1. Introduction
Cervical cancer, the second most prevalent cancer in female throughout the world, is resulted from Human papilloma virus (HPV) [
1]. HPV types associated with cervical cancer have been classified as low risk type (LR-HPV), probably high risk type (PrHR-HPV), and high risk type (HR-HPV) according to their oncogenic risk potential. This classification has been performed after evidence of etiological connection of HPV types to cervical cancer [
2]. HPV approximately has 170 agents and more than 40 of them are most commonly transmitted through sexual contact. HPV-16 and HPV-18 are the etiological agents of cervical cancer that causes 70% of all cases, resulting in death of 270,000 women per year [
3]. The cure rate of cervical cancer, however, is high when diagnosed early.
The Papanicolaou (Pap) smear and HPV-DNA tests are current HPV detection techniques and they are very significant to detect cervical cancer early. However, the traditional methods like standard tube-based PCR tests are not suitable since they require highly trained personals, intensive needs for resource and clean lab environment as well as taking turnaround times of weeks. Therefore, it is urgently needed to develop a cheap and robust device for cervical cancer.
Microfluidic integrated analysis systems are of great importance as candidates for developing the global health and controlling epidemics. The potential benefits of these devices are low cost, integration of complex functions like an entire laboratory, faster reaction time and enhanced analytical sensitivity [
4]. In microfluidic devices, these properties can be customized for portable POC systems. Hence, development of disposable, simple and reliable devices is urgently needed.
In the present study, we have developed an entirely integrated non-enzymatic paper based analytical microfluidic chip to detect cervical cancer early from HPV 16&18 reagents selectively in low LOD. The fabricated paper based POC system is composed of an extraction tool and its chemical single step buffer for combined HPV form to lyse cervical cells. Furthermore, patient samples’ DNA has been specifically hybridized with silver nanoparticles and streptavidin coated magnetically labeled primers for non-enzymatic amplification and target analyte detection. The fabricated device is a promising tool to diagnose patients and detect cervical cancer early in less than 40 min.
2. Materials and Methods
The technique by Chomczynski et al. [
5] was used to develop single step extraction and precipitation process for combined HPV 16, 18 and RNase P DNA form. To release HPV DNA for downstream amplification and detection, cervical cells were lysed. The amounts of chemical lysis, precipitation and extraction matrix were optimized. Firstly, the cells and virus particles in DNA extractions were lysed with a single step extraction buffer including Guanidinium thiocyanate (GuSCN), NaCl, Glycogen co-precipitant and alcohol. Biosystems 7500 Fast Real-Time PCR System measured the lysis and extraction buffer efficiency with amplification of combined HPV DNA to optimize and confirm experiments. E7 genes of HPV 16&18, RNase P fragments and their primers and probes were used as reference for comparison. Optimization of Q-PCR temperature and time was performed as follows: denaturation firstly was applied at 90 °C for 10 min, thereafter 95 °C for 30 s, followed by 55 °C extension for 30 s and 60 °C reading for 1.5 m.
AgNPs labels and magnetic beads (Mag-Bead) were modified with real patient samples to provide analytical detection. Streptavidin-coated magnetic microbeads and Citrate-capped AgNPs, obtained from Bangs Laboratories (Fishers, IN, USA) and Ted Pella (Redding, CA, USA) respectively, were modified with biotinylated DNA based on protocol by Alivisatos and co-workers [
6]. AgNPs and Mag-Bead modifications were also carried out with S-DNA-Bio for confirmation and they were confirmed via Agilent 8453 UV-Vis spectra (Agilent Technologies, Santa Clara, CA, USA). A potentiostat (700E, CH Instruments, Austin, TX, USA) was used to investigate and optimize electrochemical detection of the hybridized analyte. Glassy carbon (1.0 mm diameter), Ag/AgCl (KCl = 1 M) and Pt wire were used as working electrode, reference electrode and counter electrode, respectively. In addition, phosphate buffer and borate chloride electrolyte solutions were used as a medium and KMnO
4 were used as the oxidation agent.
Crooks and co-workers technique [
7] was used for fabrication of paper chip to detect combined HPV 16&18. The design and fabrication of the paper-based device was done by ColorQube 8570DN wax printer (Xerox Corporation, Norwalk, CT, USA), laser cutter (Epilog Laser, Golden, CO, USA) and 3D modelling software (SolidWorks). After printing coloured wax, providing paper with hydrophobic and hydrophilic feature, the paper was put on the heater at 120 °C for 30 s to achieve 3D hydrophobic surfaces. Then, paper was cooled to room temperatures. SolidWork was also used to design carbon past electrodes that were printed with stencil.
