abundance of annotated functional genes in both tundra communities, followed by genes annotated as “Genetic Information Processing”, “Environmental Information Processing”, and “Cellular Processes”, with significant differences in relative abundance between tussock tundra and wet sedge tundra communities for all categories except Genetic Information Processing (Figure S3). At the KEGG tier III level (sub-categories within tier II), we found significant differences in the relative abundance of most functional gene categories (15 of 22 categories) between tundra communities (ANOVA; p < 0.05 for all; Figure 2A,B). Specifically, there was significantly greater abundance for Metabolism functional genes associated with amino acids, carbohydrates, nucleotides, biosynthesis of secondary metabolites, and xenobiotics degradation in the tussock tundra community relative to the wet sedge tundra community (Figure 2B). There was also greater relative abundance of functional genes associated with folding, sorting and degradation, signal transduction, as well as cellular communities, cell motility, and transport and catabolism in the tussock tundra community. In contrast, there was greater relative abundance of functional genes associated with energy metabolism, translation, and membrane transport in the wet sedge tundra community (Figure 2B). The wet sedge tundra microbial community had significantly higher alpha diversity of functional genes than the tussock tundra community (paired t-test; p < 0.001; Figure S3; Table S4). Beta diversity results indicated significant differences in the relative abundance of functional genes between tundra communities (p < 0.001), but no difference in relative abundance between sampling time points (p = 0.8) or the interaction of tundra communities by sampling time points (PERMANOVA; p = 0.6; Figure S3; Table S5).
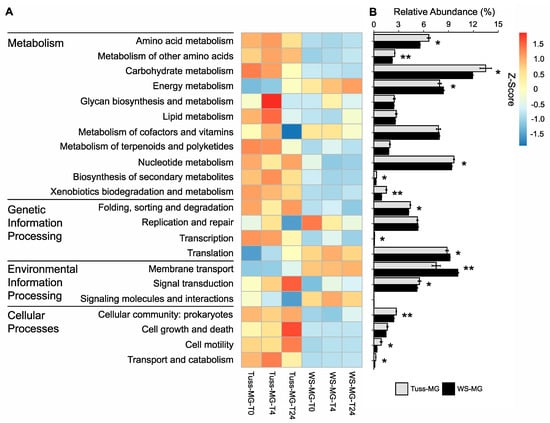
Figure 2.
Heatmap (A) and barplot (B) of genomic potential based on the mean relative abundance of GPM-normalized metagenome (MG) genes belonging to KEGG tier III functional pathways by sampling time point in mesocosm replicates of tussock tundra and wet sedge tundra communities. Z-score transformations based on the mean relative abundance of genes between sampling time points for each KEGG pathway. Error bars for each barplot represent standard deviation from the mean. Asterisks mark the significance of differences in mean relative gene abundance between tundra communities for each KEGG tier III pathway (paired t-test; ** p < 0.01; * p < 0.05).
3.5. Microbial Community Expression in Response to Rainfall
The change in microbial gene expression in response to rainfall was determined using the relative abundance of TPM-normalized metatranscriptomes annotated at KEGG tiers II-IV. Within the tussock tundra community, the relative abundance of expressed functional genes annotated to the KEGG tier II category of Metabolism was significantly greater at T0 relative to the abundance of expressed genes at T4 and T24 (ANOVA; p < 0.01 for both; Figure 3A). Following simulated rainfall, the relative abundance of expressed genes at T4 and T24 was significantly greater for the KEGG tier II category of Genetic Information Processing relative to T0 (ANOVA; p < 0.01 for both; Figure 3A). The relative abundance of expressed genes for the KEGG tier II category of Cellular Processes was also significantly greater at T4 relative to T0 (ANOVA; p < 0.01; Figure 3A) but was not significantly different at T24 relative to T0 (ANOVA; p = 0.1; Figure 3A).
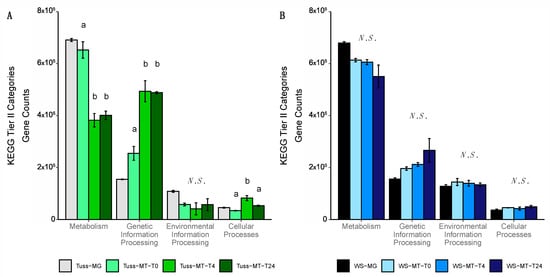
Figure 3.
Barplot of microbial gene expression based on the mean relative abundance of TPM-normalized metatranscriptome (MT) genes annotated as KEGG tier II functional pathways by sampling time point in (A) tussock tundra and (B) wet sedge tundra communities (error bars represent standard deviation of the mean). Letters indicate significant differences in mean pairwise similarity between sampling time points for each KEGG functional pathway (ANOVA; p < 0.05), with non-significant differences among sampling time points indicated as N.S. The relative abundance of GPM-normalized metagenome (MG) functional genes is provided for reference only and their abundance was not incorporated in the pairwise statistical analysis.
In contrast to the tussock tundra community, broad patterns of microbial gene expression in the wet sedge tundra community for the KEGG tier II categories of Metabolism, Genetic Information Processing, Environmental Information Processing, or Cellular Processes showed no significant change in relative abundance between any sampling time points (ANOVA; p > 0.05 for all; Figure 3B). Expressed gene abundance was greatest in the Metabolism category and included numerous anaerobic pathways with oxygen-regulated KOs for carbohydrate metabolism (K00128, K00161, K00656, K00975), amino acid metabolism (K00146), lipid metabolism (K00763), and energy metabolism, the latter of which included KOs from multiple pathways of methanogenesis (hydrogenotrophic: K00200, K00204, K00319, K00672, K01499, K13942; acetoclastic: K00193, K00625, K00925, K01895; and methylotrophic: K04480, K14080, K14081) as well as methanotrophy (K10944, K14028, K16157, K16160, K23995) (Table S8). Additional pathways with oxygen-regulated KOs included anaerobic carbon fixation (K00174, K00194, K00244, K00855, K01648), fermentation (K00169, K00016), and denitrification (K00368, K00370, K00376, K02305, K02567, K15864) (Table S8). Using DGE analysis, we found no significant differences in the expression of any oxygen-regulated KO between sampling timepoints, but there were significant increases in the expression of obligate anaerobic denitrification KOs including nitrite reductases (K00368, K00370, K15864), nitric oxide reductases (K02305, K04561), and nitrous-oxide reductase (K00376) following rainfall at T4 relative to T0 (DGE; FDR < 0.05 for all; Dataset S5). Overall, the alpha diversity of expressed functional genes was similar between sampling time points throughout the experiment (ANOVA; p > 0.5; Figure S4; Table S4). Beta diversity results indicated no significant difference in the relative abundance of expressed functional genes between sampling time points within the wet sedge tundra community (PERMANOVA; p = 0.2; Figure S4; Table S5).
Within the tussock tundra community, the relative abundance of expressed transcripts in the Metabolism KEGG tier IV pathways of glycolysis/gluconeogenesis and butanoate metabolism (tier III carbohydrates), nicotinate and nicotinamide metabolism (tier III cofactors and vitamins), and carbon metabolism, fatty acid metabolism, and degradation of aromatic compounds (tier III overview), were significantly greater at T0 relative to their abundance at T4 and T24 (ANOVA; p < 0.05 for all; Figure 4). For Genetic Information Processing, the KEGG tier IV pathway of ribosome (tier III translation) had significantly greater abundance of expressed transcripts at T24 relative to T0, but not initially following rainfall at T4 relative to T0 (ANOVA; p = 0.02, p = 0.07, respectively; Figure 4). The KEGG tier IV pathway of RNA degradation (tier III folding, sorting and degradation) had significantly greater abundance of expressed transcripts at T4 relative to T0 (ANOVA; p < 0.01; Figure 4). For Cellular Processes, the KEGG tier IV pathway of Caulobacter cell cycling (tier III cell growth and death) had significantly greater abundance of expressed transcripts at T4 relative to T0 (ANOVA; p < 0.01; Figure 4). Likewise, the relative abundance of expressed transcripts in the tier IV pathway of ferroptosis was significantly greater at T4 relative to T0 (ANOVA; p = 0.02, not shown).
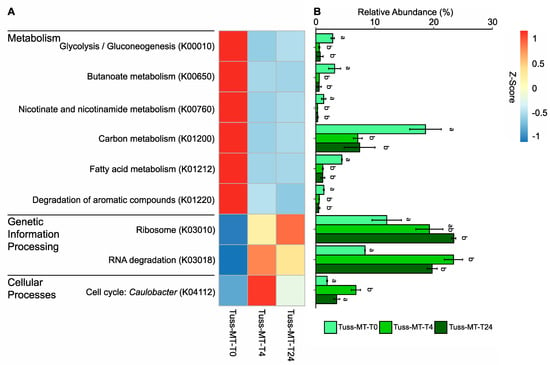
Figure 4.
Heatmap (A) and barplot (B) of microbial gene expression between sampling time points within tussock tundra based on the mean relative abundance of TPM-normalized metatranscriptome (MT) gene transcripts belonging to KEGG tier IV functional pathways. Z-score transformations based on the mean relative abundance of transcripts between sampling time points. Error bars for each barplot represent standard deviation from the mean. Letters indicate significant differences in mean pairwise similarity between sampling time points in each KEGG tier IV category (ANOVA; p < 0.05).
