Machine Learning in the Management of Lateral Skull Base Tumors: A Systematic Review
Abstract
1. Introduction
2. Materials and Methods
3. Results
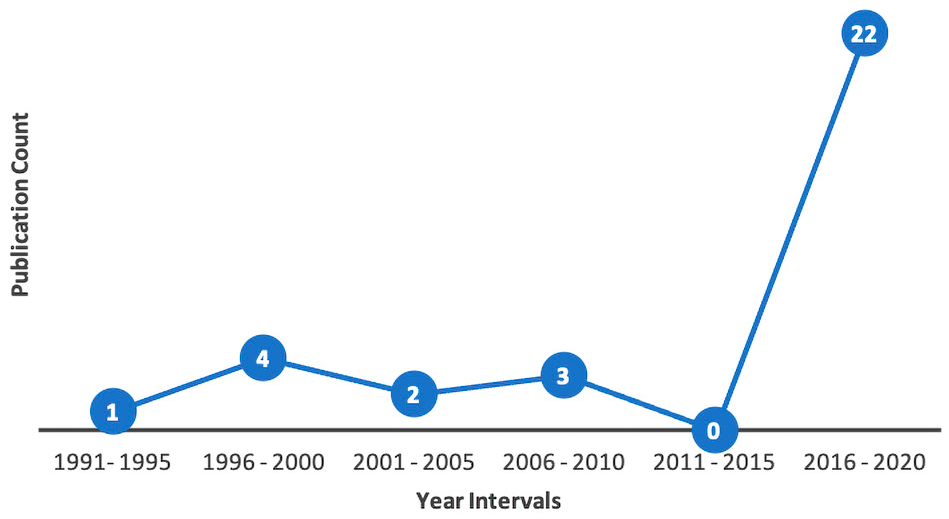
3.1. Topic Trends
3.2. Surgical Management
3.3. Disease Classification
3.4. Tumor Segmentation
3.5. Other Clinical Applications
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Deo, R.C. Machine Learning in Medicine. Circulation 2015, 132, 1920–1930. [Google Scholar] [CrossRef]
- Libbrecht, M.W.; Noble, W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef]
- Zampieri, G.; Vijayakumar, S.; Yaneske, E.; Angione, C. Machine and deep learning meet genome-scale metabolic modeling. PLoS Comput. Biol. 2019, 15, e1007084. [Google Scholar] [CrossRef]
- Houy, N.; Le Grand, F. Personalized oncology with artificial intelligence: The case of temozolomide. Artif. Intell. Med. 2019, 99, 101693. [Google Scholar] [CrossRef]
- Cruz, J.A.; Wishart, D.S. Applications of Machine Learning in Cancer Prediction and Prognosis. Cancer Inform. 2006, 2, 59–77. [Google Scholar] [CrossRef]
- Asiri, N.; Hussain, M.; Al Adel, F.; Alzaidi, N. Deep learning based computer-aided diagnosis systems for diabetic retinopathy: A survey. Artif. Intell. Med. 2019, 99, 101701. [Google Scholar] [CrossRef]
- Zheng, Y.; Hu, X. Healthcare predictive analytics for disease progression: A longitudinal data fusion approach. J. Intell. Inf. Syst. 2020, 55, 351–369. [Google Scholar] [CrossRef]
- Jagga, Z.; Gupta, D. Machine learning for biomarker identification in cancer research—developments toward its clinical application. Pers. Med. 2015, 12, 371–387. [Google Scholar] [CrossRef]
- Chowdhury, N.; Smith, T.L.; Chandra, R.; Turner, J.H. Automated classification of osteomeatal complex inflammation on computed tomography using convolutional neural networks. Int. Forum Allergy Rhinol. 2018, 9, 46–52. [Google Scholar] [CrossRef]
- Bing, D.; Ying, J.; Miao, J.; Lan, L.; Wang, D.; Zhao, L.; Yin, Z.; Yu, L.; Guan, J.; Wang, Q. Predicting the hearing outcome in sudden sensorineural hearing loss via machine learning models. Clin. Otolaryngol. 2018, 43, 868–874. [Google Scholar] [CrossRef]
- ARTIFICIAL INTELLIGENCE: Healthcare’s New Nervous System. Accenture.com. 2017. Available online: https://www.accenture.com/_acnmedia/pdf-49/accenture-health-artificial-intelligence.pdf (accessed on 24 June 2021).
