Molecular Characterisation of Post-Fire Naturally Regenerated Populations of Maritime Pine (Pinus pinaster Ait.) in the North of Portugal
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Area
2.2. Plant Material
2.3. Genomic DNA Extraction
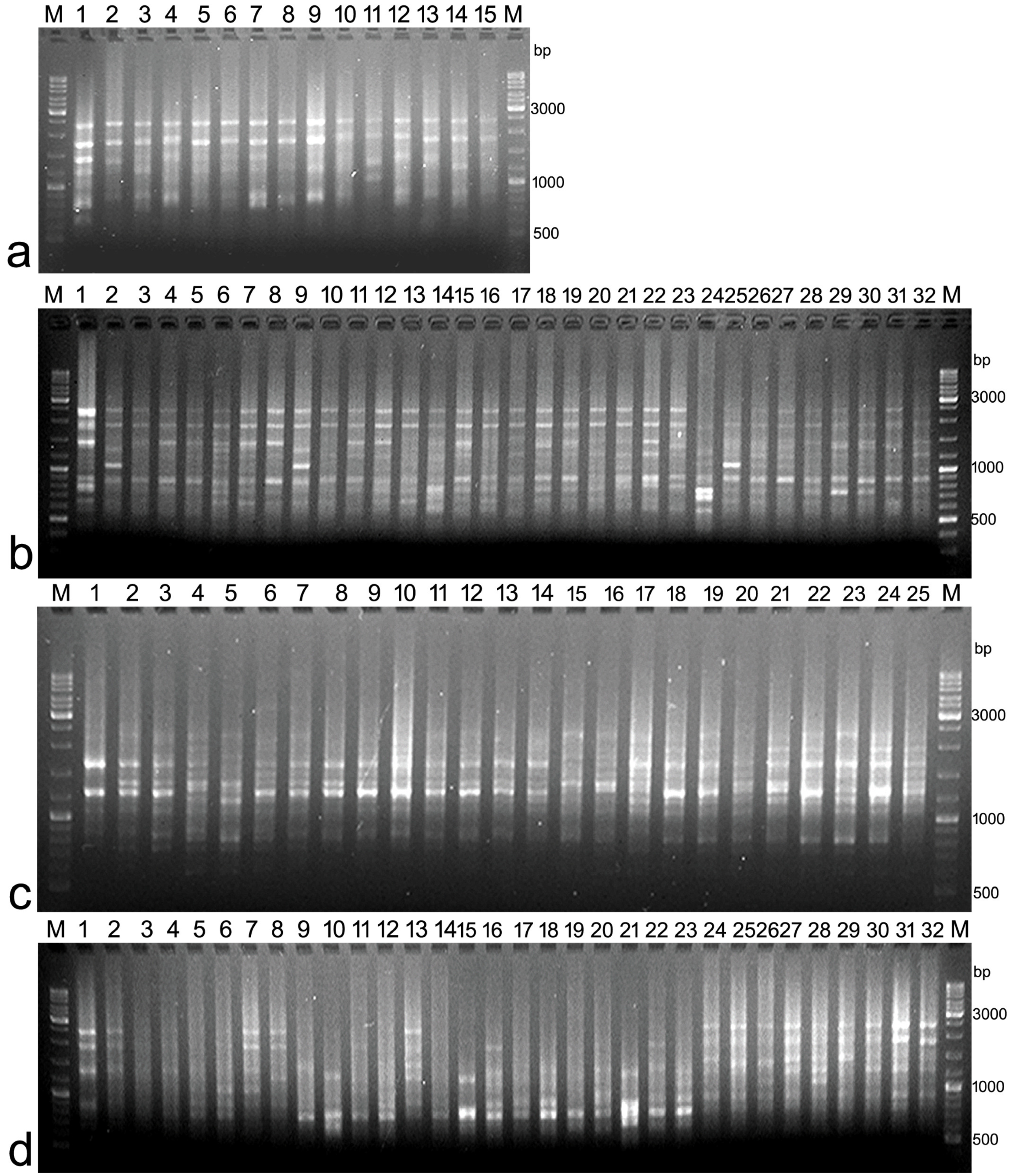
2.4. Amplification and Visualisation of ISSR Markers
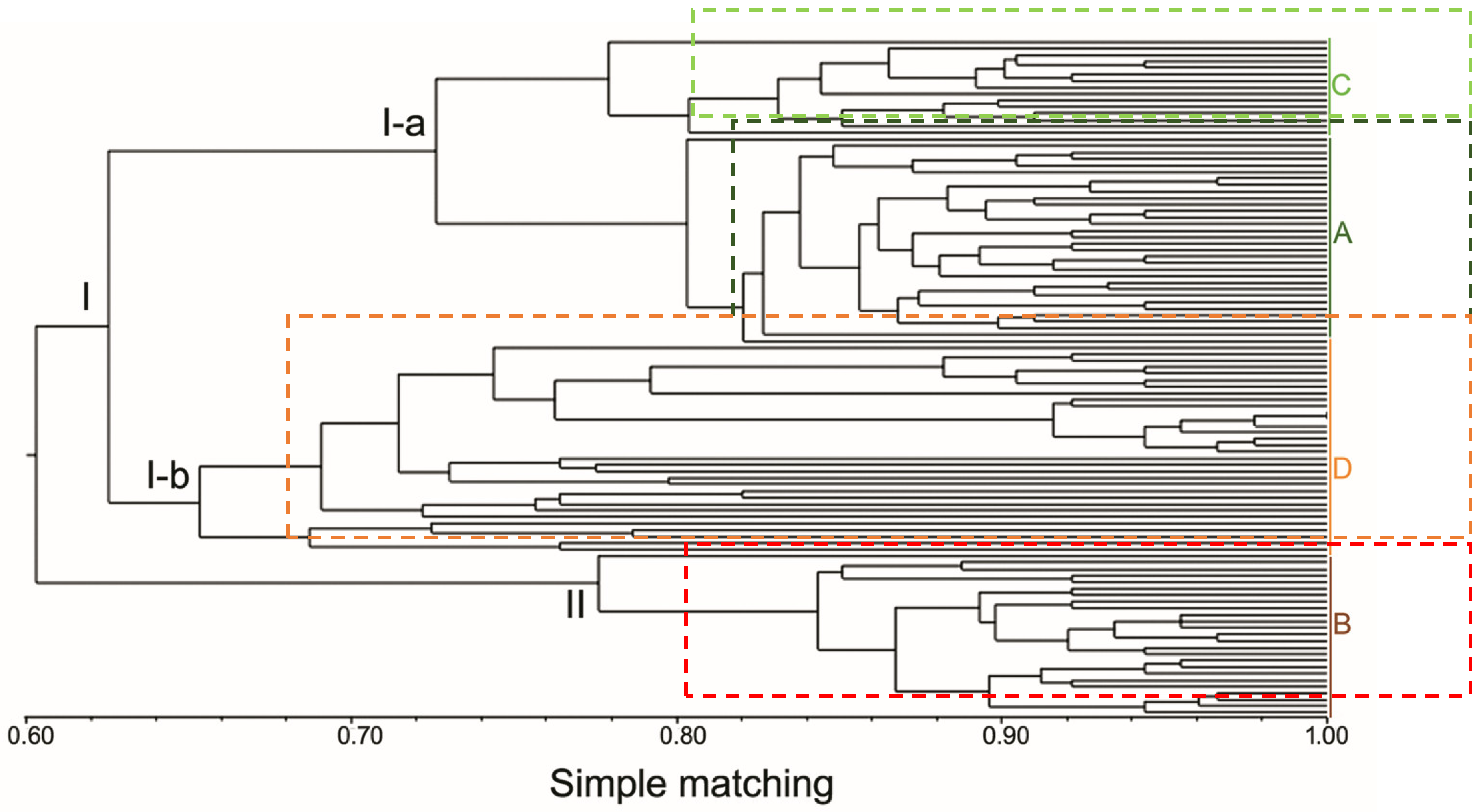
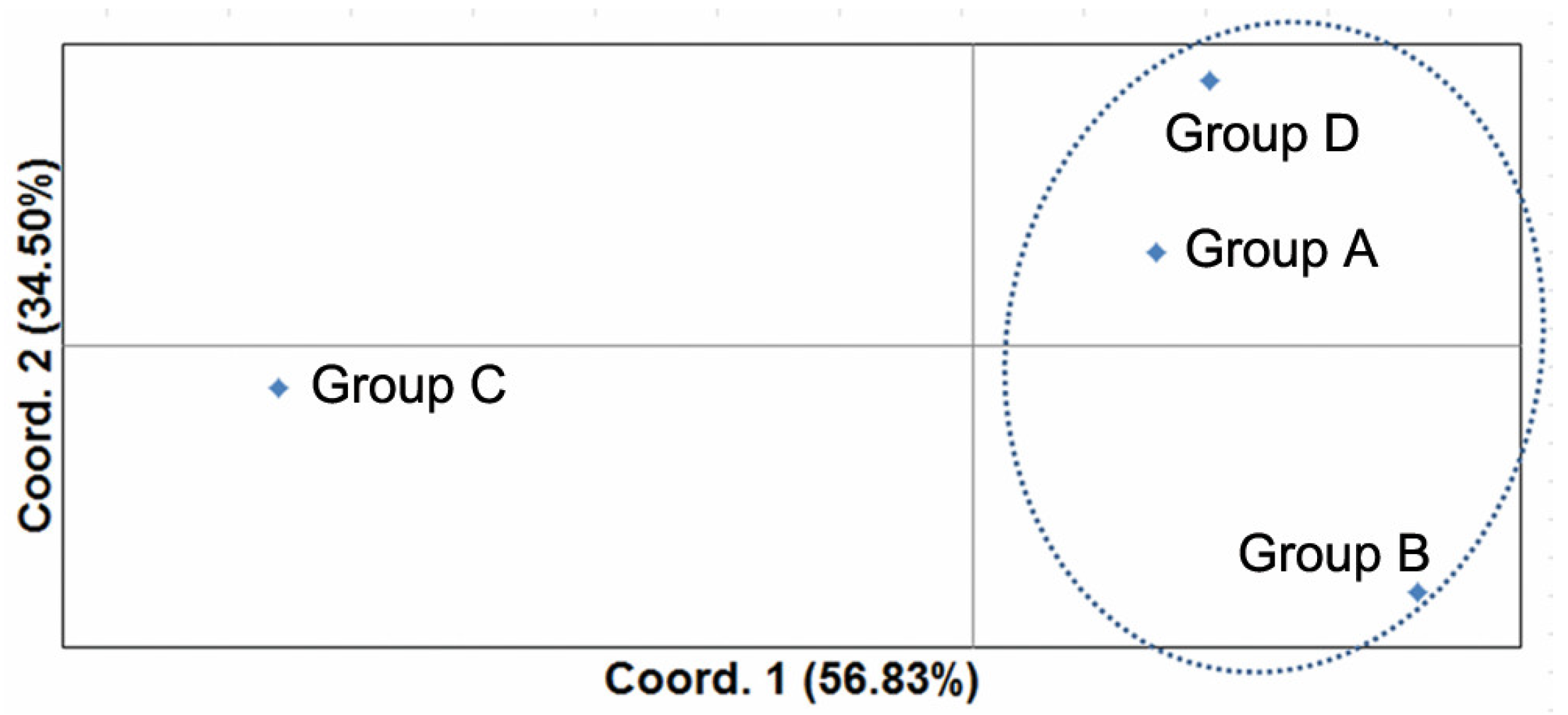
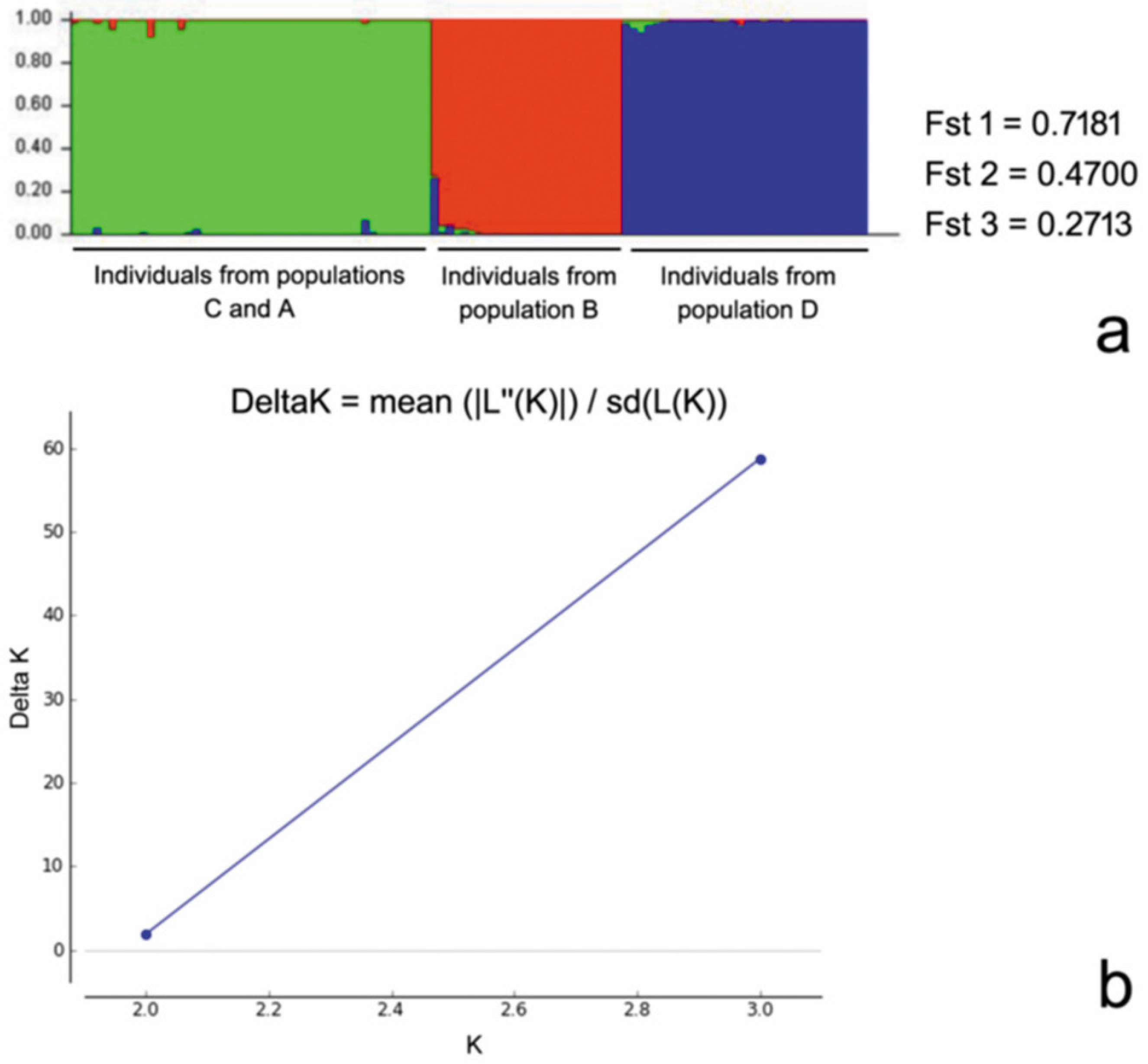
2.5. Statistical Analyses
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- IPCC. Climate Change 2022: Impacts, Adaptation, and Vulnerability; Contribution of Working Group II to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change (IPCC) 2022; Pörtner, H.-O., Roberts, D.C., Tignor, M., Poloczanska, E.S., Mintenbeck, K., Alegría, A., Craig, M., Langsdorf, S., Löschke, S., Möller, V., et al., Eds.; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2022; 3056p. [Google Scholar]
- Rajora, O.P.; Fageria, M.S.; Fitzsimmons, M. Effects of wild forest fires on genetic diversity and population structure of a boreal conifer, white spruce (Picea glauca (Moench.) Voss.): Implications for genetic resource management and adaptive potential under climate change. Forests 2023, 14, 157. [Google Scholar] [CrossRef]
- Aravanopoulos, F.A.; Panetsos, K.P.; Skaltsoyiannes, A. Genetic structure of Pinus brutia stands exposed to wild fires. Plant Ecol. 2004, 171, 175–183. [Google Scholar] [CrossRef]
- Aravanopoulos, F.A.; Panetsos, K.P. Genetics and evolution of natural Pinus brutia populations in Lesvos Island. Geotech. Sci. Issues 1998, 9, 10–19, (In Greek with English Summary). [Google Scholar]