Within significant KEGG-Metabolism categories (ANOVA; Figure 4), we found several KOs explicitly identified as oxygen-regulated [75] that were significantly differentially expressed to a lower extent at T4 relative to T0 in the tussock tundra community using DGE analysis (DGE; FDR < 0.05 for all; Table 4). This included oxygen-regulated KOs that are negatively affected by O2 availability expressed in carbohydrate metabolism (K00975, K00656), cofactor and vitamins metabolism (K00763), amino acid metabolism (K01915), and carbon metabolism (K00873, K01623, K00626) (Table 4). Only aldehyde dehydrogenase (K00128), an oxygen-regulated KO positively affected by O2 availability expressed in carbohydrate metabolism, was expressed at a significantly higher rate at T4 relative to T0 (Table 4). We also found that oxygen-regulated KOs negatively affected by O2 availability expressed in anaerobic carbon fixation (K00174, K00855), fermentation (K00169), and acetoclastic methanogenesis (K00625, K00925, K01895), were all expressed to a significantly lower extent at T4 relative to T0 in the tussock tundra community (DGE; FDR < 0.05; Table 4).

Table 4.
Differential gene expression (DGE) for oxygen-regulated KEGG orthologs (KOs). Each KO represents an oxygen-regulated gene and was included based on DGE significance within Tussock T4 vs. T0 sampling period (FDR < 0.05; Dataset S5), where a positive log-fold change (logFC) value indicates higher expression at T4 relative to T0.
From our DGE analysis, we also found that KOs related to ribosome translation (KEGG tier II Genetic Information Processing), including DNA repair proteins (K03584), RNA polymerase (K03040, K03043, K03046), pre-mRNA-processing factors (K12856, K12821), RNA-binding proteins (K12876), and more than 150 differentially expressed ribosomal protein genes (small subunit, large subunit; ko03010 path) from multiple bacterial phyla and even several species of fungi within the Ascomycetes, were all expressed to a significantly higher extent at T4 relative to T0 in the tussock tundra community (DGE; FDR < 0.05; Dataset S5). KOs related to Caulobacter cell cycling (KEGG tier II Cellular Processes), including ATP-dependent Lon protease (K01338), ATP-dependent Clp protease (K01358), and cell division proteins (K03531) also had significantly higher expression at T4 relative to T0 (DGE; FDR < 0.05; Dataset S5). KOs related to ferroptosis (KEGG tier II Cellular Processes), including an iron-regulated transporter for solute carrier family 40 (K14685), which regulates iron ion transmembrane transporter activity, also had significantly higher expression at T4 relative to T0 (DGE; FDR < 0.05; Dataset S5).
Overall, the DGE analysis indicated that the greatest differential expression of KOs was between tussock tundra and wet sedge tundra communities at each sampling time point. Specifically, 24% of KOs (out of 166,754 total KOs) were differentially expressed between tundra communities at T0 (DGE; FDR < 0.05; Table 5). Similarly, 28% and 10% of KOs were differentially expressed between tundra communities at T4 and T24, respectively (DGE; FDR < 0.05; Table 5). Within the tussock tundra community, DGE results indicated that 1.3% of KOs were differentially expressed between sampling time points T4 and T0 (out of 100,055 total KOs; FDR < 0.05; Table 5), which included the oxygen-regulated KOs (Table 4) as well as all KOs in the KEGG tier II categories of Genetic Information Processes and Cellular Processes (Dataset S5) as described above. The DGE analysis for all remaining time point comparisons in both tundra communities indicated that < 0.1% of total KOs were differentially expressed (DGE; FDR < 0.05; Table 5).

Table 5.
Summary of the differential gene expression (DGE) analysis. Values represent counts of KEGG orthologs (KOs; i.e., genes) considered to be differentially expressed if the False Discovery Rate (FDR) was < 0.05. “Higher” or “Lower” differential expression is relative to the first variable in the comparison (e.g., T4 vs. T0 summarizes higher or lower expression in T4 relative to T0).
3.6. Iron Redox Cycling Gene Expression
Within the tussock tundra community, the relative abundance of expressed obligate aerobic genes associated with iron oxidation was significantly greater at T4 relative to their abundance at T0 (ANOVA; p = 0.04; Figure 5A). The relative abundance of expressed obligate anaerobic genes associated with dissimilatory iron reduction was negligible (<1% of expressed iron gene relative abundance; Figure 5A). Furthermore, the relative abundance of dissimilatory iron reducing genes in the tussock tundra metagenome dataset was negligible (<1% of total iron gene relative abundance; Figure 5A), indicating a complete lack of dissimilatory iron reducing genes in the genomic potential of the microbial community. The mean relative abundance of expressed iron oxidation genes in the tussock tundra community was 43-fold greater than the mean relative abundance of expressed iron reduction genes when averaged across all sampling time points (paired t-test; p < 0.01; Figure 5A). In contrast, there were no significant differences in the relative abundance of expressed iron oxidation genes or iron reduction genes between sampling time points in the wet sedge tundra community (ANOVA; p = 0.2; Figure 5B). The mean relative abundance of expressed iron reduction genes in the wet sedge tundra community was 4-fold greater than the mean relative abundance of expressed iron oxidation genes when averaged across time points (paired t-test; p < 0.01; Figure 5B).
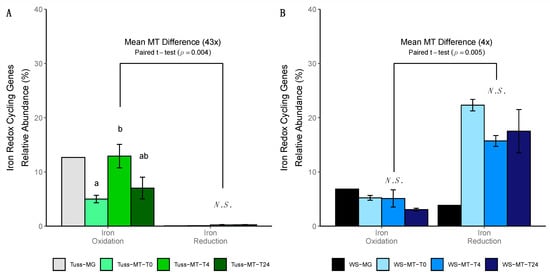
Figure 5.
Barplot of iron redox cycling gene expression based on the mean relative abundance of iron oxidation and iron reduction genes at each sampling time point in (A) tussock tundra and (B) wet sedge tundra communities (error bars represent standard deviation of the mean). Letters indicate significant differences in mean pairwise similarity between sampling time points in each iron redox cycling functional category (ANOVA; p < 0.05), with non-significant differences between sampling time points indicated as N.S. The relative abundance of each metagenome (MG) iron redox cycling functional category is provided for reference only and was not incorporated in the pairwise analysis.
3.7. Soil Incubations
The rates of microbial respiration (CO2 as ng C g−1 soil h−1) did not differ significantly between sampling time points in the tussock tundra or wet sedge tundra communities (ANOVA; p = 0.9, 0.3, respectively; Figure 6A). However, the mean rate of microbial respiration was 4.5-fold higher in tussock tundra compared to wet sedge tundra when averaged across sampling time points (paired t-test; p < 0.001; Figure 6A). In both tundra soils, the rates for CH4 production (CH4 as pg C g−1 soil h−1) measured at T0, which was incubated under anoxic conditions, were significantly higher than rates measured at T4 and T24, where soils were incubated under oxic conditions (ANOVA; p < 0.001 for all; Figure 6B). Negative values for CH4 production at T4 and T24 indicate aerobic CH4 consumption under the oxic incubation conditions (Figure 6B).
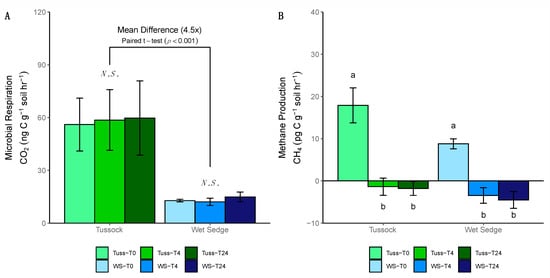
Figure 6.
Barplot of production rates for (A) microbial respiration and (B) methane (CH4) production for tussock tundra and wet sedge tundra communities (error bars represent standard deviation of the mean). Letters indicate significant differences in mean pairwise similarity between sampling time points (ANOVA, p < 0.05), with non-significant differences between sampling time points indicated as N.S. Negative values for CH4 production indicate CH4 consumption.
4. Discussion
We used rainfall to add oxygen and change redox conditions in the active layer of saturated tundra soils, and measured gene expression of the microbial community. We determined that the microbial response to rainfall was not consistent between tussock and wet sedge tundra types, and the patterns of gene expression differed due to inherent differences in microbial composition and their genomic potential. We also determined that differences in soil drainage rates during the rainfall event led to a greater increase in soil O2 concentration in the tussock tundra soil. We measured microbial gene expression at broad metabolic levels as well as from select oxygen-regulated marker genes and coupled our observed changes in microbial gene expression to the modeled changes in soil O2 concentrations following rainfall to determine the mechanistic response of the microbial communities between tundra types.
4.1. Microbial Composition Differs among Tundra Communities
Previous research conducted in the Toolik Lake region showed that microbial community composition is distinct among tundra types due to landscape-dependent differences in plant community composition and litter biochemistry [37,38], as well as physical gradients of soil drainage [77,78]. Tussock tundra is dominated by sedges (Eriophorum vaginatum) and dwarf shrubs (Betula nana, Ledum palustre) that occupy hillslopes with greater soil drainage than wet sedge tundra, which is dominated entirely by sedges (Carex chordorrhize, C. rotundata, Eriphorum aquatilis, E. angustifolium) at the base of hillslopes and valley bottoms where soil water accumulates [79]. These ecological and physical differences lead to compositionally distinct microbial communities between tussock tundra and wet sedge tundra soils, but previous studies of these communities were limited to PLFA and T-RFLP analyses [38,45], which provided limited information about the phylogenetic composition of these communities.
