Catching the Sugars: Electrochemical Aptasensors for the Detection of Cancer-Related Glycosylation Changes in Prostate-Specific Antigen †
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Apparatus
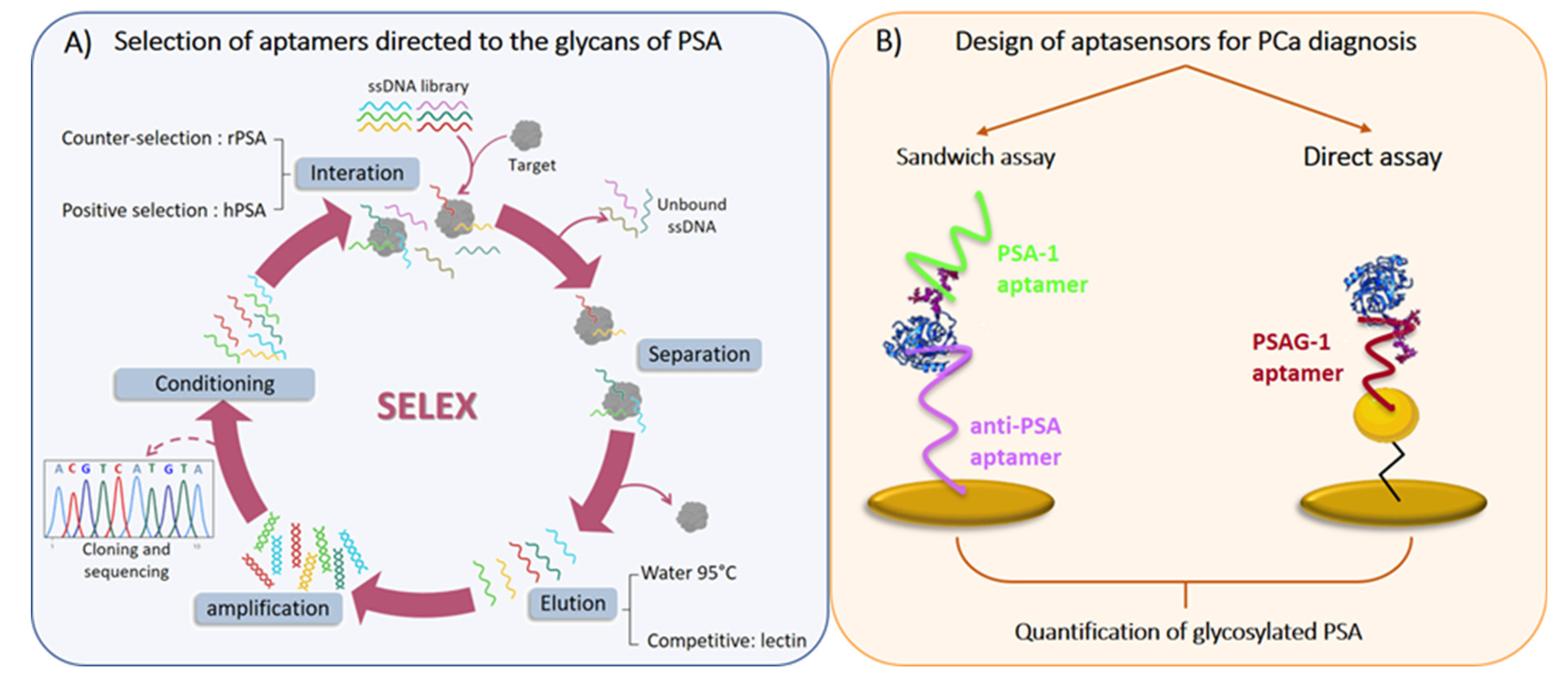
2.2. SELEX Procedure
2.3. Electrochemical Binding Assays
2.4. Deglycosyation Studies
2.5. Sandwich Aptasensor
2.6. Impedimetric Aptasensor
3. Results and Discussion
3.1. Selection and Characterization of Aptamers
3.2. Study of the Binding Site
3.3. Detection of Glycosylated PSA Using Aptasensors
Funding
Acknowledgments
Conflicts of Interest
References
- Lilja, H.; Ulmert, D.; Vickers, A.J. Prostate-specific antigen and prostate cancer: Prediction, detection and monitoring. Nat. Rev. Cancer 2008, 8, 268–278. [Google Scholar] [CrossRef]
- Lomas, D.J.; Ahmed, H.U. All change in the prostate cancer diagnostic pathway. Nat. Rev. Clin. Oncol. 2020, 17, 372–381. [Google Scholar] [CrossRef] [PubMed]
- Pinho, S.S.; Reis, C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef] [PubMed]
- Gilgunn, S.; Conroy, P.J.; Saldova, R.; Rudd, P.M.; O’Kennedy, R.J. Aberrant PSA glycosylation—A sweet predictor of prostate cancer. Nat. Rev. Urol. 2013, 10, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Fernández, A.; Miranda-Castro, R.; de-los-Santos-Álvarez, N.; Lobo-Castañón, M.J. Post-translational modifications in tumor biomarkers: The next challenge for aptamers? Anal. Bioanal. Chem. 2018, 410, 2059–2065. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Fernández, A.; Miranda-Castro, R.; de-los-Santos-Álvarez, N.; Fernández-Rodríguez, E.; Lobo-Castañón, M.J. Focusing aptamer selection on the glycan structure of prostate-specific antigen: Toward more specific detection of prostate cancer. Biosens. Bioelectron. 2019, 128, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Fernández, A.; Miranda-Castro, R.; Díaz, N.; Suárez, D.; de-los-Santos-Álvarez, N.; Lobo-Castañón, M.J. Aptamers targeting protein-specific glycosylation in tumor biomarkers: General selection, characterization and structural modeling. Chem. Sci. 2020, 11, 9402–9413. [Google Scholar] [CrossRef] [PubMed]
