Assessing the Methylation Status of Two Potential Key Factors Involved in Cervical Oncogenesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Samples Collection
2.2. DNA Isolation
2.3. HPV DNA Detection and Genotyping
2.4. Bisulfite Conversion
2.5. RNA Isolation and cDNA Synthesis
2.6. Primer Design
2.7. Quantitative Real-Time and Methylation-Specific Polymerase Chain Reaction (qRT-PCR and qMS-PCR)
2.8. Statistical Analysis
3. Results
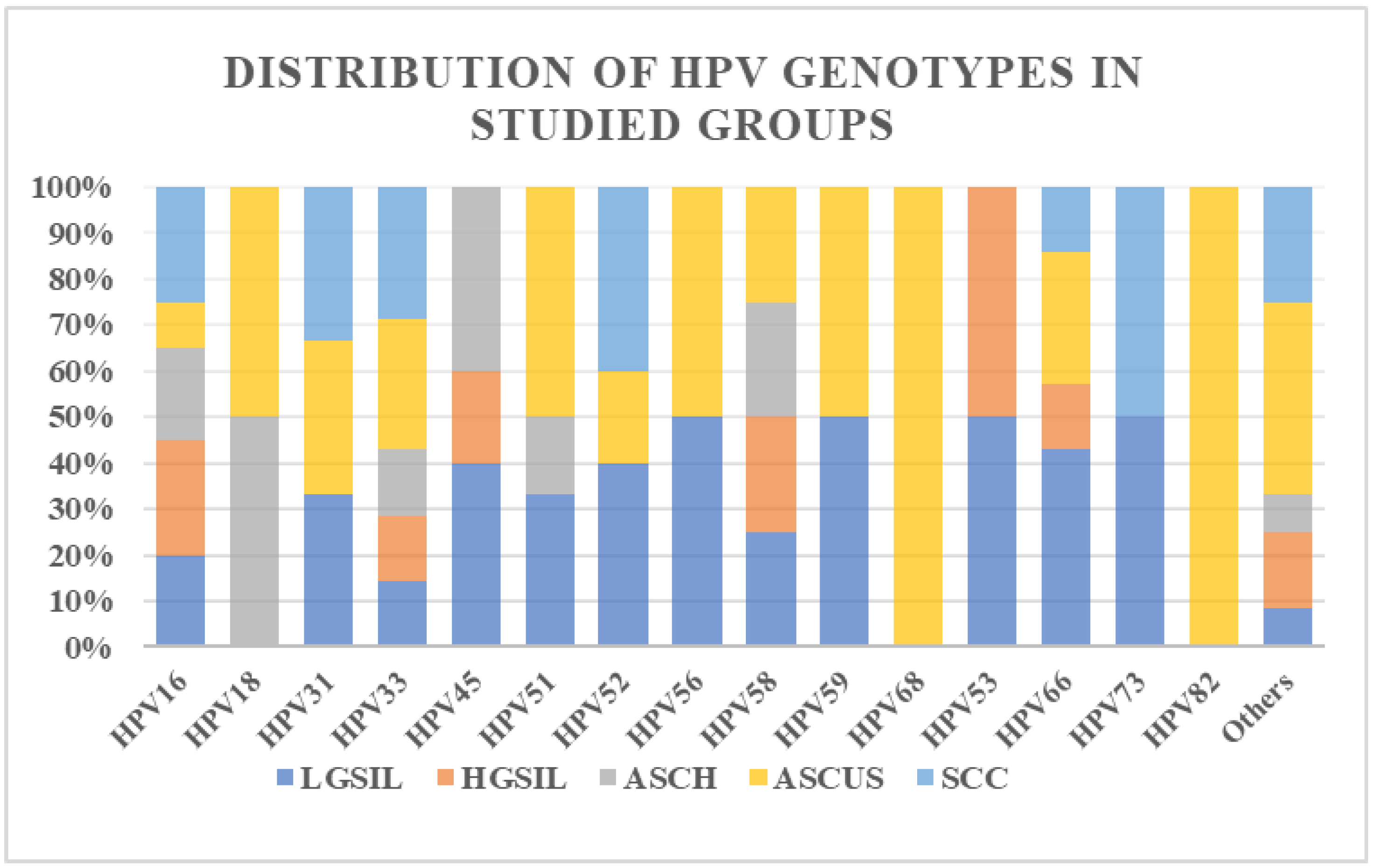
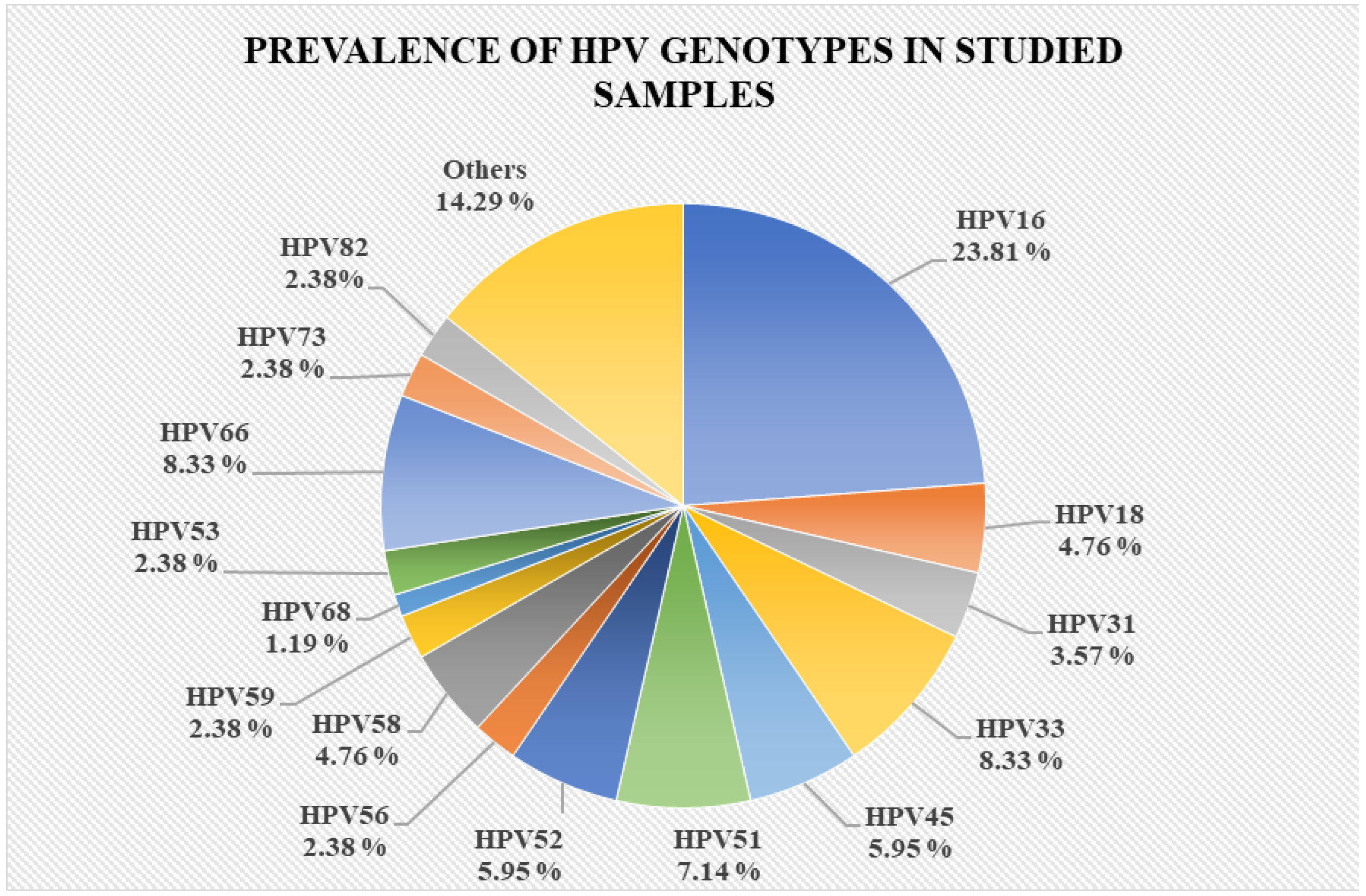
3.1. Study Group Characterization
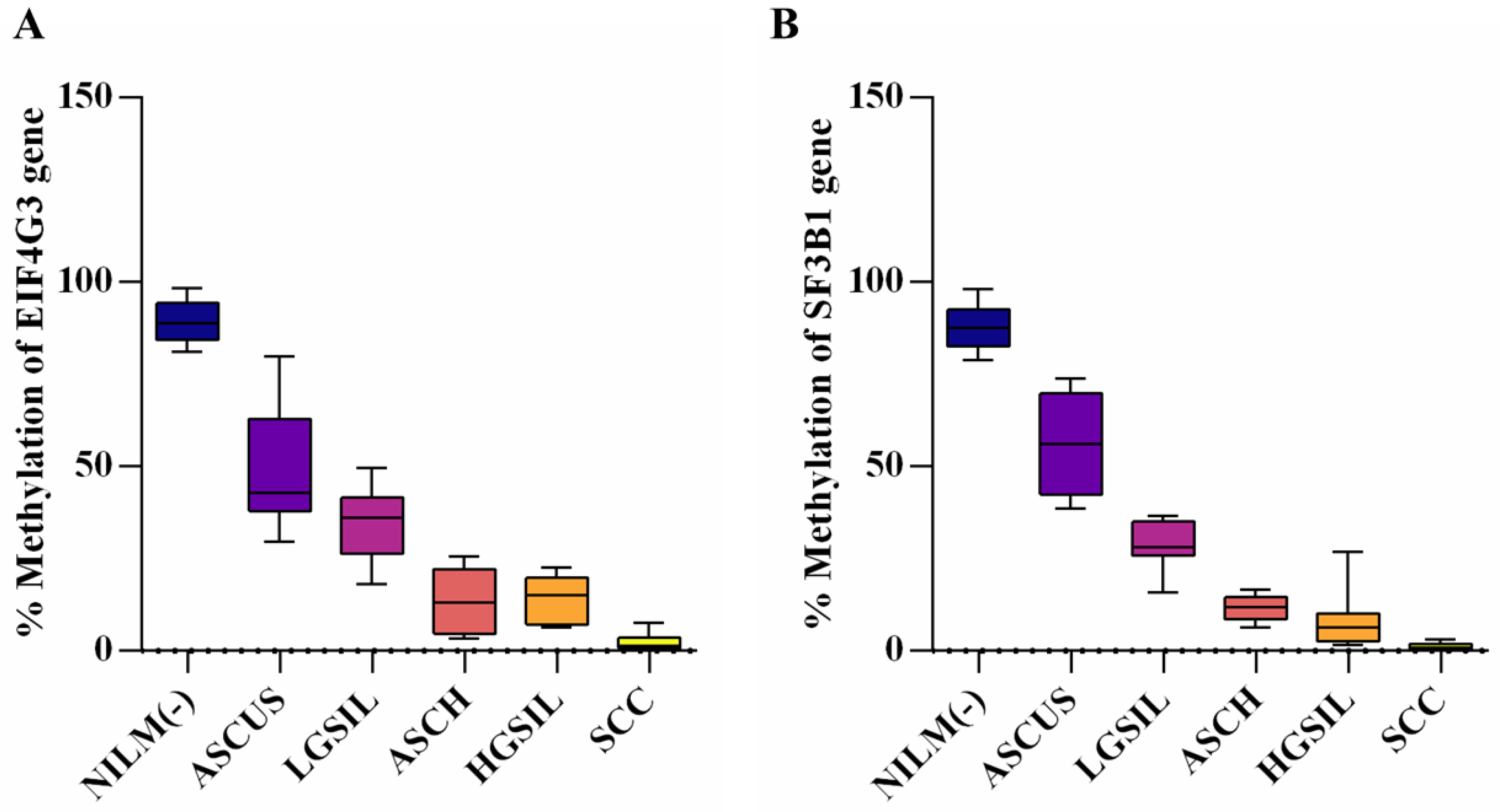
3.2. Evaluation of Promoter Methylation Status
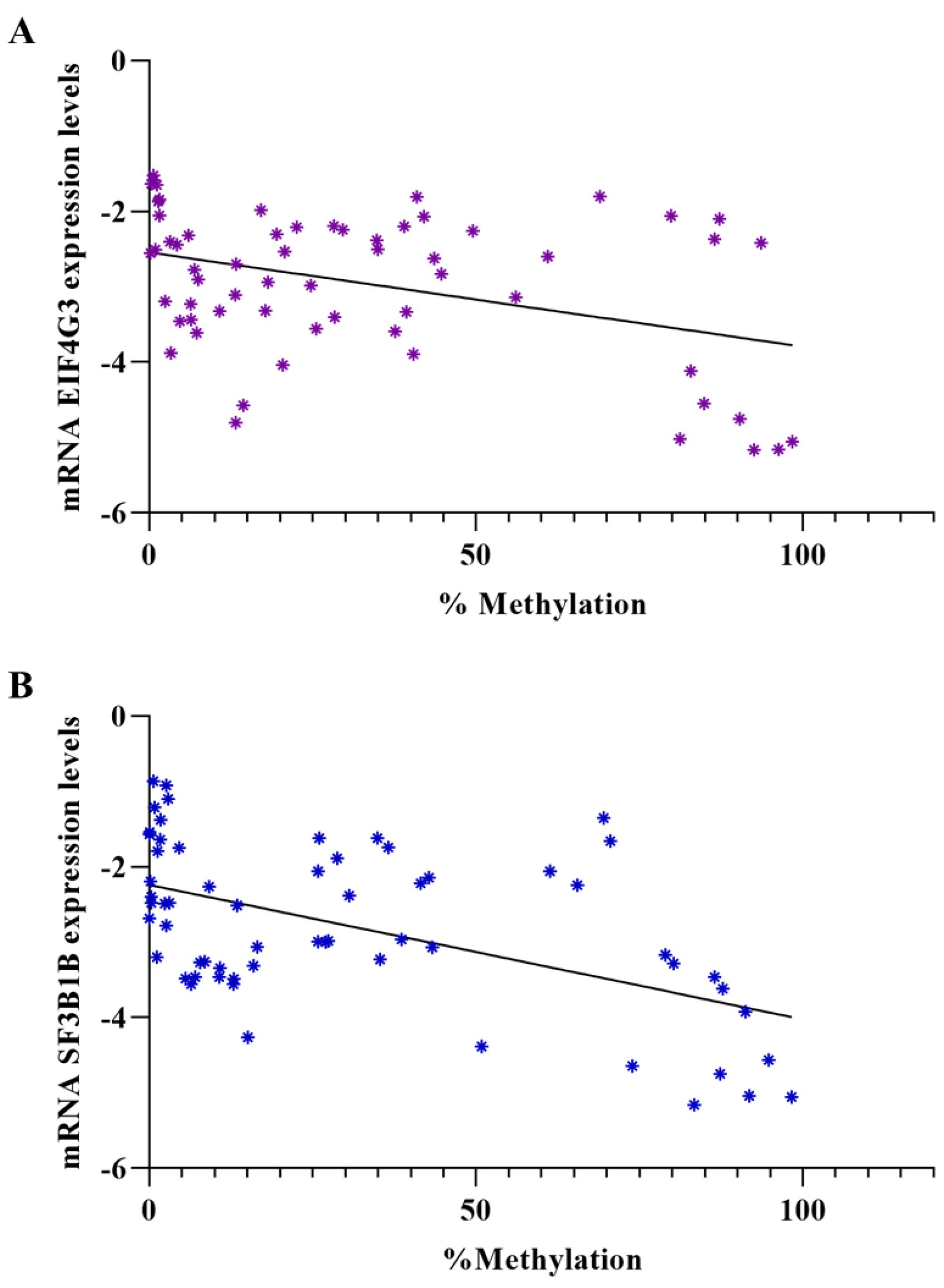
3.3. Evaluation of Gene Expression Levels in Patient Samples and Correlation between Expression Levels and Methylation Percentage
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Steenbergen, R.D.; Snijders, P.J.; Heideman, D.A.; Meijer, C.J. Clinical implications of (epi)genetic changes in HPV-induced cervical precancerous lesions. Nat. Rev. Cancer 2014, 14, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Sadia, H.; Shahwani, I.M.; Bana, K.F.M. Risk factors of cervical cancer and role of primary healthcare providers regarding PAP smears counseling: Case control study. Pak. J. Med. Sci. 2022, 38 Pt II, 998–1003. [Google Scholar] [CrossRef]
- Zhu, H.; Zhu, H.; Tian, M.; Wang, D.; He, J.; Xu, T. DNA Methylation and Hydroxymethylation in Cervical Cancer: Diagnosis, Prognosis and Treatment. Front. Genet. 2020, 11, 347. [Google Scholar] [CrossRef]