Here, higher resolution meta-omics analyses showed significant differences in the relative abundance of nearly all bacterial phyla between tussock tundra and wet sedge tundra communities (Table 3). Specifically, we found greater relative abundance of Acidobacteria in the tussock tundra community, likely related to the lower pH of the tussock tundra soil compared to wet sedge tundra soil (Table 1). This is consistent with previous studies showing strong negative correlations between Acidobacteria abundance and soil pH [80,81,82]. We also found a greater relative abundance of Alpha- and Gammaproteobacteria in tussock tundra soil that could be related to their copiotrophic life strategies because nutrient levels are normally greater in tussock tundra soils relative to the downslope wet sedge tundra soils [83], albeit with seasonal soil nitrogen limitations on microbial growth [84]. In addition, our MAGs annotated as Acidobacteria, Alpha- and Gammaproteobacteria had relatively large genomes (Table S6), which is a feature of copiotrophs that prefer nutrient-rich environments [85]. The high relative abundance of Actinobacteria in both tundra communities is likely due to the periodic (tussock tundra) or persistent (wet sedge tundra) saturation of the active layer and subsequent anoxia because Actinobacteria play an important role in the anaerobic degradation of soil organic carbon [80]. Firmicutes are also known to contribute to anaerobic soil organic carbon degradation [80] and their greater relative abundance in the wet sedge tundra community likely emphasizes the fact that wet sedge tundra soils experience more persistent soil anoxia due to longer periods of soil water ponding than in tussock tundra soils. Furthermore, we found that wet sedge tundra soils also harbored greater relative abundance of Deltaproteobacteria (Table 3), which included known anaerobic iron reducing Geobacter spp. (Table S7) that have previously been found in high abundance in the Toolik Lake region [86].
The dominant archaeal taxa were primarily methanogenic Euryarchaeota, almost exclusive to the wet sedge tundra community (Table 3; Figure 1A,B). Specifically, we found that Methanobacteria, Methanosarcina, and Methanothrix were the dominant archaeal taxa in the wet sedge tundra community (Table S7), which is consistent with previous findings in organic soils across circumpolar arctic sites [32,42,43,44,87]. Members of the Methanobacteria are hydrogenotrophic methanogens most commonly found in upper, saturated soils [42,88] while members of Methanosarcina and Methanothrix are acetoclastic methanogens that are typically the dominant archaeal taxa in organic peat soils [89,90]. The high relative abundance of both groups of methanogens in the wet sedge tundra community suggests an even greater genomic potential for CH4 production because multiple pathways of methanogenesis can be carried out depending on substrate availability in the soil. In contrast, we were only able to detect a very low abundance of archaeal taxa in the tussock tundra community, mainly Methanosarcina and Methanothrix spp. (Dataset S4). This may be due to the greater extent of soil drainage in the active layer that prevents substantial colonization by anaerobic archaeal taxa, or it may be an artifact of our soil samples being collected at the 10–20 cm depth in the organic active layer, which may have under-sampled the bulk of methanogenic taxa reportedly found in deeper mineral soil layers in arctic soils [42]. However, even the low abundance of these active acetoclastic methanogens in the tussock tundra community led to significant declines in the expression of phosphate acetyltransferase, acetate kinase, and acetyl-CoA synthetase genes (Table 4), which are essential genes in the production pathway of CH4 from acetate, following rainfall-induced soil oxidation.
Soil fungi were more abundant in the tussock tundra community than the wet sedge tundra community (Table 3; Figure 1A,B), confirming previous findings using PLFA ratios [38]. Historically, soil fungi have been under-sampled in arctic tundra [91] and we provide some of the first evidence that numerous filamentous taxa within the Ascomycetes are common in tussock tundra and absent from wet sedge tundra (Table S7). The lack of fungi in the wet sedge tundra community may be due to environmental limitations on filamentous fungi, which with few exceptions are obligate aerobes [92], and to their inability to disperse into and colonize the persistently anaerobic conditions of the wet sedge tundra soil [38]. Additionally, sedge species (Carex and Eriophorum) that dominate wet sedge tundra vegetation do not typically associate with mycorrhizal fungi whereas most dwarf shrubs in the tussock tundra plant community (Betula and Salix) form mycorrhizal associations [93]. In addition, taxa within the Leotiomycetes class (Helotiales order) such as Glarea spp. and Sclerotinia spp. found here (Dataset S4) are known to express a broad suite of functional genes that degrade cellulolytic plant litter [94]. The presence of fungi, even though their abundance in the microbial community is relatively small, likely increases the genomic potential of the tussock tundra community to degrade soil organic carbon under aerobic conditions because of their unique suite of functional genes.
Differences in the relative abundance and diversity of microbial taxa between tussock tundra and wet sedge tundra communities likely reflect landscape-dependent patterns of community composition and lifestyle strategies. For example, the composition of the tussock tundra community suggests a copiotrophic soil community dominated by relatively few bacterial phyla and filamentous fungi that prefer more frequently aerated, nutrient-rich soils. In contrast, the higher diversity and composition of the microbial community within the persistently saturated wet sedge tundra soils appear more similar to oligotrophic sediment communities measured from lakes and streams in the Toolik Lake region [37,95]. This comparison is reasonable given that the greater nutrient availability in tussock tundra soils [83] likely supports more copiotrophic life strategies prevalent among the dominant taxa found in the tussock tundra community. Likewise, wet sedge tundra borders stream and lake margins [39], where groundwater flows through the active layer of wet sedge tundra and into adjacent waterways, likely transporting the soil microbial taxa [37] that constitute the bulk of the diversity in river or sediment communities [95].
4.2. Genomic Potential Differs between Tundra Communities
Consistent with the differences in microbial composition between tundra communities, we found that the genomic potential of these distinct tundra communities differed from each other at the broad KEGG tier II categories (Figure S3C). The wet sedge tundra community had a greater relative abundance of genes belonging to the Energy Metabolism pathway, suggesting a greater genomic potential for carbon metabolism, especially methanogenesis under anoxic soil conditions. In contrast, the tussock tundra community had a greater relative abundance of most other Metabolism categories at all sampling time points during our rainfall experiment (Figure 2A,B), possibly due to the fact that the tussock tundra community had lower alpha diversity of functional genes relative to the wet sedge tundra community (Tables S4 and S5). Yet, stark differences in the relative abundance of functional genes between tundra communities are seen in the beta diversity PCoA plot (Figure S3B) where functional genes separate most strongly by tundra type independent of differences in soil redox conditions between soil sampling time points. Taken together, our results suggest that community composition and the associated functional genes (i.e., genomic potential) are distinct between tundra communities, which in turn may constrain or predict the microbial gene expression patterns following rainfall-induced redox changes in tussock tundra and wet sedge tundra soils.
4.3. Soil O2 Accumulation during Rainfall Differs among Tundra Soils
A novel aspect of our mesocosm experimental design was our ability to model (Figure S2) the rates of soil O2 accumulation introduced during the simulated rainfall event (Frain) and soil O2 consumption (Sall) during the stagnant re-acclimation phase following rainfall. Using the soil O2 accumulation rates, we estimated the final soil O2 concentration immediately following rainfall (Mf). Then, by incorporating the soil O2 consumption rates, we further estimated the final concentrations of soil O2 at the 10–20 cm sampling depth within tussock tundra and wet sedge tundra at the 4-h (Mf+4) and 24-h (Mf+24) sampling time points following the rainfall event. In our mass balance model (Figure S2), we assumed that the contribution of O2 from atmospheric diffusion into saturated soils (Fatm) to a depth of 10–20 cm was negligible following the rainfall event given that gas diffusion in water is slower than in air by a factor of 104 [96,97]. We also assumed that the extent of any atmospheric O2 introduced deep into saturated soils via diffusion through the aerenchyma tissues of dominant sedge species [98,99] would be limited to the immediate vicinity surrounding plant roots because of the slow diffusion rate of gas in water, thus limiting the role of plant-mediated increases in soil O2 concentrations. Likewise, we assumed that the contribution of soil O2 introduced through groundwater flow (Fground) or removed through soil leaching (Fleach) would be zero given that our mesocosm chambers were only open at the top and sealed at the bottom, except during the rainfall event.
Our mass balance model found that the accumulation of soil O2 introduced through rainfall differed among tundra soils, with tussock tundra soils accumulating 2.5-fold more soil O2 than wet sedge tundra soils (Table 2). This greater accumulation of soil O2 in tussock tundra occurred even though both tundra soils experienced similar rates of soil O2 accumulation and were flushed with identical volumes of DI water during the simulated rainfall event. We found that the duration of the DI water flush, which took nearly 3.5 times longer to complete in the tussock tundra soil, was the primary mechanism responsible for the greater accumulation of soil O2. Soil drainage through the active layer of tussock tundra is strongly inhibited by the dense mineral layer located below the surface organic layer. Typically, the mineral layer in tussock tundra strictly limits deeper soil drainage under field conditions and the surface organic layer remains saturated periodically following rain events due to its high water-holding capacity [100,101] until the soil water drains downslope through the organic layer into wet sedge tundra soils located at the base of hills [102]. Thus, the longer residence time of oxygenated rainwater added to the surface organic layer of tussock tundra led to greater O2 concentrations at the 10–20 cm sampling depth at the end of the rainfall event compared with the wet sedge tundra soil, which drained the oxygenated rainwater quickly through its porous organic soil. This rapid flushing of the oxygenated rainwater through the saturated wet sedge tundra mesocosm chamber is likely similar to field conditions where rainwater rapidly flushes through the saturated active layer and into an adjacent stream or lake, thus limiting O2 accumulation in the soil.