- Panetsos, K.P.; Aravanopoulos, F.A.; Skaltsoyiannes, A.B. Genetic variation of Pinus brutia in islands of the northeastern Aegean Sea. Silvae Genet. 1998, 47, 115–120. [Google Scholar]
- Lucas-Borja, M.E.; Ahrazem, O.; Candel-Pérez, D.; Moya, D.; Fonseca, T.; Tecles, E.H.; De las Heras, J.; Gómez-Gómez, L. Evaluation of fire recurrence effect on genetic diversity in maritime pine (Pinus pinaster Ait.) populations using Inter-Simple Sequence Repeat profiles. Sci. Total Environ. 2016, 572, 1322–1328. [Google Scholar] [CrossRef]
- ICNF—Instituto da Conservação da Natureza e das Florestas. IFN6—6.º Inventário Florestal Nacional 2015; Relatório Final; ICNF: Lisboa, Portugal, 2015; 284p, Available online: https://www.icnf.pt/api/file/doc/c8cc40b3b7ec8541 (accessed on 11 April 2023). (In Portuguese)
- Gómez-Vázquez, I.; Fernandes, P.M.; Arias-Rodil, M.; Barrio-Anta, M.; Castedo-Dorado, F. Using density management diagrams to assess crown fire potential in Pinus pinaster Ait. stands. Ann. For. Sci. 2014, 71, 473–484. [Google Scholar] [CrossRef]
- Stevens-Rumann, C.S.; Morgan, P. Tree regeneration following wildfires in the western US: A review. Fire Ecol. 2019, 15, 15. [Google Scholar] [CrossRef]
- Coop, J.D.; Massatti, R.T.; Schoettle, A.W. Subalpine vegetation pattern three decades after stand-replacing fire: Effects of landscape context and topography on plant community composition, tree regeneration and diversity. J. Veg. Sci. 2010, 21, 472–487. [Google Scholar] [CrossRef]
- Turner, M.G.; Whitby, T.G.; Tinker, D.B.; Romme, W. Twenty-four years after the Yellowstone fires: Are postfire lodgepole pine stands converging in structure and function? Ecology 2016, 97, 1260–1273. [Google Scholar] [CrossRef]
