Genetic Analysis of Mutant Strains of Saccharomyces cerevisiae with Defects in Mannoprotein Synthesis †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains
2.2. Extraction of Nucleic Acids
3. Results and Discussion
Identification of Mutations in MNN Genes Responsible for Synthesis of Outer Chain
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aebi, M. N-linked protein glycosylation in the ER. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2013, 1833, 2430–2437. [Google Scholar] [CrossRef] [PubMed]
- Ballou, C.E. Isolation, characterization and properties of Saccharomyces cerevisiae mnn mutants with nonconditional protein glycosylation defects. Methods Enzymol. 1990, 185, 440–470. [Google Scholar] [PubMed]
- Hernandez, L.M.; Ballou, L.; Alvarado, E.; Tsai, P.K.; Ballou, C.E. Structure of the phosphorylated N-linked oligosaccharides from the mnn9 and mnn10 mutants of Saccharomyces cerevisiae. J. Biol. Chem. 1989, 264, 13648–13659. [Google Scholar] [CrossRef]
- Jigami, Y.; Odani, T. Mannosylphosphate transfer to yeast mannan. Biochim. Biophys. Acta 1999, 1426, 335–345. [Google Scholar] [CrossRef]
- Rayner, J.C.; Munro, S. Identification of the MNN2 and MNN5 mannosyltransferases required for forming and extending the mannose branches of the outer chain mannans of Saccharomyces cerevisiae. J. Biol. Chem. 1998, 273, 26836–26843. [Google Scholar] [CrossRef] [PubMed]