- Senders, J.T.; Staples, P.C.; Karhade, A.V.; Zaki, M.M.; Gormley, W.B.; Broekman, M.L.; Smith, T.R.; Arnaout, O. Machine Learning and Neurosurgical Outcome Prediction: A Systematic Review. World Neurosurg. 2018, 109, 476–486.e1. [Google Scholar] [CrossRef]
- Crowson, M.G.; Ranisau, J.; Eskander, A.; Babier, A.; Xu, B.; Kahmke, R.R.; Chen, J.M.; Chan, T.C.Y. A contemporary review of machine learning in otolaryngology–head and neck surgery. Laryngoscope 2019, 130, 45–51. [Google Scholar] [CrossRef]
- You, E.; Lin, V.; Mijovic, T.; Eskander, A.; Crowson, M.G. Artificial Intelligence Applications in Otology: A State of the Art Review. Otolaryngol. Neck Surg. 2020, 163, 1123–1133. [Google Scholar] [CrossRef]
- Zacharaki, E.I.; Wang, S.; Chawla, S.; Yoo, D.S.; Wolf, R.; Melhem, E.R.; Davatzikos, C. Classification of brain tumor type and grade using MRI texture and shape in a machine learning scheme. Magn. Reson. Med. 2009, 62, 1609–1618. [Google Scholar] [CrossRef]
- Crowson, M.G.; Dixon, P.; Mahmood, R.; Lee, J.W.; Shipp, D.; Le, T.; Lin, V.; Chen, J.; Chan, T.C.Y. Predicting Postoperative Cochlear Implant Performance Using Supervised Machine Learning. Otol. Neurotol. 2020, 41, e1013–e1023. [Google Scholar] [CrossRef]
- Bur, A.M.; Shew, M.; New, J. Artificial Intelligence for the Otolaryngologist: A State of the Art Review. Otolaryngol. Neck Surg. 2019, 160, 603–611. [Google Scholar] [CrossRef]
- Crowson, M.G.; Lin, V.; Chen, J.M.; Chan, T.C.Y. Machine Learning and Cochlear Implantation—A Structured Review of Opportunities and Challenges. Otol. Neurotol. 2020, 41, e36–e45. [Google Scholar] [CrossRef]
- Theunissen, E.A.R.; Bosma, S.C.J.; Zuur, C.L.; Spijker, R.; Van Der Baan, S.; Dreschler, W.A.; De Boer, J.P.; Balm, A.J.M.; Rasch, C.R.N. Sensorineural hearing loss in patients with head and neck cancer after chemoradiotherapy and radiotherapy: A systematic review of the literature. Head Neck 2014, 37, 281–292. [Google Scholar] [CrossRef]
- Casasola, R. Head and neck cancer. J. R. Coll. Physicians Edinb. 2010, 40, 343–345. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Ansari, S.F.; Terry, C.; Cohen-Gadol, A.A. Surgery for vestibular schwannomas: A systematic review of complications by approach. Neurosurg. Focus 2012, 33, E14. [Google Scholar] [CrossRef] [PubMed]
- A Telian, S. Management of the small acoustic neuroma: A decision analysis. Am. J. Otol. 1994, 15, 358–365. [Google Scholar] [PubMed]
- Abouzari, M.; Bs, K.G.; Sarna, B.; Bs, P.K.; Ms, T.R.; Mostaghni, N.; Lin, H.W.; Djalilian, H.R. Prediction of vestibular schwannoma recurrence using artificial neural network. Laryngoscope Investig. Otolaryngol. 2020, 5, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Claudia, D.N.; Amico, N.; Merone, M.; Sicilia, R.; Cordelli, E.; D’Antoni, F.; Antoni, N.; Zanetti, I.B.; Valbusa, G.; Grossi, E.; et al. Tackling imbalance radiomics in acoustic neuroma. Int. J. Data Min. Bioinform. 2019, 22, 365. [Google Scholar] [CrossRef]
- D’Amico, N.C.; Sicilia, R.; Cordelli, E.; Valbusa, G.; Grossi, E.; Zanetti, I.B.; Beltramo, G.; Fazzini, D.; Scotti, G.; Iannello, G.; et al. Radiomics for Predicting CyberKnife response in acoustic neuroma: A pilot study. In Proceedings of the 2018 IEEE International Conference on Bioinformatics and Biomedicine, BIBM, Madrid, Spain, 3–6 December 2018; pp. 847–852. [Google Scholar] [CrossRef]