The greater accumulation of soil O2 in the tussock tundra soil compared to the wet sedge tundra soil at the end of the rainfall event was based solely on the accumulation rate of soil O2 introduced during the DI water flush. However, after applying the soil O2 consumption rates that were modeled from the re-acclimation phase following rainfall, we still found that the final soil O2 concentrations at 4-h and 24-h following rainfall were 2.5-fold greater in tussock tundra soil compared with wet sedge tundra soil (Table 2). This difference in soil O2 concentration between tundra soils was consistent among time points even though the rate of soil O2 consumption was greater in the tussock tundra soil than in the wet sedge tundra soil (Table 2). An important note is that the soil O2 consumptions rates are likely minimum estimates of O2 consumption from abiotic reactions and biotic processes because the rates do not account for O2 consumed during the slow diffusion of O2 into the stagnant boundary layer of the soil cores [5].
4.4. Tussock Tundra Community Responds to Rainfall-Induced Soil Oxidation
For the tussock tundra community, we identified short-term (T4) and longer-term (T24) changes in microbial gene expression (Figure 3A and Figure 4A,B) following an 8.5-fold increase in total soil O2 concentration introduced during rainfall (Table 2). These changes suggest that the increase in soil O2 stimulated microbial taxa to invest more energy towards growth by transcribing ribosomal protein genes, and to invest less in resource acquisition via membrane transport and associated metabolic pathways. These changes also suggest a shift in expression from lower-energy anaerobic metabolic pathways to higher energy aerobic pathways (Figure 4A,B; Table 4). DGE analysis identified elevated expression of more than 150 ribosomal protein genes across multiple bacterial and fungal taxa, and of genes for DNA repair proteins, RNA polymerase, pre-mRNA-processing factors, and RNA-binding proteins (Dataset S5). This functional gene response to rainfall was similar to the response observed in a previous study investigating the effects of sunlight photo-alteration of dissolved organic matter (DOM) leached from tussock tundra soils on microbial gene expression [103]. This study concluded that photo-chemical changes in DOM made available easily metabolized compounds that cause microbes to enter lag phase or log phase growth. This idea that rainfall and soil oxygenation releases compounds that boost microbial growth in tussock tundra is further supported by results from the abiotic response cores [5] showing that the composition of dissolved organic carbon (DOC) changed during flushing. In these cores there was a significant decrease in the fluorescence index of the DOC as the volume of water flushed through the soil increased, indicating that the DOC exported was likely more aromatic [104], which could stimulate an increase in microbial metabolism that leads to rapid growth.
The increase in expression of genes for microbial growth following rainfall was coupled with a subsequent decrease in the expression of genes for anaerobic methane metabolism associated with acetoclastic methanogenesis such as phosphate acetyltransferase, acetate kinase, and acetyl-CoA synthetase (Table 4). There were also significant declines in the expression of genes for anaerobic carbon fixation and fermentation (Table 4). We found lower expression for glucose-1-phosphate adenylyltransferase (Table 4), which has been considered the rate-limiting step of bacterial glycogen biosynthesis [105] or the first step of glycogen hydrolysis [106]. Glycogen can serve as an alternative carbon source for bacterial survival in carbon-depleted anaerobic environments [106] or to prolong the lifespan of microorganisms while dormant [107]. Thus, the lower expression of glucose-1-phosphate adenylyltransferase, an oxygen-regulated gene negatively affected by O2 availability, following rainfall provides further evidence that the increase in soil oxygenation made available more easily metabolized carbon substrates (e.g., aromatic DOC) [103] such that microbes no longer needed to synthesize and degrade glycogen, or possibly that microbial cells may have started to leave their dormant state. We found lower expression for formate C-acetyltransferase (Table 4) in multiple Actinobacterial taxa, which is another oxygen-regulated gene negatively affect by O2 availability that helps regulate anaerobic glucose metabolism [108]. Furthermore, we found lower expression for pyruvate kinase (Table 4), a negatively affected oxygen-regulated gene involved in glycolysis that helps regulate anaerobic cell metabolism [109]. In contrast, we found higher expression for aldehyde dehydrogenase (NAD+) (Table 4), an oxygen-regulated gene positively affected by O2 availability that catalyzes the oxidation of aldehydes into carboxylic acids [110] or promotes the production of acetate from acetaldehyde under aerobic conditions [111]. The change in expression (higher or lower) of each of these marker genes that are positively or negatively regulated by O2 availability following rainfall is consistent with our expectations given the rapid increase in soil O2 introduced during the rainfall event.
We also investigated the microbial expression of aerobic Fe(II) oxidation by iron oxidizing bacteria as further support that our rainfall event stimulated gene expression by aerobic bacteria in the tussock tundra community. Previous studies showed that iron oxidizing Betaproteobacteria such as Leptothrix and Gallionella spp. are found in abundance in the Toolik Lake region [86]. Here, we found evidence of their ribosomal genes in our metagenomic and metatranscriptomic datasets (Dataset S4). These iron oxidizing bacteria use Fe(II) as their primary energy source to fix CO2 into cellular biomass by reducing dissolved O2 to water [112]. Thus, the greater expression of aerobic iron oxidizing genes following our simulated rainfall event in tussock tundra (T4; Figure 5A) is also consistent with our expectations for aerobic gene expression following the rapid increase in soil O2 availability introduced during our rainfall event.
Our metatranscriptomic analysis also revealed an increase in ferroptosis gene expression following rainfall in tussock tundra (Figure 4). Ferroptosis is a form of regulated cell death that results from the iron-dependent accumulation of lipid peroxides associated with reactive oxygen species [113]. Previous research has found that the overproduction of hydroxyl radical is significantly associated with lipid peroxidation in ferroptosis [114]. Here, it is likely that gene expression for ferroptosis is directly linked with the overproduction of hydroxyl radical given that hydroxyl radical production associated with the geochemical oxidation of Fe(II) can increase in tussock tundra following rainfall [5]. However, little is known about the regulation of ferroptosis in soil environments or even how it is regulated within microbial cells regardless of environment [115]. While we find this potential association between hydroxyl radical generation and ferroptosis expression after rainfall interesting, more studies need to be done to determine any direct link between these two processes.
4.5. Wet Sedge Tundra Community Maintains Anaerobic Gene Expression Following Rainfall
Results previously reported from the abiotic response cores [5] indicate that the amount of soil O2 introduced during rainfall was sufficient to increase geochemical (abiotic) oxidation reactions in wet sedge tundra soils, although these geochemical reactions occurred at a lower rate than in tussock tundra soils. Here, we were unable to detect a significant increase in the relative abundance of expressed genes involved in aerobic respiration (Dataset S5) or bacterial iron oxidation (Figure 5B) following rainfall in wet sedge tundra regardless of the 4-fold increase in total soil O2 concentration introduced during the rainfall event (Table 2). Furthermore, we were unable to detect a significant change in the relative abundance of any expressed gene pathways (Figure 3B), oxygen-regulated genes (Table S8; Dataset S5), or iron redox cycling genes (Figure 5B) following rainfall. Active microbial taxa continued to express genes that degrade organic compounds and inorganic nutrients through anaerobic respiration or fermentation pathways (Dataset S3), and actually increased their expression of numerous anaerobic denitrification genes following rainfall (Dataset S5). This suggests that the redox conditions in the soil environment were comparatively poor for microbial growth because they promoted anaerobic microbial taxa to allocate more energy towards resource acquisition, which microbes only do when resources are scarce [116].
The lack of microbial response to rainfall in our wet sedge cores may be due to an insufficient accumulation of oxygenated rainwater during the rainfall event because it drained through the porous organic soil too quickly. In addition, rainwater O2 may have been consumed too rapidly by abiotic reactions before microbes could respond and shift their expression to promote aerobic respiration pathways. For example, the higher pH in wet sedge tundra (6.12 ± 0.06; Table 1) likely promotes the abiotic oxidation of Fe(II) over biotic processes [112], which could account for the observed rates of Fe(II) oxidation in the abiotic response cores [5]. However, we observed gene expression for the aerobic oxidation of Fe(II) in the wet sedge tundra community (Figure 5B), which suggests that some soil O2 was available and consumed through biotic processes throughout the experiment. It is also plausible that energetic constraints caused by the low availability of dissolved O2 and alternative electron acceptors maintained anaerobic metabolism and fermentation pathways throughout the rainfall event cf. [117]. The high abundance and diversity of oligotrophic, and mainly anaerobic microbial taxa in the wet sedge tundra community may also have constrained a community-wide shift from anaerobic metabolism following soil oxidation. This argument applies well to the short timeframe of the rainfall-induced oxidation event, where our soil model implies that the bulk of the oxygen in rainwater was consumed back to pre-rainfall anoxic concentrations within several days. As such, the oligotrophic community in wet sedge tundra soils may be poorly adapted to respond rapidly to periodic bursts of soil oxygenation introduced during rainfall.
We also observed that the relative expression of genes for Fe(III) reduction by iron reducing bacteria did not decline following our rainfall event, with expression for Fe(III) reduction occurring at 4-fold greater abundance relative to the expression for Fe(II) oxidation averaged across sampling time points (Figure 5B). Emerson et al. [86] found iron reducing Geobacter spp. (Deltaproteobacteria) in high abundance in the Toolik Lake region. We also found evidence of Geobacter spp. ribosomal genes in our metagenomic and metatranscriptomic datasets, but only in the wet sedge tundra community (Table S7). These iron reducing bacteria use Fe(III) as a terminal electron acceptor to carry out anaerobic respiration coupled to the mineralization of organic carbon under anoxic conditions [118]. The consistent expression of anaerobic iron reducing genes before and after rainfall, and in greater abundance relative to the aerobic iron oxidizing genes in the wet sedge tundra community further implies that rainfall does not appear to alter community-wide anaerobic gene expression regardless of the rapid increase in soil O2 availability introduced during the rainfall event.