- Fang, J.; Zhang, H.; Jin, S. Epigenetics and cervical cancer: From pathogenesis to therapy. Tumour Biol. 2014, 35, 5083–5093. [Google Scholar] [CrossRef] [PubMed]
- Hesselink, A.T.; Heideman, D.A.; Steenbergen, R.D.; Coupé, V.M.; Overmeer, R.M.; Rijkaart, D.; Berkhof, J.; Meijer, C.J.; Snijders, P.J. Combined promoter methylation analysis of CADM1 and MAL: An objective triage tool for high-risk human papillomavirus DNA-positive women. Clin. Cancer Res. 2011, 17, 2459–2465. [Google Scholar] [CrossRef]
- Eijsink, J.J.; Lendvai, Á.; Deregowski, V.; Klip, H.G.; Verpooten, G.; Dehaspe, L.; de Bock, G.H.; Hollema, H.; van Criekinge, W.; Schuuring, E.; et al. A four-gene methylation marker panel as triage test in high-risk human papillomavirus positive patients. Int. J. Cancer 2012, 130, 1861–1869. [Google Scholar] [CrossRef]
- Botezatu, A.; Goia-Rusanu, C.D.; Iancu, I.V.; Huica, I.; Plesa, A.; Socolov, D.; Ungureanu, C.; Anton, G. Quantitative analysis of the relationship between microRNA-124a, -34b and -203 gene methylation and cervical oncogenesis. Mol. Med. Rep. 2011, 4, 121–128. [Google Scholar] [CrossRef]
- Lorincz, A.T. Virtues and weaknesses of DNA methylation as a test for cervical cancer prevention. Acta Cytol. 2016, 60, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Fudulu, A.; Diaconu, C.C.; Iancu, I.V.; Plesa, A.; Albulescu, A.; Bostan, M.; Socolov, D.G.; Stoian, I.L.; Balan, R.; Anton, G.; et al. Exploring the Role of E6 and E7 Oncoproteins in Cervical Oncogenesis through MBD2/3-NuRD Complex Chromatin Remodeling. Genes 2024, 15, 560. [Google Scholar] [CrossRef] [PubMed]