- Cha, D.; Shin, S.H.; Kim, S.H.; Choi, J.Y.; Moon, I.S. Machine learning approach for prediction of hearing preservation in vestibular schwannoma surgery. Sci. Rep. 2020, 10, 1–6. [Google Scholar] [CrossRef]
- Dang, S.; Manzoor, N.F.; Chowdhury, N.; Tittman, S.M.; Yancey, K.L.; Monsour, M.A.; O’Malley, M.R.; Rivas, A.; Haynes, D.S.; Bennett, M.L. Investigating Predictors of Increased Length of Stay After Resection of Vestibular Schwannoma Using Machine Learning. Otol. Neurotol. 2021, 42, e584–e592. [Google Scholar] [CrossRef]
- George-Jones, N.A.; Wang, K.; Wang, J.; Hunter, J.B. Prediction of Vestibular Schwannoma Enlargement After Radiosurgery Using Tumor Shape and MRI Texture Features. Otol. Neurotol. 2020, 42, e348–e354. [Google Scholar] [CrossRef]
- Langenhuizen, P.P.J.H.; Sebregts, S.H.P.; Zinger, S.; Leenstra, S.; Verheul, J.B.; With, P.H.N. Prediction of transient tumor enlargement using MRI tumor texture after radiosurgery on vestibular schwannoma. Med Phys. 2020, 47, 1692–1701. [Google Scholar] [CrossRef]
- Langenhuizen, P.P.J.H.; Zinger, S.; Leenstra, S.; Kunst, H.P.M.; Mulder, J.J.S.; Hanssens, P.E.J.; de With, P.H.N.; Verheul, J.B. Radiomics-Based Prediction of Long-Term Treatment Response of Vestibular Schwannomas Following Stereotactic Radiosurgery. Otol. Neurotol. 2020, 41, e1321–e1327. [Google Scholar] [CrossRef]
- Langenhuizen, P.; Van Gorp, H.; Zinger, S.; Verheul, J.; Leenstra, S.; De With, P.H.N. Dose distribution as outcome predictor for Gamma Knife radiosurgery on vestibular schwannoma. Proc. SPIE 2019, 10950, 109504C. [Google Scholar] [CrossRef]
- Langenhuizen, P.P.J.H.; Legters, M.J.W.; Zinger, S.; Verheul, J.; De With, P.N.; Leenstra, S. MRI textures as outcome predictor for Gamma Knife radiosurgery on vestibular schwannoma. Proc. SPIE 2018, 10575, 105750H. [Google Scholar] [CrossRef]
- Lee, S.; Seo, S.-W.; Hwang, J.; Seol, H.J.; Nam, D.-H.; Lee, J.-I.; Kong, D.-S. Analysis of risk factors to predict communicating hydrocephalus following gamma knife radiosurgery for intracranial schwannoma. Cancer Med. 2016, 5, 3615–3621. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.-C.; Wu, C.-C.; Lee, C.-C.; Huang, H.-E.; Lee, W.-K.; Chung, W.-Y.; Wu, H.-M.; Guo, W.-Y.; Wu, Y.-T.; Lu, C.-F. Prediction of pseudoprogression and long-term outcome of vestibular schwannoma after Gamma Knife radiosurgery based on preradiosurgical MR radiomics. Radiother. Oncol. 2020, 155, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Ta, N.H. ENT in the context of global health. Ann. R. Coll. Surg. Engl. 2019, 101, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Nouraei, S.; Huys, Q.; Chatrath, P.; Powles, J.; Harcourt, J. Screening patients with sensorineural hearing loss for vestibular schwannoma using a Bayesian classifier. Clin. Otolaryngol. 2007, 32, 248–254. [Google Scholar] [CrossRef]
- Juhola, M. On Machine Learning Classification of Otoneurological Data. Stud. Health Technol. Inform. 2008, 136, 211–216. [Google Scholar] [CrossRef]
- Juhola, M.; Laurikkala, J.; Viikki, K.; Kentala, E.; Pyykkö, I. Classification of patients on the basis of otoneurological data by using Kohonen networks. Acta Otolaryngol. Suppl. 2001, 545, 50–52. [Google Scholar] [CrossRef]
- Kentala, E.; Pyykkö, I.; Viikki, K.; Juhola, M. Production of diagnostic rules from a neurotologic database with decision trees. Ann. Otol. Rhinol. Laryngol. 2000, 109, 170–176. [Google Scholar] [CrossRef]