Taken together, rainfall events in wet sedge tundra may serve only to maintain persistently saturated soil conditions that promote anaerobic metabolism and CH4 production. The unaltered expression of anaerobic and fermentative pathways following an increase in soil O2 concentration introduced during rainfall, coupled with the high diversity and abundance of methanogenic archaea active within these soils provides support for this interpretation. Additionally, the combination of warmer soil temperatures driven by an increase in rainfall and the concurrent increase in active layer depth with persistently saturated conditions throughout the active layer profile has led to an increase in CH4 emissions by ~30% [21]. Here, we add further evidence that rainfall will likely enhance CH4 production from low-lying permafrost regions such as wet sedge tundra by maintaining saturated soil conditions rather than altering the highly reduced soil redox conditions through an increase in dissolved O2 availability.
4.6. Relating Microbial Expression to Rates of Respiration and CH4 Production
Our results showed no correlation between aerobic microbial gene expression and rates of microbial respiration (CO2 production) following rainfall in tussock tundra soils. Specifically, the consistent rates of microbial respiration before and after rainfall (Figure 6A) did not reflect the increase we observed in aerobic microbial gene expression in the tussock tundra community following rainfall (Figure 4). This result is not unexpected because previous studies have also been unable to directly link microbial dynamics and carbon emissions [119], likely because respiration is the product of many processes in microbial communities. Here, the artificial conditions of the respiration rate incubations (i.e., N2 gas for anoxic conditions, compressed air for oxic conditions) were intended to replicate field conditions, similar to previous incubation studies [38], but it is possible that other conditions in these incubations such as total O2 availability and soil moisture differed from conditions in the soil mesocosms where microbial gene expression was sampled. It should be noted that our measured rates of CO2 production (Figure 6A) were similar to rates previously measured from soil waters at our study sites (19.6 ± 3.0 ng C g−1 soil h−1) [36]. Additionally, the greater rate of CO2 production in the tussock tundra soil compared with the wet sedge tundra soil was consistent with the ~3.4-fold greater release of carbon from aerobic soils compared with anaerobic soils across circumpolar arctic sites [120].
Our measured rates of CH4 production under anoxic incubation conditions (Figure 6B) were also comparable with previous rates (37.2 ± 51.8 pg C g−1 soil h−1) [36]. These results support a common interpretation that CH4 was produced by methanogenic archaea in the anoxic environment but likely consumed by CH4-oxidizing bacteria in the oxidized environment. We have evidence of gene expression from multiple pathways of methanogenesis associated with methanogenic archaea as well as evidence of gene expression from methanotrophy pathways associated with Actinobacteria and multiple classes of Proteobacteria in both tundra communities (Table S8; Dataset S3). However, the greater production of CH4 under anoxic conditions at T0 from the tussock tundra soil compared to the wet sedge tundra soil (Figure 6B) is inconsistent with the lower relative abundance of methanogenic archaea in the tussock tundra community compared to the wet sedge tundra community (Table 3) or with the broader suite of methanogenic pathways available to the diverse composition of methanogenic archaea within the wet sedge tundra (Dataset S3). It is possible that the rates of CH4 production measured from our tussock tundra soil incubations (and to some extent measured in our wet sedge tundra soils) represent the natural release of trapped CH4 following disturbance to the soil [3], rather than just biological production under the saturated, anoxic soil conditions in this study.
5. Conclusions
Results from this study suggest that rainfall differentially affected soil redox conditions between tundra soil types due to inherent differences in soil drainage rates between tussock tundra and wet sedge tundra. Measurements and our soil oxygen model indicate that rainfall increased the dissolved O2 concentration among tundra soils, but the extent of soil oxygenation was 2.5-fold greater in tussock tundra compared with wet sedge tundra. The substantial difference in soil oxygenation between tundra types during rainfall appears to be the primary mechanism controlling the microbial response following rainfall in both the tussock tundra and wet sedge tundra communities. Gene expression from active microbial taxa in the tussock tundra community experienced a significant shift following rainfall from anaerobic metabolic pathways to aerobic pathways of Fe(II) oxidation and an increase in ribosome transcription that indicates microbial growth. In contrast, gene expression from active microbial taxa in the wet sedge tundra community remained consistent for anaerobic metabolism, fermentation pathways, and Fe(III) reduction before and after rainfall, regardless of the increase in dissolved O2 introduced by the rainfall event. As such, the rates of microbial respiration were greater in the tussock tundra community but did not correlate directly with our observed changes in aerobic microbial gene expression. The rates of CH4 production and its consumption under oxidizing conditions were similar among tundra soils but did not correlate proportionally with the greater abundance of methanogenic archaea and their diverse suite of methanogenic pathways in the wet sedge tundra community, possibly due to dissimilar soil conditions between the mesocosm chambers and the incubation jars. Validation of our metatranscriptomic results for gene expression involved in methanogenesis via quantitative PCR (qPCR) may provide clarity on the relationship between changes in methanogenic gene expression and measured rates of CH4 production in this experiment.
We conclude that differences in the extent of soil oxygenation introduced during rainfall and their impact on soil redox conditions clearly control whether microbial communities will shift gene expression from anaerobic metabolism to aerobic microbial growth, or simply continue to express anaerobic genes typical of anoxic soil conditions. Here, the microbial response in the tussock tundra community was consistent with our hypothesis that rainfall-induced soil oxidation would alter anaerobic gene expression and promote aerobic microbial activity, in large part due to the greater abundance of aerobic bacterial and fungal taxa and their suite of aerobic functional genes. In contrast, oxygen from rainfall does not appear to accumulate long enough, or to a sufficient concentration, in wet sedge tundra soils to induce a community-wide shift from anaerobic metabolism; rainfall may serve only in maintaining persistently saturated conditions that could potentially enhance CH4 production due to the high diversity of methanogenic archaea inhabiting these low-lying permafrost soils across the arctic landscape.
Supplementary Materials
The following are available online at https://www.mdpi.com/2571-8789/5/1/17/s1, Supplementary Material and Methods, Figure S1: Conceptual figure, Figure S2: Mass balance model, Figure S3: Metagenomic functional diversity, Figure S4: Metatranscriptomic functional diversity, Table S1: 16S sequencing summary, Table S2: DNA sequencing summary, Table S3: RNA sequencing summary, Table S4: Alpha diversity summary, Table S5: Beta diversity summary, Table S6: Metagenome assembled genomes (MAGs) summary, Table S7: Relative abundance of the active microbial taxa, Table S8: Oxygen-regulated functional KEGG orthologs (KOs), Dataset S1: 16S rRNA OTU taxonomy and relative abundance, Dataset S2: Metagenome functional KO annotations, Dataset S3: Metatranscriptome functional KO annotations, Dataset S4: Metagenome and metatranscriptome taxonomy and relative abundance, Dataset S5: Differential gene expression (DGE) results from metatranscriptome analysis.
Author Contributions
Conceptualization, K.J.R. and G.W.K.; methodology, K.J.R. and G.W.K.; formal analysis, K.J.R., B.C.C. and G.W.K.; data curation, K.J.R.; writing—original draft preparation, K.J.R.; writing—review and editing, K.J.R., B.C.C. and G.W.K.; funding acquisition, G.W.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Science Foundation (NSF), grant numbers DEB-1637459, DEB-1754835, and OPP-1936769. Additional block grant funding was provided to K.J.R. by the Department of Ecology and Evolutionary Biology, University of Michigan.
Data Availability Statement
Raw read sequences for 16S rRNA amplicons, metagenomes, and metatranscriptomes were deposited in the Sequence Read Archive of NCBI and are publicly accessible through BioProject accession PRJNA666429. Rendered code for full reproducibility of the bioinformatic and statistical analyses is available online at Supplementary Data.