- Strand, E.K.; Satterberg, K.L.; Hudak, A.T.; Byrne, J.; Khalyani, A.H.; Smith, A. Does burn severity affect plant community diversity and composition in mixed conifer forests of the United States Intermountain West one decade post fire? Fire Ecol. 2019, 15, 25. [Google Scholar] [CrossRef]
- Garzón, M.B.; Alía, R.; Robson, T.M.; Zavala, M.A. Intra-specific variability and plasticity influence potential tree species distributions under climate change. Glob. Ecol. Biogeogr. 2011, 20, 766–778. [Google Scholar] [CrossRef]
- Kavaliauskas, D.; Fussi, B.; Westergren, M.; Aravanopoulos, F.; Finzgar, D.; Baier, R.; Alizoti, P.; Bozic, G.; Avramidou, E.; Konnert, M.; et al. The interplay between forest management practices, Genetic monitoring, and other long-term monitoring systems. Forests 2018, 9, 133. [Google Scholar] [CrossRef]
- Gonçalves, A.C.; Fonseca, T.F. Influence management and disturbances on the regeneration of forest stands. Front. For. Glob. Chang.—Forest Manag. 2023, 6, 1123215. [Google Scholar] [CrossRef]
- Ribeiro, S.; Gaspar, M.J.; Lima-Brito, J.; Fonseca, T.; Soares, P.; Cerveira, A.; Fernandes, P.M.; Louzada, J.; Carvalho, A. Impact of fire recurrence and induced water stress on seed germination and root mitotic cell cycle of Pinus pinaster Aiton. Forests 2023, 14, 78. [Google Scholar] [CrossRef]
- Correia, I.; Santos, L.; Faria, C.; Nóbrega, C.; Almeida, M.H.; David, T. Cone to seedling-variation between Pinus pinaster provenances from contrasting altitudes. For. Sci. 2014, 60, 724–732. [Google Scholar] [CrossRef]