- Kentala, E.; Pyykkö, I.; Laurikkala, J.; Juhola, M. Discovering diagnostic rules from a neurotologic database with genetic algorithms. Ann. Otol. Rhinol. Laryngol. 1999, 108, 948–954. [Google Scholar] [CrossRef]
- Laurikkala, J.P.S.; Kentala, E.L.; Juhola, M.; Pyykkö, I.V. A novel machine learning program applied to discover otological diagnoses. Scand. Audiol. Suppl. 2001, 52, 100–102. [Google Scholar] [CrossRef]
- Miettinen, K.; Juhola, M. Classification of otoneurological cases according to Bayesian probabilistic models. J. Med Syst. 2008, 34, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Viikki, E.K.K. Decision tree induction in the diagnosis of otoneurological diseases. Med. Inform. Internet Med. 1999, 24, 277–289. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-C.; Guo, W.-Y.; Chung, W.-Y.; Wu, H.-M.; Lin, C.-J.; Lee, C.-C.; Liu, K.-D.; Yang, H.-C. Magnetic resonance imaging characteristics and the prediction of outcome of vestibular schwannomas following Gamma Knife radiosurgery. J. Neurosurg. 2017, 127, 1384–1391. [Google Scholar] [CrossRef] [PubMed]
- Dickson, S.; Thomas, B.T.; Goddard, P. Using Neural Networks to Automatically Detect Brain Tumours in MR Images. Int. J. Neural Syst. 1997, 8, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.-K.; Wu, C.-C.; Lee, C.-C.; Lu, C.-F.; Yang, H.-C.; Huang, T.-H.; Lin, C.-Y.; Chung, W.-Y.; Wang, P.-S.; Wu, H.-M.; et al. Combining analysis of multi-parametric MR images into a convolutional neural network: Precise target delineation for vestibular schwannoma treatment planning. Artif. Intell. Med. 2020, 107, 101911. [Google Scholar] [CrossRef]
- Bs, N.A.G.; Wang, K.; Wang, J.; Hunter, J.B. Automated Detection of Vestibular Schwannoma Growth Using a Two-Dimensional U-Net Convolutional Neural Network. Laryngoscope 2020, 131, E619–E624. [Google Scholar] [CrossRef]
- Shapey, J.; Wang, G.; Dorent, R.; Dimitriadis, A.; Li, W.; Paddick, I.; Kitchen, N.; Bisdas, S.; Saeed, S.R.; Ourselin, S.; et al. An artificial intelligence framework for automatic segmentation and volumetry of vestibular schwannomas from contrast-enhanced T1-weighted and high-resolution T2-weighted MRI. J. Neurosurg. 2021, 134, 171–179. [Google Scholar] [CrossRef]
- Lee, C.-C.; Lee, W.-K.; Wu, C.-C.; Lu, C.-F.; Yang, H.-C.; Chen, Y.-W.; Chung, W.-Y.; Hu, Y.-S.; Wu, H.-M.; Wu, Y.-T.; et al. Applying artificial intelligence to longitudinal imaging analysis of vestibular schwannoma following radiosurgery. Sci. Rep. 2021, 11, 1–10. [Google Scholar] [CrossRef]
- Neves, C.A.; Tran, E.D.; Kessler, I.M.; Blevins, N.H. Fully automated preoperative segmentation of temporal bone structures from clinical CT scans. Sci. Rep. 2021, 11, 1–11. [Google Scholar] [CrossRef]
- Uetani, H.; Nakaura, T.; Kitajima, M.; Yamashita, Y.; Hamasaki, T.; Tateishi, M.; Morita, K.; Sasao, A.; Oda, S.; Ikeda, O.; et al. A preliminary study of deep learning-based reconstruction specialized for denoising in high-frequency domain: Usefulness in high-resolution three-dimensional magnetic resonance cisternography of the cerebellopontine angle. Neuroradiology 2020, 63, 63–71. [Google Scholar] [CrossRef]
- Windisch, P.; Weber, P.; Fürweger, C.; Ehret, F.; Kufeld, M.; Zwahlen, D.; Muacevic, A. Implementation of model explainability for a basic brain tumor detection using convolutional neural networks on MRI slices. Neuroradiology 2020, 62, 1515–1518. [Google Scholar] [CrossRef] [PubMed]