Acknowledgments
We thank R. Cory, A. Trusiak, J. Dobkowski, C. Cook, L. Triebergs, J. Evans, D. Smith, J. Payet, and researchers, technicians, and support staff of the Arctic LTER project and Toolik Field Station for assistance. Additional thanks to the University of Michigan Advanced Genomics Core, Microbiome Core, and Advanced Research Computing Technology Services.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hugelius, G.; Strauss, J.; Zubrzycki, S.; Harden, J.W.; Schuur, E.A.G.; Ping, C.L.; Schirrmeister, L.; Grosse, G.; Michaelson, G.J.; Koven, C.D.; et al. Estimated stocks of circumpolar permafrost carbon with quantified uncertainty ranges and identified data gaps. Biogeosciences 2014, 11, 6573–6593. [Google Scholar] [CrossRef]
- Gorham, E. Northern peatlands: Role in the carbon cycle and probable responses to climatic warming. Ecol. Appl. 1991, 1, 182–195. [Google Scholar] [CrossRef]
- Margesin, R. Permafrost soils. In Soil Biology; Margesin, R., Ed.; Springer: Berlin/Heidelberg, Germany, 2019; Volume 16. [Google Scholar]
- Zona, D. Long-term effects of permafrost thaw. Nature 2016, 537, 625–626. [Google Scholar] [CrossRef][Green Version]
- Trusiak, A.; Treibergs, L.A.; Kling, G.W.; Cory, R.M. The controls of iron and oxygen on hydroxyl radical (OH) production in soils. Soil. Syst. 2019, 3, 1. [Google Scholar] [CrossRef]
- Herndon, E.M.; Kinsman-Costello, L.; Godsey, S. Biogeochemical cycling of redox-sensitive elements in permafrost-affected ecosystems. In Biogeochemical Cycles: Ecological Drivers and Environmental Impact, Geophysical Monograph; Dontsova, K., Balogh-Brunstad, Z., Le Roux, G., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 2020. [Google Scholar]
- Bubier, J.L.; Moore, T.R.; Bellisario, L.; Comer, N.T.; Crill, P.M. Ecological controls on methane emissions from a northern peatland complex in the zone of discontinuous permafrost, Manitoba, Canada. Glob. Biogeochem. Cycles. 1995, 9, 455–470. [Google Scholar] [CrossRef]
- Turetsky, M.R.; Treat, C.C.; Waldrop, M.P.; Waddington, J.M.; Harden, J.W.; McGuire, A.D. Short-term response of methane fluxes and methanogen activity to water table and soil warming manipulations in an Alaskan peatland. J. Geophys. Res.-Biogeo. 2008, 113, G00A10. [Google Scholar] [CrossRef]
- Olefeldt, D.; Turetsky, M.R.; Crill, P.M.; McGuire, A.D. Environmental and physical controls on northern terrestrial methane emissions across permafrost zones. Glob. Chang. Biol. 2013, 19, 589–603. [Google Scholar] [CrossRef]
- Miller, P.C.; Kendall, R.; Oechel, W.C. Simulating carbon accumulation in northern ecosystems. Simulation 1983, 40, 119–131. [Google Scholar] [CrossRef]
- McGuire, A.D.; Anderson, L.G.; Christensen, T.R.; Dallimore, S.; Guo, L. Sensitivity of the carbon cycle in the arctic to climate change. Ecol. Monogr. 2009, 79, 523–555. [Google Scholar] [CrossRef]
- Oechel, W.C.; Hastings, S.J.; Vourlitis, G.; Jenkins, M.; Riechers, G.; Grulke, N. Recent change of Arctic tundra ecosystems from a net carbon dioxide sink to a source. Nature 1993, 361, 520–523. [Google Scholar] [CrossRef]
- Euskirchen, E.S.; Bret-Harte, M.S.; Shaver, G.R.; Edgar, C.W.; Romanovsky, V.E. Long-term release of carbon dioxide from arctic tundra ecosystems in Alaska. Ecosystems 2017, 20, 960–974. [Google Scholar] [CrossRef]
- Osterkamp, T.E.; Romanovsky, V.E. Evidence for warming and thawing of discontinuous permafrost in Alaska. Permafr. Periglac. 1999, 10, 17–37. [Google Scholar] [CrossRef]
- Brown, J.G.; Hinkel, K.M.; Nelson, F.E. The circumpolar active layer monitoring (CALM) program: Research designs and initial results. Polar. Geogr. 2000, 24, 165–258. [Google Scholar] [CrossRef]
- Jorgenson, M.T.; Shur, Y.L.; Pullman, E.R. Abrupt increase in permafrost degradation in Arctic Alaska. Geophys. Res. Lett. 2006, 33, D22103. [Google Scholar] [CrossRef]
- Kumar, M.; Want, R.; Link, T.E. Effects of more extreme precipitation regimes on maximum seasonal snow water equivalent. Geophys. Res. Lett. 2012, 39, 1–6. [Google Scholar] [CrossRef]
- Spence, C.; Phillips, R.W. Refining understanding of hydrological connectivity in a boreal catchment. Hydrol. Process. 2015, 29, 3491–3505. [Google Scholar] [CrossRef]
- Bintanja, R.; Andry, O. Towards a rain-dominated Arctic. Nat. Clim. Chang. 2017, 7, 263–268. [Google Scholar] [CrossRef]
- Douglas, T.A.; Turetsky, M.R.; Koven, C.D. Increased rainfall stimulates permafrost thaw across a variety of interior Alaskan boreal ecosystems. Npj. Clim. Atmos. Sci. 2020, 28, 1–7. [Google Scholar] [CrossRef]
- Neumann, R.B.; Moorber, C.J.; Lundquist, J.D.; Turner, J.C.; Waldrop, M.P.; McFarland, J.W.; Euskirchen, E.S.; Edgar, C.W.; Turetsky, M.R. Warming effects of spring rainfall increase methane emissions from thawing permafrost. Geophys. Res. Lett. 2019, 46, 1393–1401. [Google Scholar] [CrossRef]
- Schuur, E.A.G.; Vogel, J.G.; Crummer, K.G.; Lee, H.; Sickman, J.O.; Osterkamp, T.E. The effect of permafrost thaw on old carbon release and net carbon exchange from tundra. Nature 2009, 459, 556–559. [Google Scholar] [CrossRef]
- Grosse, G.; Romanovsky, V.; Jorgenson, T.; Anthony, K.W.; Brown, J.; Overduin, P.P. Vulnerability and feedbacks of permafrost to climate change. Eos. Trans. Am. Geophys. Union. 2011, 92, 73–74. [Google Scholar] [CrossRef]
- Mackelprang, R.; Waldrop, M.P.; DeAngelis, K.M.; David, M.M.; Chavarria, K.L.; Blazewicz, S.J.; Rubin, E.M.; Jansson, J.K. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature 2011, 480, 368–371. [Google Scholar] [CrossRef]
- Xue, K.; Yuan, M.M.; Shi, Z.J.; Qin, Y.; Deng, Y.; Cheng, L.; Wu, L.; He, Z.; Van Nostrand, J.D.; Bracho, R.; et al. Tundra soil carbon is vulnerable to rapid microbial decomposition under climate change. Nat. Clim. Chang. 2016, 6, 595–600. [Google Scholar] [CrossRef]
- Wickland, K.P.; Striegl, R.G.; Neff, J.C.; Sachs, T. Effects of permafrost melting on CO2 and CH4 exchange of a poorly drained black spruce lowland. J. Geophys. Res. Biogeo. 2006, 111, G2. [Google Scholar] [CrossRef]
- Merbold, L.; Kutsch, W.L.; Corradi, C.; Kolle, O.; Rebmann, C.; Stoy, P.C.; Schulze, E.D. Artificial drainage and associated carbon fluxes (CO2/CH4) in a tundra ecosystem. Glob. Chang. Biol. 2009, 15, 2599–2614. [Google Scholar] [CrossRef]
- Huemmrich, K.F.; Kinoshita, G.; Gamon, J.A.; Houston, S.; Kwon, H.; Oechel, W.C. Tundra carbon balance under varying temperature and moisture regimes. J. Geophys. Res. 2010, 115, 1–8. [Google Scholar] [CrossRef]
- Zona, D.; Lipson, D.A.; Paw, U.K.T.; Oberbauer, S.F.; Olivas, P.; Gioli, B.; Oechel, W.C. Increased CO2 loss from vegetated drained lake tundra ecosystems due to flooding. Glob. Biogeochem. Cycles 2012, 26, 1–16. [Google Scholar] [CrossRef]
- Elberling, B.; Michelsen, A.; Schadel, C.; Schuur, E.A.G.; Christiansen, H.H.; Berg, L.; Tamstorf, M.P.; Sigsgaard, C. Long-term CO2 production following permafrost thaw. Nat. Clim. Chang. 2013, 3, 890–894. [Google Scholar] [CrossRef]
- Lawrence, D.M.; Koven, C.D.; Swenson, S.C.; Riley, W.J.; Slater, A.G. Permafrost thaw and resulting soil moisture changes regulate projected high-latitude CO2 and CH4 emissions. Environ. Res. Lett. 2015, 10, 094011. [Google Scholar] [CrossRef]
- Lipson, D.A.; Haggerty, J.M.; Srinivas, A.; Raab, T.K.; Sathe, S.; Dinsdale, E.A. Metagenomic insights into anaerobic metabolism along an arctic peat soil profile. PLoS ONE 2013, 8, e64659. [Google Scholar] [CrossRef]
- Ivanova, A.A.; Wegner, C.E.; Kim, Y.; Liesack, W.; Dedysh, S.N. Identification of microbial populations driving biopolymer degradation in acidic peatlands by metatranscriptomic analysis. Mol. Ecol. 2016, 25, 4818–4835. [Google Scholar] [CrossRef]
- Kolb, S. Aerobic methanol-oxidizing bacteria in soil. Fems. Microbiol. Lett. 2009, 300, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Walker, M.D.; Walker, D.A.; Auerback, N.A. Plant communities of a tussock tundra landscape in the Brooks Range foothills, Alaska. J. Veg. Sci. 1994, 5, 843–866. [Google Scholar] [CrossRef]
- Judd, K.E.; Kling, G.W. Production and export of dissolved C in arctic tundra mesocosms: The roles of vegetation and water flow. Biogeochemistry 2002, 60, 213–234. [Google Scholar] [CrossRef]