3. Results and Discussion
Figure 1 illustrates modification of Double Stranded DNA (DS DNA) of extracted patient sample with primers after denaturation process where biotin and thiol ended primers have been labeled with AgNPs and magnetic beads. The novel technique implemented in this experiment differs compared to the previous methods in that specific primers of HPV 16&18 DNAs are labeled with specific reagents in accordance with this method. Optimum parameters were obtained by altering molarity and volume of DS DNA and its primers. 100 μM 200 μL AgNPs-Primer, 100 μM 100 μL AgNPs-Primer, 10 μM 200 μL AgNPs-Primer and 10 μM 100 μL AgNPs-Primer were compared for 2.65 pM model analyte.
In this work, thiol and biotin labeled primers with various concentrations were modified with AgNPs and magnetic microbeads (
Figure 1a,b). As seen in
Figure 1c,d, oligonucleotides were first heated at 95 °C for denaturation and then cooled and annealed with primers. For accurate detection, the model analyte was finally washed by using magnet to remove remaining from environment. In order to detect lower model analyte concentrations, the hybridization of DNA with reagent labeled primers were modified with 200 μL 100 μM AgNPs-primer and 530 fM of HPV model analyte was measured. As seen in
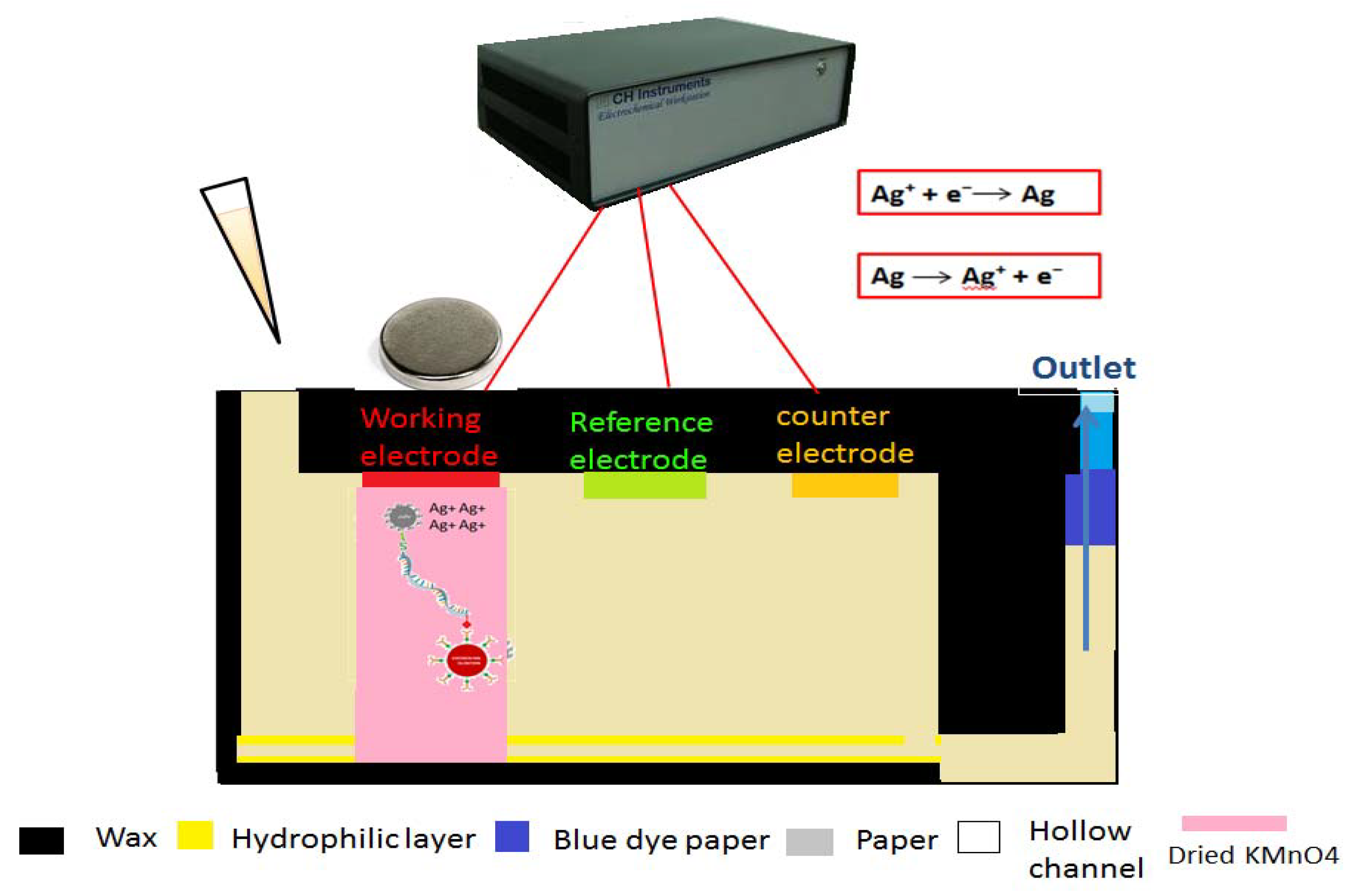
Figure 2, involving electrochemical detection method, the hybridized analyte is detected on the surface of working electrode with the help of a magnet. Firstly, the buffer containing model analyte added to the inlet is concentrated at the working electrode. After saturation of the sink, a moveable piece of paper, containing dried KMnO
4−, is sent through the working electrode. The dissolved MnO
4− oxidizes AgNPs to Ag
+ ions. After that, negative potential is applied for 200 s to electrodeposit Ag