- Kügler, D.; Sehring, J.; Stefanov, A.; Stenin, I.; Kristin, J.; Klenzner, T.; Schipper, J.; Mukhopadhyay, A. i3PosNet: Instrument pose estimation from X-ray in temporal bone surgery. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-S.; Park, H.; Hong, S.H.; Chung, W.-H.; Cho, Y.-S.; Moon, I.J. Predicting cochlear dead regions in patients with hearing loss through a machine learning-based approach: A preliminary study. PLoS ONE 2019, 14, e0217790. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, J.E.; Laurell, G.; Rask-Andersen, H.; Bergquist, J.; Eriksson, P.O. The proteome of perilymph in patients with vestibular schwannoma. A possibility to identify biomarkers for tumor associated hearing loss? PLoS ONE 2018, 13, e0198442. [Google Scholar] [CrossRef]
- Pinto, A.; Faiz, O.; Davis, R.; Almoudaris, A.; Vincent, C. Surgical complications and their impact on patients’ psychosocial well-being: A systematic review and meta-analysis. BMJ Open 2016, 6, e007224. [Google Scholar] [CrossRef]
- Jencks, S.F.; Williams, M.V.; Coleman, E.A. Rehospitalizations among Patients in the Medicare Fee-for-Service Program. N. Engl. J. Med. 2009, 360, 1418–1428. [Google Scholar] [CrossRef]
- Hernandez-Meza, G.; McKee, S.; Carlton, D.; Yang, A.; Govindaraj, S.; Iloreta, A. Association of Surgical and Hospital Volume and Patient Characteristics with 30-Day Readmission Rates. JAMA Otolaryngol. Neck Surg. 2019, 145, 328–337. [Google Scholar] [CrossRef]
- Graboyes, E.; Yang, Z.; Kallogjeri, D.; Diaz, J.A.; Nussenbaum, B. Patients Undergoing Total Laryngectomy. JAMA Otolaryngol. Neck Surg. 2014, 140, 1157–1165. [Google Scholar] [CrossRef]
- Ferrandino, R.; Garneau, J.; Roof, S.; Pacheco, C.; Poojary, P.; Saha, A.; Chauhan, K.; Miles, B. The national landscape of unplanned 30-day readmissions after total laryngectomy. Laryngoscope 2017, 128, 1842–1850. [Google Scholar] [CrossRef]
- Dziegielewski, P.T.; Boyce, B.; Manning, A.; Agrawal, A.; Old, M.; Ozer, E.; Teknos, T.N. Predictors and costs of readmissions at an academic head and neck surgery service. Head Neck 2015, 38, E502–E510. [Google Scholar] [CrossRef]
- Bur, A.; Brant, J.; Mulvey, C.L.; Nicolli, E.A.; Brody, R.M.; Fischer, J.P.; Cannady, S.B.; Newman, J.G. Association of Clinical Risk Factors and Postoperative Complications With Unplanned Hospital Readmission After Head and Neck Cancer Surgery. JAMA Otolaryngol. Neck Surg. 2016, 142, 1184–1190. [Google Scholar] [CrossRef] [PubMed]
- Goel, A.N.; Raghavan, G.; John, M.A.S.; Long, J.L. Risk Factors, Causes, and Costs of Hospital Readmission After Head and Neck Cancer Surgery Reconstruction. JAMA Facial Plast. Surg. 2019, 21, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Kripalani, S.; Theobald, C.N.; Anctil, B.; Vasilevskis, E.E. Reducing hospital readmission rates: Current strategies and future directions. Annu. Rev. Med. 2014, 65, 471–485. [Google Scholar] [CrossRef]
- van Walraven, C.; Wong, J.; Forster, A.J. LACE+ index: Extension of a validated index to predict early death or urgent read-mission after hospital discharge using administrative data. Open Med. 2012, 6, 1–11. [Google Scholar]
- How Two Health Systems Use Predictive Analytics to Reduce Readmissions | Managed Healthcare Executive. Available online: https://www.managedhealthcareexecutive.com/view/how-two-health-systems-use-predictive-analytics-reduce-readmissions (accessed on 18 August 2022).