- Santos, L.; Capelo, J.; Tavares, M. Germination patterns of soil seed banks in relation to fire in Portuguese littoral pine forest vegetation. Fire Ecol. 2010, 6, 1–15. [Google Scholar] [CrossRef]
- Alvarez, R.; Valbuena, L.; Calvo, L. Influence of tree age on seed germination response to environmental factors and inhibitory substances in Pinus pinaster. Int. J. Wildland Fire 2005, 14, 277–284. [Google Scholar] [CrossRef]
- Cipriano, J.; Carvalho, A.; Fernandes, C.; Gaspar, M.J.; Pires, J.; Bento, J.; Roxo, L.; Louzada, J.; Lima-Brito, J. Evaluation of genetic diversity of Portuguese Pinus sylvestris L. populations based on molecular data and inferences about the future use of this germplasm. J. Genet. 2013, 92, e41–e48. [Google Scholar] [CrossRef]
- Dias, A.; Lemos, M.; Pavia, I.; Gaspar, M.J.; Silva, M.E.; Louzada, J.L.; Lima-Brito, J.; Carvalho, A. Genetic characterization of Portuguese allochthonous populations of Pinus nigra using ISSRs and SCoTs and extrapolation of their infraspecific taxonomy. Physiol. Mol. Biol. Plant 2019, 25, 799–805. [Google Scholar] [CrossRef]
- Carvalho, A.; Matos, M.; Lima-Brito, J.; Guedes-Pinto, H.; Benito, C. DNA fingerprint of F1 interspecific hybrids from the Triticeae tribe using ISSRs. Euphytica 2005, 143, 93–99. [Google Scholar] [CrossRef]
- Rohlf, F.J. NTSYS—Numerical Taxonomy and Multivariate Analysis System; Version 2.2, Getting Started Guide; Applied Biostatistics Inc., Exeter Publishing: Setauket, NY, USA, 1998. [Google Scholar]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individual. Genetics 1978, 89, 583–590. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- McDermott, J.M.; McDonald, B.A. Gene flow in plant pathosystems. Annu. Rev. Phytopathol. 1993, 31, 353–373. [Google Scholar] [CrossRef]