- Ballou, L.; Alvarado, E.; Tsai, P.K.; Dell, A.; Ballou, C.E. Protein glycosylation defects in the Saccharomyces cerevisiae mnn7 mutant class. Support for the stop signal proposed for regulation of outer chain elongation. J. Biol. Chem. 1989, 264, 11857–11864. [Google Scholar] [CrossRef]
- Jungmann, J.; Rayner, J.C.; Munro, S. The Saccharomyces cerevisiae protein Mnn10p/Bed1p is a subunit of a Golgi mannosyltransferase complex. J. Biol. Chem. 1999, 274, 6579–6585. [Google Scholar] [CrossRef] [PubMed]
- Lussier, M.; Sdicu, A.-M.; Bussey, H. The KTR and MNN1 mannosyltransferase families of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1999, 1426, 323–334. [Google Scholar] [CrossRef]
- Yip, C.L.; Welch, S.K.; Klebl, F.; Gilbert, T.; Seidel, P.; Grant, F.J.; O’Hara, P.J.; MacKay, V.L. Cloning and analysis of the Saccharomyces cerevisiae MNN9 and MNN1 genes required for complex glycosylation of secreted proteins. Proc. Natl. Acad. Sci. USA 1994, 91, 2723–2727. [Google Scholar] [CrossRef] [PubMed]
- Corbacho, I.; Olivero, I.; Hernandez, L.M. Identification of the MNN3 gene of Saccharomyces cerevisiae. Glycobiology 2010, 20, 1336–1340. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Rapid isolation of yeast DNA. Cold Spring Harbor Protoc. 2016, 2006, prot4039. [Google Scholar] [CrossRef] [PubMed]
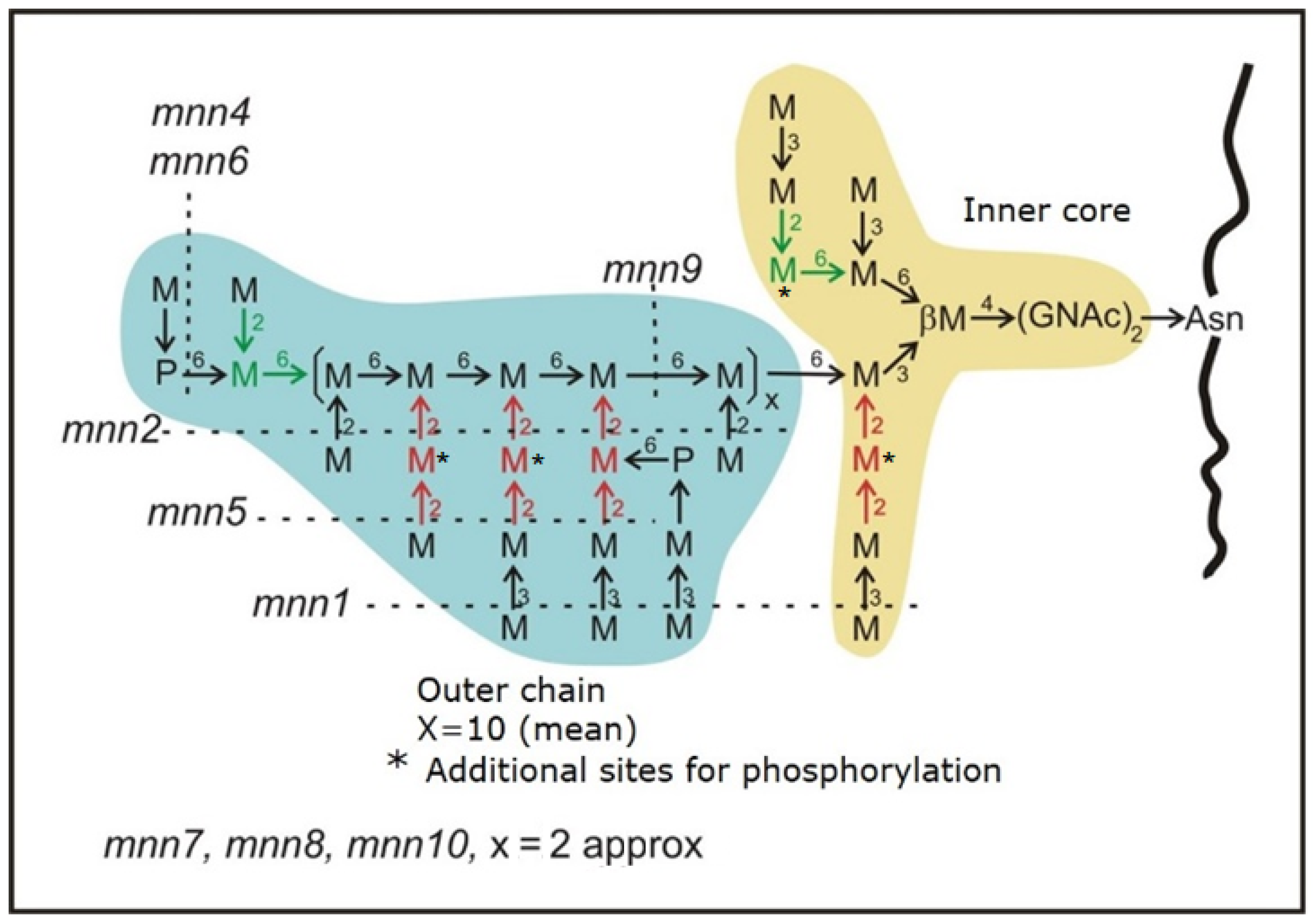
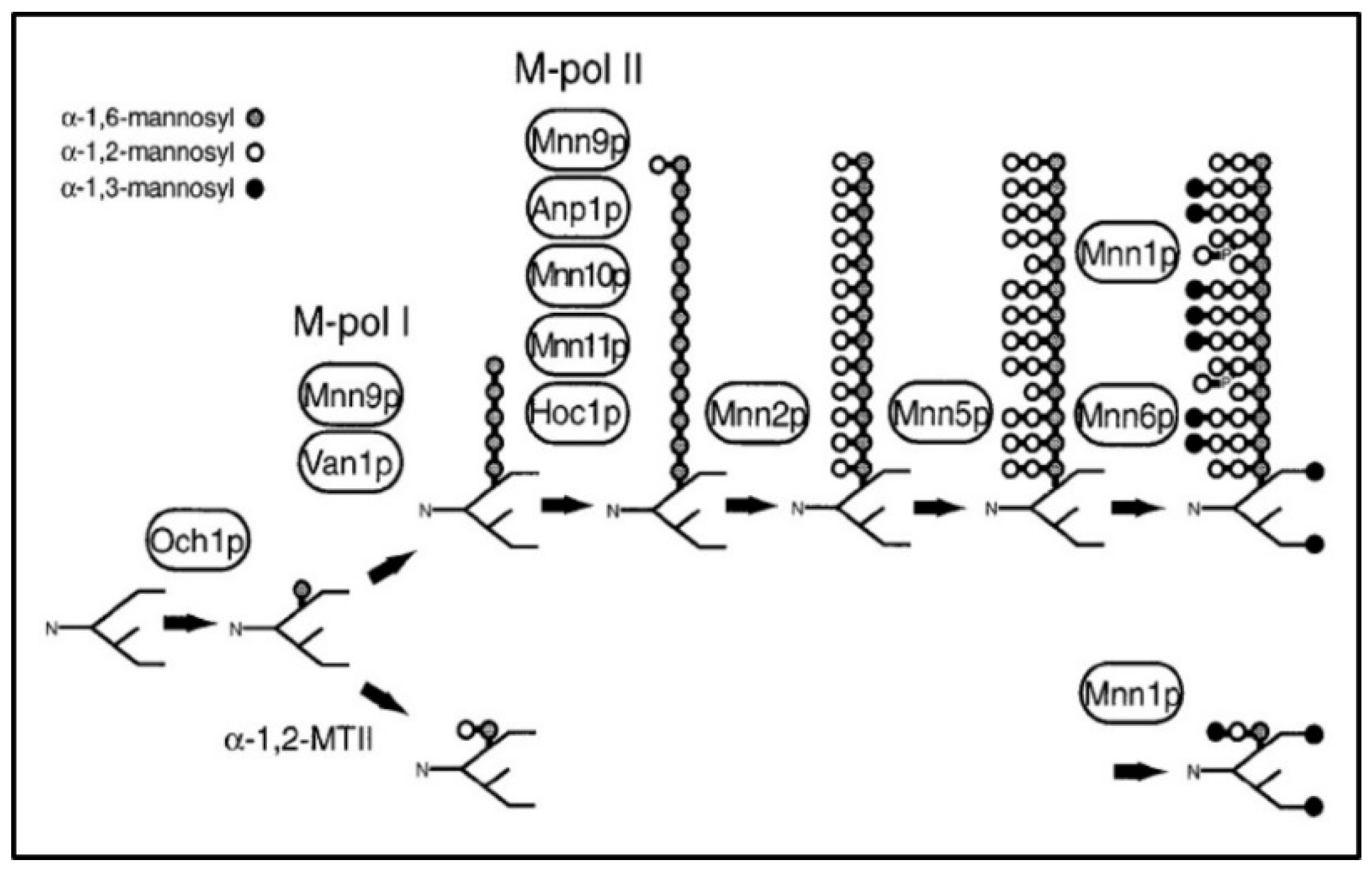
| GENE | Systematic Name | Location in Chromosome | Mutation in DNA | Effect in Protein | Position in Protein |
|---|---|---|---|---|---|
| MNN1 | YER001W | 153,520…155,808 | G > A | C > Y | 697 |