- Li, L.C.; Dahiya, R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics 2002, 18, 1427–1431. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Fackler, M.J.; McVeigh, M.; Mehrotra, J.; Blum, M.A.; Lange, J.; Lapides, A.; Garrett, E.; Argani, P.; Sukumar, S. Quantitative multiplex methylation-specific PCR assay for the detection of promoter hypermethylation in multiple genes in breast cancer. Cancer Res. 2004, 64, 4442–4452. [Google Scholar] [CrossRef]
- Cuzick, J.; Arbyn, M.; Sankaranarayanan, R.; Tsu, V.; Ronco, G.; Mayrand, M.H.; Dillner, J.; Meijer, C.J. Overview of human papillomavirus-based and other novel options for cervical cancer screening in developed and developing countries. Vaccine 2008, 26 (Suppl. 10), K29–K41. [Google Scholar] [CrossRef]
- Cuzick, J.; Bergeron, C.; von Knebel Doeberitz, M.; Gravitt, P.; Jeronimo, J.; Lorincz, A.T.; J L M Meijer, C.; Sankaranarayanan, R.; J F Snijders, P.; Szarewski, A. New technologies and procedures for cervical cancer screening. Vaccine 2012, 30 (Suppl. 5), F107–F116. [Google Scholar] [CrossRef] [PubMed]
- Lleras, R.A.; Smith, R.V.; Adrien, L.R.; Schlecht, N.F.; Burk, R.D.; Harris, T.M.; Childs, G.; Prystowsky, M.B.; Belbin, T.J. Unique DNA methylation loci distinguish anatomic site and HPV status in head and neck squamous cell carcinoma. Clin. Cancer Res. 2013, 19, 5444–5455. [Google Scholar] [CrossRef] [PubMed]