- Yeh, F.C.; Yang, R.C.; Boyle, T.B.J.; Ye, Z.H.; Mao, J.X. POPGENE Version 1.32, the User-Friendly Shareware for Population Genetic Analysis; Molecular Biology and Biotechnology Centre, University of Alberta: Edmonton, KY, USA, 1999. [Google Scholar]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Wen, X.; Falush, D. Documentation for STRUCTURE Software: Version 2.3; University of Chicago: Chicago, IL, USA, 2010; Available online: https://docplayer.net/4695550-Documentation-for-structure-software-version-2-3.html (accessed on 31 March 2023).
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure: Extensions to linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualising STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. CLUMPAK: A program for identifying clustering modes andpackaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef]
- Reis, E. Estatística Multivariada Aplicada; Edições Sílabo, Lda.: Lisboa, Portugal, 1997. (In Portuguese) [Google Scholar]
- Aparício, B.A.; Santos, J.A.; Freitas, T.R.; Sá, A.C.L.; Pereira, J.M.C.; Fernandes, P.M. Unravelling the efect of climate change on fire danger and fire behaviour in the Transboundary Biosphere Reserve of Meseta Ibérica (Portugal-Spain). Clim. Change 2022, 173, 5. [Google Scholar] [CrossRef]
- Pausas, J.G. Evolutionary fire ecology: Lessons learned from pines. Trends Plant Sci. 2015, 20, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, A.C.; Sousa, A.M.O. The Fire in the Mediterranean Region: A Case Study of Forest Fires in Portugal. In Mediterranean Identities—Environment, Society, Culture; Fuerst-Bjelis, B., Ed.; IntechOpen: Rijeka, Croatia, 2017; Chapter 13; pp. 305–335. [Google Scholar] [CrossRef]