- AlQuraishi, M. AlphaFold at CASP13. Bioinformatics 2019, 35, 4862–4865. [Google Scholar] [CrossRef] [PubMed]
- Cireşan, D.C.; Meier, U.; Gambardella, L.M.; Schmidhuber, J. Deep, Big, Simple Neural Nets for Handwritten Digit Recognition. Neural Comput. 2010, 22, 3207–3220. [Google Scholar] [CrossRef] [PubMed]
- Tschuchnig, M.E.; Oostingh, G.J.; Gadermayr, M. Generative Adversarial Networks in Digital Pathology: A Survey on Trends and Future Potential. Patterns 2020, 1, 100089. [Google Scholar] [CrossRef]
- Li, Y.; Rao, S.; Solares, J.R.A.; Hassaine, A.; Ramakrishnan, R.; Canoy, D.; Zhu, Y.; Rahimi, K.; Salimi-Khorshidi, G. BEHRT: Transformer for Electronic Health Records. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Rolston, J.D.; Han, S.J.; Chang, E.F. Systemic inaccuracies in the National Surgical Quality Improvement Program database: Implications for accuracy and validity for neurosurgery outcomes research. J. Clin. Neurosci. 2017, 37, 44–47. [Google Scholar] [CrossRef]
- Zhang, Z.; Beck, M.W.; Winkler, D.A.; Huang, B.; Sibanda, W.; Goyal, H.; on behalf of AME Big-Data Clinical Trial Collaborative Group. Opening the black box of neural networks: Methods for interpreting neural network models in clinical applications. Ann. Transl. Med. 2018, 6, 216. [Google Scholar] [CrossRef]
- McGrath, H.; Li, P.; Dorent, R.; Bradford, R.; Saeed, S.; Bisdas, S.; Ourselin, S.; Shapey, J.; Vercauteren, T. Manual segmentation versus semi-automated segmentation for quantifying vestibular schwannoma volume on MRI. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 1445–1455. [Google Scholar] [CrossRef] [PubMed]


| Author, Year, References | Aim | Algorithm(s) | Outcomes |
|---|---|---|---|
| Abouzari et al., 2020 [24] | Predicting the recurrence of vestibular schwannoma using artificial neural network compared to logistic regression. | Artificial neural network | Artificial neural network was superior to logistic regression in predicting the recurrence of vestibular schwannoma with an accuracy of 0.70. |
| Claudia et al., 2019 [25] | Using machine-learning radiomics to predict response to CyberKnife treatment of vestibular schwannoma. | Decision tree Random forest XGBoost | Machine-learning radiomics predicted response to CyberKnife treatment of vestibular schwannoma with an accuracy of 0.92. |
| D’Amico et al., 2018 [26] | Computing quantitative biomarkers from MRI to predict CyberKnife treatment response on vestibular schwannoma. | Decision tree | Treatment response was predicted with an accuracy of 0.85 using a machine-learning based radiomic pipeline. |
| Cha et al., 2020 [27] | Predicting hearing preservation following surgery in patients with vestibular schwannoma. | Support vector machine Gradient boosting machine Deep neural network Random forest | Hearing preservation was predicted with most accurately by deep neural network with an accuracy of 0.9 |
| Dang et al., 2021 [28] | Elucidation of risk factors contributing to increased length of stay after vestibular schwannoma surgery. | Random forest | Preoperative tumor volume and dimensions, coronary artery disease, hypertension, major complications, and operative time were significant predictive factors for prolonged length of stay |
| George-Jones et al., 2020 [29] | Predicting post-stereotactic surgery enlargement of vestibular schwannoma. | Support vector machine | Enlargement was predicted with an overall AUC of 0.75. |
| Langenhuizen et al., 2020 [30] | Prediction of tumor progression after stereotactic surgery of vestibular schwannoma using MRI texture. | Support vector machine | Machine learning achieved an AUC of 0.93 and an accuracy of 0.77 for prediction of tumor progression. |
| Langenhuizen et al., 2020 [31] | Prediction of transient tumor enlargement after vestibular schwannoma radiosurgery using MRI textures. | Support vector machine | A maximum AUC of 0.95, sensitivity of 0.82, and specificity of 0.89 were achieved for prediction. |
| Langenhuizen et al., 2019 [32] | Predicting the influence of dose distribution on the treatment response of gamma knife radiosurgery on vestibular schwannoma. | Support vector machine | 3D histogram of oriented gradients features correlate with treatment outcomes (AUC = 0.79, TPR = 0.80, TNR = 0.75, with support vector machine) |