- Wentzensen, N.; Sherman, M.E.; Schiffman, M.; Wang, S.S. Utility of methylation markers in cervical cancer early detection: Appraisal of the state-of-the-science. Gynecol. Oncol. 2009, 112, 293–299. [Google Scholar] [CrossRef]
- Sahasrabuddhe, V.V.; Luhn, P.; Wentzensen, N. Human papillomavirus and cervical cancer: Biomarkers for improved prevention efforts. Future Microbiol. 2011, 6, 1083–1098. [Google Scholar] [CrossRef] [PubMed]
- Bonde, J.; Floore, A.; Ejegod, D.; Vink, F.J.; Hesselink, A.; van de Ven, P.M.; Valenčak, A.O.; Pedersen, H.; Doorn, S.; Quint, W.G.; et al. Methylation markers FAM19A4 and miR124-2 as triage strategy for primary human papillomavirus screen positive women: A large European multicenter study. Int. J. Cancer 2021, 148, 396–405. [Google Scholar] [CrossRef]
- Dippmann, C.; Schmitz, M.; Wunsch, K.; Schütze, S.; Beer, K.; Greinke, C.; Ikenberg, H.; Hoyer, H.; Runnebaum, I.B.; Hansel, A.; et al. Triage of hrHPV-positive women: Comparison of two commercial methylation-specific PCR assays. Clin. Epigenetics 2020, 12, 171. [Google Scholar] [CrossRef]
- Burdier, F.R.; Waheed, D.E.; Nedjai, B.; Steenbergen, R.D.M.; Poljak, M.; Baay, M.; Vorsters, A.; Van Keer, S. DNA methylation as a triage tool for cervical cancer screening—A meeting report. Prev. Med. Rep. 2024, 41, 102678. [Google Scholar] [CrossRef] [PubMed]
- Yoshimi, A.; Abdel-Wahab, O. Molecular Pathways: Understanding and Targeting Mutant Spliceosomal Proteins. Clin. Cancer Res. 2017, 23, 336–341. [Google Scholar] [CrossRef] [PubMed]