- Keeley, J.E. Ecology and evolution of pine life histories. Ann. For. Sci. 2012, 69, 445–453. [Google Scholar] [CrossRef]

| Primer Name | Sequence 5′→3′ |
|---|---|
| UBC 825 | ACA CAC ACA CAC ACA CT |
| UBC 826 | ACA CAC ACA CAC ACA CC |
| UBC 846 | CAC ACA CAC ACA CAC ART * |
| UBC 849 | GTG TGT GTG TGT GTG TYA * |
| UBC 850 | GTG TGT GTG TGT GTG TYC * |
| UBC 855 | ACA CAC ACA CAC ACA CYT * |
| UBC 856 | ACACACACACACACACYA * |
| Percentage of ISSR Polymorphism within the Group of Individuals Originating from: | |||||||
|---|---|---|---|---|---|---|---|
| Population C (control) | Population B | ||||||
| Primer | T | P | % P | Primer | T | P | % P |
| 825 | 7 | 6 | 85.71 | 825 | 6 | 4 | 66.67 |
| 826 | 4 | 1 | 25.00 | 826 | 7 | 4 | 57.14 |
| 846 | 9 | 5 | 55.56 | 846 | 8 | 5 | 62.50 |
| 849 | 8 | 8 | 100 | 849 | 8 | 7 | 87.50 |
| 850 | 9 | 6 | 66.67 | 850 | 11 | 6 | 54.55 |
| 855 | 6 | 6 | 100 | 855 | 7 | 7 | 100 |
| 856 | 8 | 8 | 100 | 856 | 8 | 7 | 87.50 |
| Total | 51 | 40 | 78.43 | Total | 55 | 40 | 72.73 |
| Population A | Population D | ||||||
| Primer | T | P | % P | Primer | T | P | % P |
| 825 | 9 | 7 | 77.78 | 825 | 15 | 15 | 100 |
| 826 | 6 | 3 | 50.00 | 826 | 11 | 11 | 100 |
| 846 | 9 | 7 | 77.78 | 846 | 12 | 12 | 100 |
| 849 | 4 | 3 | 75.00 | 849 | 9 | 9 | 100 |
| 850 | 11 | 9 | 81.82 | 850 | 12 | 11 | 91.67 |
| 855 | 10 | 9 | 90.00 | 855 | 13 | 5 | 38.46 |
| 856 | 5 | 4 | 80.00 | 856 | 13 | 13 | 100 |
| Total | 54 | 42 | 77.78 | Total | 85 | 76 | 89.41 |
| Percentage of ISSR Polymorphism among Individuals | Genetic Diversity Parameters (Mean ± Standard Error) Per Group of Individuals with Origin in: | ||||||