- Judd, K.E.; Crump, B.C.; Kling, G.W. Environmental drivers control eco- system function in bacteria through changes in community composition. Ecology 2006, 87, 2068–2079. [Google Scholar] [CrossRef]
- Zak, D.R.; Kling, G.W. Microbial community composition and function across an arctic tundra landscape. Ecology 2006, 87, 1659–1670. [Google Scholar] [CrossRef]
- Hobbie, J.E. A Changing Arctic: Ecological Consequences for Tundra, Streams, and Lakes; Kling, G.W., Ed.; Oxford University Press: Oxford, UK, 2014. [Google Scholar]
- Ward, C.P.; Cory, R.M. Chemical composition of dissolved organic matter draining permafrost soils. Geochim. Cosmochim. Acta 2015, 167, 63–79. [Google Scholar] [CrossRef]
- Tas, N.; Prestat, E.; Wang, S.; Wu, Y.; Ulrich, C.; Kneafsey, T.; Tringe, S.G.; Torn, M.S.; Hubbard, S.S.; Jansson, J.K. Landscape topography structures the soil microbiome in arctic polygonal tundra. Nat. Commun. 2018, 9, 777. [Google Scholar] [CrossRef]
- Deng, J.; Gu, Y.; Zhang, J.; Xue, K.; Qin, Y.; Yuan, M.; Yin, H.; He, Z.; Wu, L.; Schuur, E.A.G.; et al. Shifts of tundra bacterial and archaeal communities along a permafrost thaw gradient in Alaska. Mol. Ecol. 2015, 24, 222–234. [Google Scholar] [CrossRef]
- Hultman, J.; Waldrop, M.P.; Mackelprang, R.; David, M.M.; McFarland, J.; Blazewicz, S.J.; Harden, J.; Turetsky, M.R.; McGuire, A.D.; Shah, M.B.; et al. Multi-omics of permafrost, active layer and thermokarst bog soil microbiomes. Nature 2015, 521, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, B.M.; Kim, M.; Kim, Y.; Byun, E.; Yang, J.W.; Ahn, J.; Lee, Y.K. Variations in bacterial and archaeal communities along depth profiles of Alaskan soil cores. Sci. Rep. 2018, 8, 504. [Google Scholar] [CrossRef]
- McMahon, S.K.; Wallenstein, M.D.; Schimel, J.P. A cross-seasonal comparison of active and total bacterial community composition in arctic tundra soil using bromodeoxyuridine labeling. Soil. Biol. Biochem. 2011, 43, 287–295. [Google Scholar] [CrossRef]
- Jansson, J.K.; Hofmockel, K.S. The soil microbiome—From metagenomics to metaphenomics. Curr. Opin. Microbiol. 2018, 43, 162–168. [Google Scholar] [CrossRef]
- Page, S.E.; Logan, J.R.; Cory, R.M.; McNeill, K. Evidence for dissolved organic matter as the primary source and sink of photochemically produced hydroxyl radical in arctic surface waters. Environ. Sci. Proc. Impacts 2014, 16, 807–822. [Google Scholar] [CrossRef] [PubMed]
- Trusiak, A.; Treibergs, L.A.; Kling, G.W.; Cory, R.M. The role of iron and reactive oxygen species in the production of CO2 in arctic soil water. Geochim. Cosmochim. Acta 2018, 224, 80–95. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic. Acids. Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahe, F. VSEARCH: A versatile open source tool for metagenomics. Peer. J. 2016, 4, e2584. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microb. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Bushnell, B. BBMap Short-Read. Aligner and Other Bioinformatics Tools. 2015. Available online: https://sourceforge.net/projects/bbmap/ (accessed on 10 March 2021).
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with bowtie. Nat. Methods 2012, 9, 357–359. [Google Scholar]
- Eren, A.M.; Esen, O.C.; Quince, C.; Vineis, J.H.; Morrison, H.G.; Sogin, M.L.; Delmont, T.O. Anvi’o: An advanced analysis and visualization platform for ‘omics data. Peer. J. 2015, 3, e1319. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. Bmc. Bioinform. 2010, 11, 1–11. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Campbell, J.H.; O’Donoghue, P.; Alisha, G.C.; Schwientek, P.; Sczyrba, A.; Woyke, T.; Söll, D.; Podar, M. UGA is an additional glycine codon in unculutured SR1 bacteria from the human microbiota. Proc. Natl. Acad. Sci. USA 2013, 110, 5540–5545. [Google Scholar] [CrossRef]
- Rinke, C.; Schwientek, P.; Sczyrba, A.; Ivanova, N.N.; Anderson, I.J.; Cheng, J.F.; Darling, A.; Malfatti, S.; Swan, B.K.; Gies, E.A. Insights into phylogeny and coding potential of microbial dark matter. Nature 2013, 499, 431–437. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Wagner, G.P.; Kin, K.; Lynch, V.J. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory. Biosci. 2012, 131, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Alneberg, J.; Bjarnason, B.S.; De Bruijn, I.; Schirmer, M.; Quick, J.; Ijaz, U.Z.; Lahti, L.; Loman, N.J.; Andersson, A.F.; Quince, C. Binning metagenomic contigs by coverage and composition. Nat. Methods 2014, 11, 1144–1146. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.D.; Froula, J.; Egan, R.; Wang, Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. Peer. J. 2015, 3, e1165. [Google Scholar] [CrossRef]
- Sieber, C.M.K.; Probst, A.J.; Sharrar, A.; Thomas, B.C.; Hess, M.; Tringe, S.G.; Banfield, J.F. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 2018, 3, 836–843. [Google Scholar] [CrossRef]
- Parks, D.H.; Chuvochina, M.; Chaumeil, P.A.; Rinke, C.; Mussig, A.J.; Hugenholtz, P. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat. Biotechnol. 2020, 38, 1079–1086. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package version 2.5–5. 2019. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 10 March 2021).
- R Core Development Team. R: A Language and Environment for Statistical Computing; R Core Development Team: Vienna, Austria, 2017. [Google Scholar]
- Villanueva, R.A.M.; Chen, Z.J. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Kolde, R.; Kolde, M.R. Pheatmap: Pretty Heatmaps. 2015. Available online: https://cran.r-project.org/web/packages/pheatmap/index.html (accessed on 10 March 2021).
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome. Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Kieft, B.; Li, Z.; Bryson, S.; Crump, B.C.; Hettich, R.; Pan, C.; Mayali, X.; Mueller, R.S. Microbial community structure-function relationships in Yaquina Bay estuary reveal spatially distinct carbon and nitrogen cycling capacities. Front. Microbiol. 2018, 9, 1282. [Google Scholar] [CrossRef]
- Wu, H.; Moore, E. Association analysis of the general environmental conditions and prokaryotes’ gene distributions in various functional groups. Genomics 2010, 96, 27–38. [Google Scholar] [CrossRef]
- Garber, A.I.; Nealson, K.H.; Okamoto, A.; McAllister, S.M.; Chan, C.S.; Barco, R.A.; Merino, N. FeGenie: A comprehensive Tool for the Identification of Iron Genes and Iron Gene Neighborhoods in Genome and Metagenome Assemblies. Front. Microbiol. 2020, 11, 37. [Google Scholar] [CrossRef]
- Giblin, A.E.; Nadelhoffer, K.J.; Shaver, G.R.; Laundre, J.A.; McKerrow, A.J. Biogeochemical diversity along a river toposequence in arctic Alaska. Ecol. Monogr. 1991, 61, 415–435. [Google Scholar] [CrossRef]
- Nadelhoffer, K.J.; Giblin, A.E.; Shaver, G.R.; Laundre, J. Effects of temperature and organic matter quality on element mineralization in six arctic soils. Ecology 1991, 72, 242–253. [Google Scholar] [CrossRef]
- Gebauer, R.L.E.; Tenhunen, J.D.; Reynolds, J.F. Soil aeration in relation to soil physical properties, nitrogen availability and root characteristics within an arctic watershed. Plant Soil 1996, 178, 37–48. [Google Scholar] [CrossRef]
- Tveit, A.T.; Schwacke, R.; Svenning, M.M.; Urich, T. Organic carbon transformations in high-Arctic peat soils: Key functions and microorganisms. ISME J. 2013, 7, 299–311. [Google Scholar] [CrossRef]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef] [PubMed]
- Rousk, J.; Baath, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef]
- Chapin, F.S., III; Fetcher, N.; Kielland, K.; Everett, K.R.; Linkins, A.E. Productivity and nutrient cycling of Alaskan tundra: Enhancement with flowing soil water. Ecology 1988, 69, 693–702. [Google Scholar] [CrossRef]
- Sistla, S.A.; Asao, S.; Schimel, J.P. Detecting microbial N-limitation in tussock tundra soil: Implications for Arctic soil organic carbon cycling. Soil. Biol. Biochem. 2012, 55, 78–84. [Google Scholar] [CrossRef]
- Lauro, F.M.; McDougald, D.; Thomas, T.; Williams, T.J.; Egan, S.; Rice, S.; DeMaere, M.Z.; Ting, L.; Ertan, H.; Johnson, J. The genomic basis of trophic strategy in marine bacteria. Proc. Natl. Acad. Sci. USA 2009, 106, 15527–15533. [Google Scholar] [CrossRef]