- Visconte, V.; Makishima, H.; Maciejewski, J.P.; Tiu, R.V. Emerging roles of the spliceosomal machinery in myelodysplastic syndromes and other hematological disorders. Leukemia 2012, 26, 2447–2454. [Google Scholar] [CrossRef]
- Zhou, Z.; Gong, Q.; Wang, Y.; Li, M.; Wang, L.; Ding, H.; Li, P. The biological function and clinical significance of SF3B1 mutations in cancer. Biomark. Res. 2020, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Malcovati, L.; Stevenson, K.; Papaemmanuil, E.; Neuberg, D.; Bejar, R.; Boultwood, J.; Bowen, D.T.; Campbell, P.J.; Ebert, B.L.; Fenaux, P.; et al. SF3B1-mutant MDS as a distinct disease subtype: A proposal from the International Working Group for the Prognosis of MDS. Blood 2020, 136, 157–170. [Google Scholar] [CrossRef]
- Cazzola, M.; Rossi, M.; Malcovati, L. Associazione Italiana per la Ricerca sul Cancro Gruppo Italiano Malattie Mieloproliferative Biologic and clinical significance of somatic mutations of SF3B1 in myeloid and lymphoid neoplasms. Blood 2013, 121, 260–269. [Google Scholar] [CrossRef]
- Zhao, L.-P.; Daltro de Oliveira, R.; Marcault, C.; Soret, J.; Gauthier, N.; Verger, E.; Maslah, N.; Roux, B.; Parquet, N.; Dosquet, C.; et al. SF3B1 mutations in the Driver Clone Increase the Risk of Evolution to Myelofibrosis in Patients with Myeloproliferative Neoplasms (MPN). Blood 2020, 136, 1. [Google Scholar] [CrossRef]