+. Finally, the potential is swept from negative to positive potential and a change in current is measured as a result of oxidation.
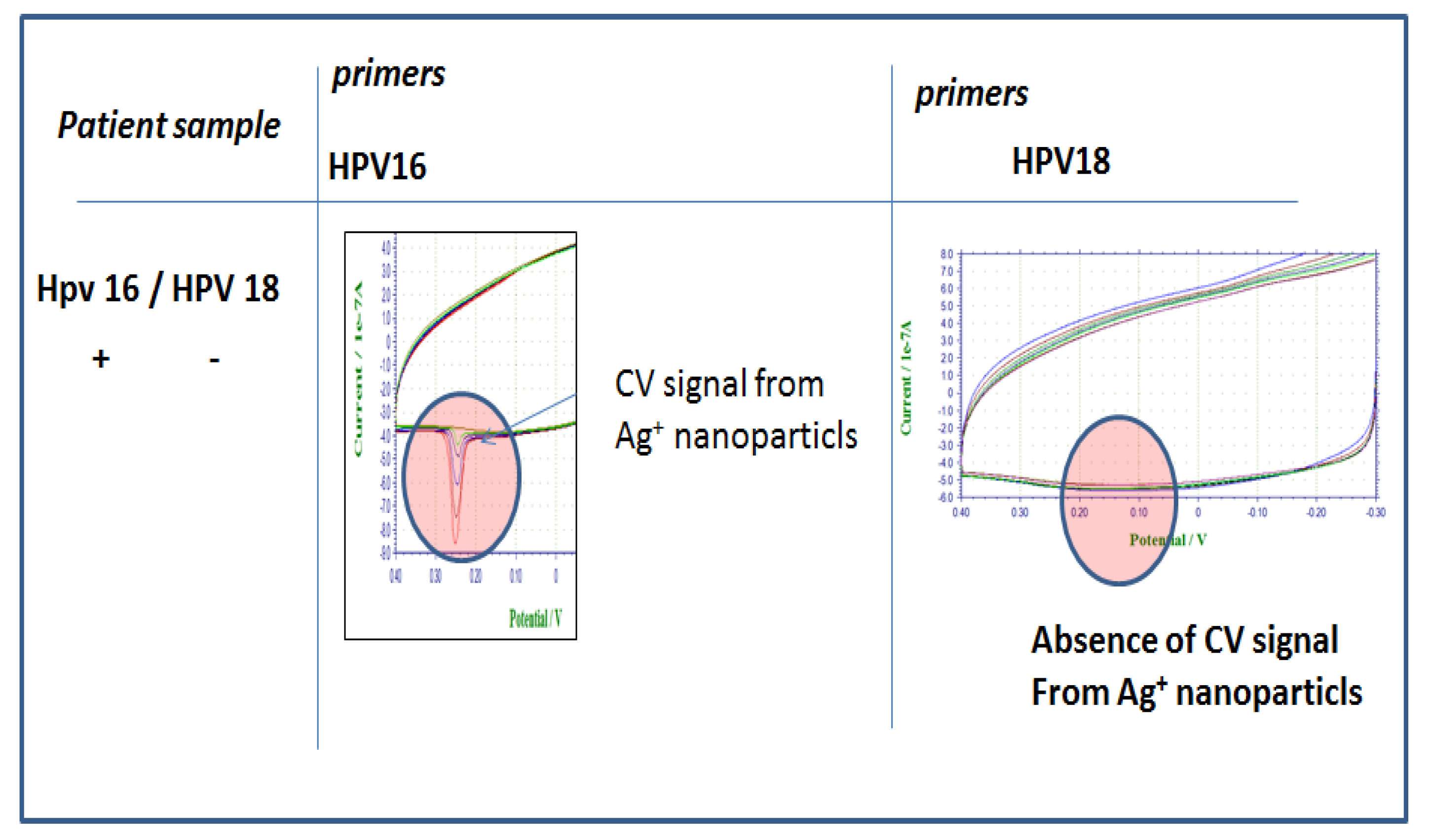
Figure 3 shows electrochemical analysis of various concentrations of model analyte. There is a linear correlation between model analyte concentration and peaks as expected and the LOD was detected as 530 fM. The detection takes less than 10 min. The selectivity of HPV 16 DNA modified with Ag and Mag-Bead labeled HPV 16&18 primers, respectively, is illustrated in
Figure 4. While HPV16 gave specific peaks with its own labeled primers; in the presence of HPV 18 primers no peak was seen. In addition, no peak occurred in the presence of non-specific primers. Using the primers specific to the target patient DNA and labeling them provide a desired selectivity in the detection of different types of HPV in patient samples.
4. Conclusions
In the present study, a new technique involving a paper based single step extraction platform and electroanalytical device for POC application of HPV 16&18 was proposed to diagnose cervical cancer early. Sensitive, robust, user friendly and cost efficient, the analytical device provides non-enzymatic and analytical amplification and detection technologies. Innovative paper based single step extraction, easy nanoparticle labelled primer hybridization and sending the sample onto device are the main reasons making our device simple. The proposed device saves significant time (15–20 min), making it attractive for HPV POC detections.
Funding
This research was supported and funded by a grant from The Scientific and Technological Research Council of Turkey (TUBITAK) [2214/A] and The Center for Future Technologies in Cancer Care, Boston, MA, as well as Scientific Research Project Council of Izmir Katip Celebi University [2014-1-PhD-48].