- Emerson, D.; Scott, J.J.; Benes, J.; Bowden, W.B. Microbial iron oxidation in the Arctic tundra and its implications for biogeochemical cycling. Appl. Environ. Microbiol. 2015, 81, 8066–8075. [Google Scholar] [CrossRef]
- Kim, H.M.; Lee, M.J.; Jung, J.Y.; Hwang, C.Y.; Kim, M.; Ro, H.M.; Chun, J.; Lee, Y.K. Vertical distribution of bacterial community is associated with the degree of soil organic matter decomposition in the active layer of moist acidic tundra. J. Microbiol. 2016, 54, 713–723. [Google Scholar] [CrossRef]
- Conrad, R.; Schutz, H.; Babbel, M. Temperature limitation on hydrogen turnover and methanogenesis in anoxic paddy soil. Fems. Microbiol. Ecol. 1987, 3, 281–289. [Google Scholar] [CrossRef]
- Metje, M.; Frenzel, P. Methanogenesis and methanogenic pathways in a peat from subarctic permafrost. Environ. Microbiol. 2007, 9, 954–964. [Google Scholar] [CrossRef]
- Tveit, A.T.; Urich, T.; Frenzei, P.; Svenning, M.M. Metabolic and trophic interactions modulate methane production by arctic peat microbiota in response to warming. Proc. Natl. Acad. Sci. USA 2015, 112, E2507–E2516. [Google Scholar] [CrossRef] [PubMed]
- Malard, L.A.; Pearce, D.A. Microbial diversity and biogeography in Arctic soils. Environ. Microbiol. Rep. 2018, 10, 611–625. [Google Scholar] [CrossRef] [PubMed]
- Alexander, M. Introduction to Soil Microbiology, 2nd ed.; John Wiley and Sons: New York, NY, USA, 1976. [Google Scholar]
- Hobbie, J.E.; Hobbie, E.A.; Drossman, H.; Conte, M.; Weber, J.C.; Shamhart, J.; Weinrobe, M. Mycorrhizal fungi supply nitrogen to host plants in Arctic tundra and boreal forests: 15N is the key signal. Can. J. Microbiol. 2009, 55, 84–94. [Google Scholar] [CrossRef]
- Boberg, J.B.; Ihrmark, K.; Lindahl, B.D. Decomposing capacity of fungi commonly detected in Pinus sylvestris needle litter. Fungal. Ecol. 2011, 4, 110–114. [Google Scholar] [CrossRef]
- Crump, B.C.; Amaral-Zettler, L.A.; Kling, G.W. Microbial diversity in arctic freshwaters is structured by inoculation of microbes from soils. ISME J. 2012, 6, 1629–1639. [Google Scholar] [CrossRef]
- Moldrup, P.; Olesen, T.; Gamst, J.; Schjonning, P.; Yamaguchi, T.; Rolston, D. Predicting the gas diffusion coefficient in repacked soil: Water-induced linear reduction model. Soil. Sci. Soc. Am. J. 2000, 64, 1588–1594. [Google Scholar] [CrossRef]
- Thorbjorn, A.; Moldrup, P.; Blendstrup, H.; Komatsu, T.; Rolston, D.E. A gas diffusivity model based on air-, solid-, and water-phase resistance in variably saturated soil. Vadose. Zone. J. 2008, 7, 1276–1286. [Google Scholar] [CrossRef]
- Armstrong, W. Oxygen diffusion from the roots of some British bog plants. Nature 1964, 204, 801–802. [Google Scholar] [CrossRef]
- King, J.Y.; Reeburgh, W.S.; Regli, S.K. Methane emission and transport by arctic sedges in Alaska: Results of a vegetation removal experiment. J. Geophys. Res. Atmos. 1998, 103, 29083–29092. [Google Scholar] [CrossRef]
- Stieglitz, M.; Shaman, J.; McNamara, J.; Engel, V.; Shanley, J.; Kling, G.W. An approach to understanding hydrologic connectivity on the hillslope and the implications for nutrient transport. Glob. Biogeochem. Cycles. 2003, 17, 1105. [Google Scholar] [CrossRef]
- O’Connor, M.T.; Cardenas, M.B.; Ferencz, S.B.; Wu, Y.; Neilson, B.T.; Chen, J.; Kling, G.W. Empirical models for predicting water and heat flow properties of permafrost soils. Geophys. Res. Lett. 2020, 47, 1–10. [Google Scholar] [CrossRef]
- Neilson, B.T.; Cardenas, M.B.; O’Connor, M.T.; Rasmussen, M.T.; King, T.V.; Kling, G.W. Groundwater flow and exchange across the land surface explain carbon export patterns in continuous permafrost watersheds. Geophys. Res. Lett. 2018, 45, 7596–7605. [Google Scholar] [CrossRef]
- Nalven, S.G.; Ward, C.P.; Payet, J.P.; Cory, R.M.; Kling, G.W.; Sharpton, T.J.; Sullivan, C.M.; Crump, B.C. Experimental metatranscriptomics reveals the costs and benefits of dissolved organic matter photo-alteration for freshwater microbes. Environ. Microbiol. 2020, 22, 3505–3521. [Google Scholar] [CrossRef] [PubMed]
- McKnight, D.M.; Boyer, E.W.; Westerhoff, P.K.; Doran, P.T.; Kulbe, T.; Andersen, D.T. Spectrofluorometric characterization of dissolved organic matter for indication of precursor organic material and aromaticity. Limnol. Oceanogr. 2001, 46, 38–48. [Google Scholar] [CrossRef]
- Shen, L.; Preiss, J. Biosynthesis of bacterial glycogen: I. Purification and properties of the adenosine diphosphoglucose pyrophosphorylase of Arthrobacter species NRRLB1973. J. Biol. Chem. 1965, 240, 2334–2340. [Google Scholar] [CrossRef]
- Sekar, K.; Linker, S.M.; Nguyen, J.; Grunhagen, A.; Stocker, R.; Sauer, U. Bacterial glycogen provides short-term benefits in changing environments. Appl. Environ. Microb. 2020, 86, 9. [Google Scholar] [CrossRef]
- Lennon, J.T. Microbial life deep underfoot. Mbio 2020, 11, e03201–e03219. [Google Scholar] [CrossRef]
- Jia, X.; Xi, B.; Li, M.; Liu, D.; Hou, J.; Hao, Y.; Meng, F. Metaproteomic analysis of the relationship between microbial community phylogeny, function and metabolic activity during biohydrogen-methane coproduction under short-term hydrothermal pretreatment from food waste. Bioresour. Technol. 2017, 245, 1030–1039. [Google Scholar] [CrossRef] [PubMed]
- Veith, N.; Feldman-Salit, A.; Cojocaru, V.; Henrich, S.; Kummer, U.; Wade, R.C. Organism-adapted specificity of the allosteric regulation of pyruvate kinase in lactic acid bacteria. Plos. Comput. Biol. 2013, 9, e1003159. [Google Scholar] [CrossRef]
- Marchitti, S.A.; Brocker, C.; Stagos, D.; Vasiliou, V. Non-P450 aldehyde oxidizing enzymes: The aldehyde dehydrogenase superfamily. Expert. Opin. Drug. Met. 2008, 4, 697–720. [Google Scholar] [CrossRef]
- Nosova, T.; Jokelainen, K.; Kaihovaara, P.; Jousimies-Somer, H.; Siitonen, A.; Heine, R.; Salaspuro, M. Aldehyde dehydrogenase activity and acetate production by aerobic bacteria representing the normal flora of human large intestine. Alcohol Alcohol. 1996, 31, 555–564. [Google Scholar] [CrossRef]
- Weber, K.A.; Achenbach, L.A.; Coates, J.D. Microorganisms pumping iron: Anaerobic microbial iron oxidation and reduction. Nat. Rev. Microbiol. 2006, 4, 752–763. [Google Scholar] [CrossRef] [PubMed]
- Stockwell, B.R.; Friedmann Angeli, J.P.; Bayir, H.; Bush, A.I.; Conrad, M.; Dixon, S.J.; Fulda, S.; Gascon, S.; Hatzios, S.K.; Kagan, V.E.; et al. Ferroptosis: A regulated cell death nexus linking metabolism, redox biology, and disease. Cell 2017, 171, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Shi, W.; Li, X.; Hu, Y.; Yu, F.; Ma, H. Ferroptosis accompanied by OH generation and cytoplasmic viscosity increase revealed via dual-functioning fluorescence probe. J. Am. Chem. Soc. 2019, 141, 18301–18307. [Google Scholar] [CrossRef] [PubMed]
- Conrad, M.; Kagan, V.E.; Bayir, H.; Pagnussat, G.C.; Head, B.; Traber, M.G.; Stockwell, B.R. Regulation of lipid peroxidation and ferroptosis in diverse species. Genes Dev. 2018, 32, 602–619. [Google Scholar] [CrossRef] [PubMed]
- Ratledge, C. (Ed.) Biochemistry of Microbial Degradation; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1994. [Google Scholar]
- Brune, A.; Frenzel, P.; Cypionka, H. Life at the oxic-anoxic interface: Microbial activities and adaptations. FEMS. Microbiol. Rev. 2000, 24, 691–710. [Google Scholar] [CrossRef]
- Emerson, D.; Revsbech, N. Investigation of an iron-oxidizing microbial mat community located near Aarhus, Denmark: Laboratory studies. Appl. Environ. Microbiol. 1994, 60, 4032–4038. [Google Scholar] [CrossRef]
- Doherty, S.J.; Barbato, R.A.; Grandy, A.S.; Thomas, W.K.; Monteux, S.; Dorrepaal, E.; Johansson, M.; Ernakovich, J.G. The transition from stochastic to deterministic bacterial community assembly during permafrost thaw succession. Front. Microbiol. 2020, 11, 596589. [Google Scholar] [CrossRef] [PubMed]
- Schadel, C.; Bader, M.K.F.; Schuur, E.A.; Biasi, C.; Bracho, R.; Capek, P.; De Baets, S.; Diakova, K.; Ernakovich, J.; Estop-Aragones, C.; et al. Potential carbon emissions dominated by carbon dioxide from thawed permafrost soils. Nat. Clim. Chang. 2016, 6, 950–953. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).