- Yoshida, K.; Sanada, M.; Shiraishi, Y.; Nowak, D.; Nagata, Y.; Yamamoto, R.; Sato, Y.; Sato-Otsubo, A.; Kon, A.; Nagasaki, M.; et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011, 478, 64–69. [Google Scholar] [CrossRef]
- Patnaik, M.M.; Lasho, T.L.; Finke, C.M.; Hanson, C.A.; Hodnefield, J.M.; Knudson, R.A.; Ketterling, R.P.; Pardanani, A.; Tefferi, A. Spliceosome mutations involving SRSF2, SF3B1, and U2AF35 in chronic myelomonocytic leukemia: Prevalence, clinical correlates, and prognostic relevance. Am. J. Hematol. 2013, 88, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Simmler, P.; Ioannidi, E.I.; Mengis, T.; Marquart, K.F.; Asawa, S.; Van-Lehmann, K.; Kahles, A.; Thomas, T.; Schwerdel, C.; Aceto, N.; et al. Mutant SF3B1 promotes malignancy in PDAC. eLife 2023, 12, e80683. [Google Scholar] [CrossRef] [PubMed]
- Alsafadi, S.; Houy, A.; Battistella, A.; Popova, T.; Wassef, M.; Henry, E.; Tirode, F.; Constantinou, A.; Piperno-Neumann, S.; Roman-Roman, S.; et al. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat. Commun. 2016, 7, 10615. [Google Scholar] [CrossRef] [PubMed]
- Popli, P.; Richters, M.M.; Chadchan, S.B.; Kim, T.H.; Tycksen, E.; Griffith, O.; Thaker, P.H.; Griffith, M.; Kommagani, R. Splicing factor SF3B1 promotes endometrial cancer progression via regulating KSR2 RNA maturation. Cell Death Dis. 2020, 11, 842. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Sun, F.; Handel, M.A. Nuclear localization of EIF4G3 suggests a role for the XY body in translational regulation during spermatogenesis in mice. Biol. Reprod. 2018, 98, 102–114. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Primer Sequence | Amplicon Size (bp) | Temperature (°C) | Reference |