- Savory, N.; Abe, K.; Sode, K.; Ikebukuro, K. Selection of DNA aptamer against prostate specific antigen using a genetic algorithm and application to sensing. Biosens. Bioelectron. 2010, 26, 1386–1391. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Fernández, A.; Miranda-Castro, R.; de-los-Santos-Álvarez, N.; Lobo-Castañón, M.J.; Estrela, P. Impedimetric aptamer-based glycan PSA score for discrimination of prostate cancer from other prostate diseases. Biosens. Bioelectron. 2020. submitted. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Díaz-Fernández, A.; Miranda-Castro, R.; Estrela, P.; de-los-Santos-Álvarez, N.; Lobo-Castañón, M.J. Catching the Sugars: Electrochemical Aptasensors for the Detection of Cancer-Related Glycosylation Changes in Prostate-Specific Antigen. Proceedings 2020, 60, 47. https://doi.org/10.3390/IECB2020-07023
Díaz-Fernández A, Miranda-Castro R, Estrela P, de-los-Santos-Álvarez N, Lobo-Castañón MJ. Catching the Sugars: Electrochemical Aptasensors for the Detection of Cancer-Related Glycosylation Changes in Prostate-Specific Antigen. Proceedings. 2020; 60(1):47. https://doi.org/10.3390/IECB2020-07023
Chicago/Turabian StyleDíaz-Fernández, Ana, Rebeca Miranda-Castro, Pedro Estrela, Noemí de-los-Santos-Álvarez, and María Jesús Lobo-Castañón. 2020. "Catching the Sugars: Electrochemical Aptasensors for the Detection of Cancer-Related Glycosylation Changes in Prostate-Specific Antigen" Proceedings 60, no. 1: 47. https://doi.org/10.3390/IECB2020-07023
APA StyleDíaz-Fernández, A., Miranda-Castro, R., Estrela, P., de-los-Santos-Álvarez, N., & Lobo-Castañón, M. J. (2020). Catching the Sugars: Electrochemical Aptasensors for the Detection of Cancer-Related Glycosylation Changes in Prostate-Specific Antigen. Proceedings, 60(1), 47. https://doi.org/10.3390/IECB2020-07023