Acknowledgments
We would like to thank Prof. Catherine Klapperich and her Lab group at Boston University; Richard M. Crooks’ group, particularly Josephine Cunningham, for their collaboration and Jacqueline C. Linnes for her support and helpful discussions.
References
- Ferlay, J.; Soerjomataram, I.; Ervik, M.; Eser, S.; Mathers, C.; Rebelo, M. GLOBOCAN 2012 v1. 0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. 2013; International Agency for Research on Cancer: Lyon, France, 2014. [Google Scholar]
- Leto, M.D.G.P.; Júnior, S.; Porro, A.M.; Tomimori, J. Human papillomavirus infection: Etiopathogenesis, molecular biology and clinical manifestations. Anais Bras. Dermatol. 2011, 86, 306–317. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Shin, H.R.; Bray, F.; Forman, D.; Mathers, C.; Parkin, D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer 2010, 127, 2893–2917. [Google Scholar] [CrossRef] [PubMed]
- Sia, S.K.; Kricka, L.J. Microfluidics and point-of-care testing. Lab Chip 2008, 8, 1982–1983. [Google Scholar] [CrossRef] [PubMed]
- Chomczynski, P.; Sacchi, N. The single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction: Twenty-something years on. Nat. Protoc. 2006, 1, 581. [Google Scholar] [CrossRef] [PubMed]
- Sönnichsen, C.; Reinhard, B.M.; Liphardt, J.; Alivisatos, A.P. A molecular ruler based on plasmon coupling of single gold and silver nanoparticles. Nat. Biotechnol. 2005, 23, 741. [Google Scholar] [CrossRef] [PubMed]
- Scida, K.; Cunningham, J.C.; Renault, C.; Richards, I.; Crooks, R.M. Simple, sensitive, and quantitative electrochemical detection method for paper analytical devices. Anal. Chem. 2014, 86, 6501–6507. [Google Scholar] [CrossRef] [PubMed]
| Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).