|---|---|---|---|---|
| EIF4G3 M_F | TTTTTAGTAGTTTTCGGAAAGAGTC | 163 | 58 | This study |
| EIF4G3 M_R | GATAAATTTTCTTCACTCAACGAA | |||
| EIF4G3 U_F | TTTAGTAGTTTTTGGAAAGAGTTGA | 163 | 56 | |
| EIF4G3 U_R | CCAATAAATTTTCTTCACTCAACAAA | |||
| SF3B1 M_F | TAAGGATTTTACGGTTCGGTTC | 182 | 58 | |
| SF3B1 M_R | CTAAAAACTACACTCTACGCGTACG | |||
| SF3B1 U_F | TTTAAGGATTTTATGGTTTGGTTTG | 182 | 58 | |
| SF3B1 U_R | AAAAACTACACTCTACACATACACC | |||
| EIF4G3_F | ACAGAATGCAGGTCCAACCA | 93 | 60 | [10] |
| EIF4G3_R | GGCCTCTGGAAAAACGGAGA | |||
| SF3B1_F | AAAAGCATAGGCGGACCATGA | 70 | 60 | |
| SF3B1_R | GGGGTTTTCCCTCCATCTGC | |||
| GAPDH | CCATCTTCCAGGAGCGAGATCCCT | 60 | ||
| GAPDH | TGAGCCCCAGCCTTCTTCATGGT |
| EIF4G3 (Mean ± SD) | p-Value * | SF3B1 (Mean ± SD) | p-Value | |
|---|---|---|---|---|
| NILM (−) | 89.36 ± 5.771 | - | 87.99 ± 6.160 | - |
| ASCUS | 49.59 ± 16.160 | <0.0001 | 55.79 ± 13.770 | <0.0001 |
| LGSIL | 34.20 ± 10.170 | <0.0001 | 28.73 ± 6.110 | <0.0001 |
| ASCH | 13.08 ± 8.597 | <0.0001 | 11.66 ± 3.432 | <0.0001 |
| HGSIL | 14.21 ± 6.084 | <0.0001 | 8.19 ± 7.523 | <0.0001 |
| SCC | 2.34 ± 2.296 | <0.0001 | 1.09 ± 1.094 | <0.0001 |
| EIF4G3 (Mean ± SD) | p-Value * | SF3B1 (Mean ± SD) | p-Value | |
|---|---|---|---|---|
| NILM (−) | −4.369 ± 0.810 | − | −4.201 ± 0.791 | - |
| ASCUS | −3.458 ± 0.919 | 0.0545 | −3.500 ± 0.351 | 0.0831 |