| Primer | T | P | % P | Population | h | I | |
| 825 | 15 | 15 | 100 | C | 0.47 ± 0.004 | 0.66 ± 0.005 | |
| 826 | 11 | 11 | 100 | A | 0.49 ± 0.002 | 0.68 ± 0.002 | |
| 846 | 12 | 12 | 100 | B | 0.48 ± 0.002 | 0.67 ± 0.003 | |
| 849 | 10 | 10 | 100 | D | 0.48 ± 0.002 | 0.68 ± 0.002 | |
| 850 | 14 | 14 | 100 | Mean | 0.48 ± 0.002 | 0.67 ± 0.002 | |
| 855 | 14 | 14 | 100 | ||||
| 856 | 13 | 13 | 100 | ||||
| Total | 89 | 89 | 100 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carvalho, A.; Ribeiro, S.; Gaspar, M.J.; Fonseca, T.; Lima-Brito, J. Molecular Characterisation of Post-Fire Naturally Regenerated Populations of Maritime Pine (Pinus pinaster Ait.) in the North of Portugal. Fire 2024, 7, 88. https://doi.org/10.3390/fire7030088
Carvalho A, Ribeiro S, Gaspar MJ, Fonseca T, Lima-Brito J. Molecular Characterisation of Post-Fire Naturally Regenerated Populations of Maritime Pine (Pinus pinaster Ait.) in the North of Portugal. Fire. 2024; 7(3):88. https://doi.org/10.3390/fire7030088
Chicago/Turabian StyleCarvalho, Ana, Stéphanie Ribeiro, Maria João Gaspar, Teresa Fonseca, and José Lima-Brito. 2024. "Molecular Characterisation of Post-Fire Naturally Regenerated Populations of Maritime Pine (Pinus pinaster Ait.) in the North of Portugal" Fire 7, no. 3: 88. https://doi.org/10.3390/fire7030088
APA StyleCarvalho, A., Ribeiro, S., Gaspar, M. J., Fonseca, T., & Lima-Brito, J. (2024). Molecular Characterisation of Post-Fire Naturally Regenerated Populations of Maritime Pine (Pinus pinaster Ait.) in the North of Portugal. Fire, 7(3), 88. https://doi.org/10.3390/fire7030088