| Langenhuizen et al., 2018 [33] | Using MRI texture feature analysis to predict vestibular schwannoma gamma knife radiosurgery treatment outcomes. | Support vector machine Decision tree | Treatment outcomes were predicted with an accuracy of 0.85 using machine learning. |
| Lee et al., 2016 [34] | Predicting risk factors leading to communicating hydrocephalus following gamma knife radiosurgery for vestibular schwannoma. | K-nearest neighbors classifier Support vector machine Decision tree Random forest AdaBoost Naïve bayes Linear discriminant analysis Gradient boosting machine | Age, tumor volume, and tumor origin are significant predictors of communicating hydrocephalus. Developing communicating HCP following gamma knife radiosurgery is most likely if the tumor was of vestibular origin and had a volume ≥13.65 cm3. |
| Telian et al., 1994 [23] | Management of vestibular schwannoma between 5–15 mm | Decision tree | Most important factor in determining to proceed with surgery is the probability of tumor growth. |
| Yang et al., 2020 [35] | Prediction of progression/outcome of vestibular schwannoma after gamma knife radiosurgery using MRI data | Support vector machine | Machine learning predicted long-term outcome and transient pseudoprogression with an accuracy of 0.88 and 0.85, respectively. |
| Author, Year, References | Aim | Algorithm(s) | Outcomes |
|---|---|---|---|
| Juhola et al., 2008 [38] | Classification of otoneurological diseases including vestibular schwannoma, benign positional vertigo, Menière’s disease, sudden deafness, traumatic vertigo, and vestibular neuritis given patient attributes. | K-nearest neighbors classifier Discriminant analysis Naïve bayes K-means clustering Decision trees Neural networks Kohonen networks | Discriminant analysis performed the best with an average accuracy of 0.96. |
| Juhola et al., 2001 [39] | Classification of otoneurological diseases including vestibular schwannoma, benign positional vertigo, Menière’s disease, sudden deafness, traumatic vertigo, and vestibular neuritis given patient attributes. | Kohonen networks | The model attained a maximum accuracy of 0.98 for classification of overrepresented pathologies. |
| Kentala et al., 2000 [40] | Classification of otoneurological diseases including vestibular schwannoma, benign positional vertigo, Menière’s disease, sudden deafness, traumatic vertigo, and vestibular neuritis given patient attributes. | Decision tree | The decision tree achieved an accuracy between 0.94 and 1 according to different pathologies. |
| Kentala et al., 1999 [41] | Classification of otoneurological diseases including vestibular schwannoma, benign positional vertigo, Menière’s disease, sudden deafness, traumatic vertigo, and vestibular neuritis given patient attributes. | Genetic algorithm | The genetic algorithm attained an accuracy of 0.80. |
| Laurikkala et al., 2001 [42] | Classification of otoneurological diseases including vestibular schwannoma, benign positional vertigo, Menière’s disease, sudden deafness, traumatic vertigo, and vestibular neuritis given patient attributes. | Genetic algorithm | The machine learning model attained an accuracy of 0.90. |
| Miettinen et al., 2008 [43] | Classification of otoneurological diseases including vestibular schwannoma, benign positional vertigo, Menière’s disease, sudden deafness, traumatic vertigo, and vestibular neuritis given patient attributes. | Bayesian classifier | The Bayesian classifier attained an accuracy of 0.97. |
| Viikki et al., 1999 [44] | Classification of otoneurological diseases including vestibular schwannoma, benign positional vertigo, Menière’s disease, sudden deafness, traumatic vertigo, and vestibular neuritis given patient attributes. | Decision tree | An average accuracy of over 0.95 was achieved. |
| Nouraei et al., 2007 [37] | Identification of vestibular schwannoma cases from a population suspected to harbor the tumor. | Bayesian classifier | The machine learning algorithm achieved an AUC of 0.80 for classification. |
| Author, Year, References | Aim | Algorithm(s) | Outcomes |
|---|---|---|---|
| Dickson et al., 1997 [46] | Detection of vestibular schwannoma on MRI scans. | Artificial neural network | The neural network attained a false negative rate of 0 and false positive rate of 0.086. |