| LGSIL | −2.305 ± 0.415 | <0.0001 | −2.672 ± 1.099 | 0.0021 |
| ASCH | −2.935 ± 0.597 | 0.0011 | −2.383 ± 0.686 | <0.0001 |
| HGSIL | −2.969 ± 0.677 | 0.0021 | −2.663 ± 0.658 | 0.0005 ** |
| SCC | −2.314 ± 0.675 | <0.0001 ** | −1.694 ± 0.668 | <0.0001 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fudulu, A.; Bostan, M.; Iancu, I.V.; Pleșa, A.; Albulescu, A.; Stoian, I.L.; Socolov, D.G.; Anton, G.; Botezatu, A. Assessing the Methylation Status of Two Potential Key Factors Involved in Cervical Oncogenesis. Reports 2024, 7, 71. https://doi.org/10.3390/reports7030071
Fudulu A, Bostan M, Iancu IV, Pleșa A, Albulescu A, Stoian IL, Socolov DG, Anton G, Botezatu A. Assessing the Methylation Status of Two Potential Key Factors Involved in Cervical Oncogenesis. Reports. 2024; 7(3):71. https://doi.org/10.3390/reports7030071
Chicago/Turabian StyleFudulu, Alina, Marinela Bostan, Iulia Virginia Iancu, Adriana Pleșa, Adrian Albulescu, Irina Liviana Stoian, Demetra Gabriela Socolov, Gabriela Anton, and Anca Botezatu. 2024. "Assessing the Methylation Status of Two Potential Key Factors Involved in Cervical Oncogenesis" Reports 7, no. 3: 71. https://doi.org/10.3390/reports7030071
APA StyleFudulu, A., Bostan, M., Iancu, I. V., Pleșa, A., Albulescu, A., Stoian, I. L., Socolov, D. G., Anton, G., & Botezatu, A. (2024). Assessing the Methylation Status of Two Potential Key Factors Involved in Cervical Oncogenesis. Reports, 7(3), 71. https://doi.org/10.3390/reports7030071