| MNN2 | YBR015C | 267,710…269,503 | G > A | G > D | 359 |
| MNN5 | YJL186W | 80,155…81,915 | G > A | W > stop signal | 561 |
| MNN6 | YPL053C | 457,118…458,458 | Insertion AA | Frameshift | 149 |
| MNN9 | YPL050C | 460,779…461,966 | G > A | W > stop signal | 16 |
| MNN10 | YDR245W | 952,800…953,981 | G > A | W > stop signal | 279 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gil, P.; Martínez, A.; Palencia, E.; Velázquez, R.; Ramírez, M.; Hernández, L.M. Genetic Analysis of Mutant Strains of Saccharomyces cerevisiae with Defects in Mannoprotein Synthesis. Proceedings 2021, 70, 18. https://doi.org/10.3390/foods_2020-07735
Gil P, Martínez A, Palencia E, Velázquez R, Ramírez M, Hernández LM. Genetic Analysis of Mutant Strains of Saccharomyces cerevisiae with Defects in Mannoprotein Synthesis. Proceedings. 2021; 70(1):18. https://doi.org/10.3390/foods_2020-07735
Chicago/Turabian StyleGil, Patricia, Alberto Martínez, Elena Palencia, Rocío Velázquez, Manuel Ramírez, and Luis Miguel Hernández. 2021. "Genetic Analysis of Mutant Strains of Saccharomyces cerevisiae with Defects in Mannoprotein Synthesis" Proceedings 70, no. 1: 18. https://doi.org/10.3390/foods_2020-07735
APA StyleGil, P., Martínez, A., Palencia, E., Velázquez, R., Ramírez, M., & Hernández, L. M. (2021). Genetic Analysis of Mutant Strains of Saccharomyces cerevisiae with Defects in Mannoprotein Synthesis. Proceedings, 70(1), 18. https://doi.org/10.3390/foods_2020-07735