| George-Jones et al., 2020 [29] | Segmentation of vestibular schwannoma from T1W with contrast MRI scans. | U-Net | The model achieved an interclass correlation coefficient of 0.99. |
| Lee et al., 2021 [50] | Segmentation of vestibular schwannoma from MRI scans. | U-Net | The model performed with an average dice score of 0.90. |
| Lee et al., 2020 [47] | Segmentation of vestibular schwannoma from multiparametric MRI scans. | U-Net | The U-Net delineated vestibular schwannoma with a dice score of 0.90 ± 0.05. |
| Neves et al., 2021 [51] | Segmentation of temporal bone structures from CT scans. | AH-Net U-Net ResNet | The model’s performed with dice scores of 0.91, 0.85, 0.75, and 0.86 for inner ear, ossicles, facial nerve, and the sigmoid sinus, respectively. |
| Shapey et al., 2021 [49] | Segmentation of vestibular schwannoma from T2W and T1W with contrast MRI scans. | Convolutional neural network | By employing a computational attention module, the algorithm attained a dice score of 0.93 and 0.94 for T1W and T2W, respectively. |
| Uetani et al., 2020 [52] | Denoising of MRI for high spatial resolution-MR cisternography for cerebellopontine angle legions via deep learning-based reconstruction. | Convolutional neural network | Images reconstructed with deep learning-based reconstruction had higher image quality (p < 0.001) due to reduced image noise while maintaining contrast and sharpness. |
| Windisch et al., 2020 [53] | Segmentation of vestibular schwannoma or glioblastoma from T1W, T2W, and T1W with contrast MRI scans with a focus on the explainability. | Convolutional neural network | The model achieved an accuracy of 0.93 while the Grad-CAM software also showed it correctly focused on tumor loci. |
| Author, Year, References | Aim | Algorithm(s) | Outcomes |
|---|---|---|---|
| Chang et al., 2019 [55] | Prediction of cochlear dead region prevalence given various sensorineural hearing loss patient data. | Decision tree Random forest | The random forest and the classification tree were capable of predicting cochlear dead regions by an accuracy of 0.82 and 0.93, respectively, while illustrating strong predictive factors for cochlear dead region prevalence. |
| Rasmussen et al., 2018 [56] | Elucidation of perilymph proteins associated to vestibular schwannoma related hearing loss and tumor diameter. | Random forest | A perilymph protein, alpha-2-HS-glycoprotein (P02765) was determined to be an independent variable for predicting tumor-associated hearing loss. |
| Kügler et al., 2020 [54] | Creation of a convolutional neural network-powered pose estimation method capable of high-precision estimation of poses for application to temporal bone surgery. | Convolutional neural network | The instrument was found to estimate the pose, or an estimation of location of surgical instrument using an X-ray image, with an error <0.05 mm on synthetic data. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsutsumi, K.; Soltanzadeh-Zarandi, S.; Khosravi, P.; Goshtasbi, K.; Djalilian, H.R.; Abouzari, M. Machine Learning in the Management of Lateral Skull Base Tumors: A Systematic Review. J. Otorhinolaryngol. Hear. Balance Med. 2022, 3, 7. https://doi.org/10.3390/ohbm3040007
Tsutsumi K, Soltanzadeh-Zarandi S, Khosravi P, Goshtasbi K, Djalilian HR, Abouzari M. Machine Learning in the Management of Lateral Skull Base Tumors: A Systematic Review. Journal of Otorhinolaryngology, Hearing and Balance Medicine. 2022; 3(4):7. https://doi.org/10.3390/ohbm3040007
Chicago/Turabian StyleTsutsumi, Kotaro, Sina Soltanzadeh-Zarandi, Pooya Khosravi, Khodayar Goshtasbi, Hamid R. Djalilian, and Mehdi Abouzari. 2022. "Machine Learning in the Management of Lateral Skull Base Tumors: A Systematic Review" Journal of Otorhinolaryngology, Hearing and Balance Medicine 3, no. 4: 7. https://doi.org/10.3390/ohbm3040007
APA StyleTsutsumi, K., Soltanzadeh-Zarandi, S., Khosravi, P., Goshtasbi, K., Djalilian, H. R., & Abouzari, M. (2022). Machine Learning in the Management of Lateral Skull Base Tumors: A Systematic Review. Journal of Otorhinolaryngology, Hearing and Balance Medicine, 3(4), 7. https://doi.org/10.3390/ohbm3040007








