Abstract
Identification of the causative pathogen in infectious diseases is important for surveillance and to guide treatment. In low- and middle-income countries (LMIC), conventional culture and identification methods, including biochemical methods, are reference-standard. Biochemical methods can lack sensitivity and specificity and have slow turnaround times, causing delays in definitive therapy. Matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI–TOF MS) is a rapid and accurate diagnostic method. Most studies comparing MALDI–TOF MS and biochemical methods are from high-income countries, with few reports from LMIC with tropical climates. The aim of this study was to assess the performance of MALDI–TOF MS compared to conventional methods in the Philippines. Clinical bacterial or fungal isolates were identified by both MALDI–TOF MS and automated (VITEK2) or manual biochemical methods in the San Lazaro Hospital, Metro Manila, the Philippines. The concordance between MALDI–TOF MS and automated (VITEK2) or manual biochemical methods was analyzed at the species and genus levels. In total, 3530 bacterial or fungal isolates were analyzed. The concordance rate between MALDI–TOF MS and biochemical methods was 96.2% at the species level and 99.9% at the genus level. Twenty-three isolates could not be identified by MALDI–TOF MS. In this setting, MALDI–TOF MS was accurate compared with biochemical methods, at both the genus and the species level. Additionally, MALDI–TOF MS improved the turnaround time for results. These advantages could lead to improved infection management and infection control in low- and middle-income countries, even though the initial cost is high.
1. Introduction
When giving treatment for bacterial infection with antibiotics, accurate identification of the causative pathogen is essential to guide their appropriate use. There are several ways to identify causative bacteria and fungi, including biochemical methods, antigen and gene detection techniques [1]. Biochemical methods, by manual tests and/or using automated equipment such as VITEK2, have been the reference-standard for the identification of bacteria in resource-limited settings. The VITEK2 system can identify bacteria automatically by reading fluorescence, turbidity and colorimetric signals. Biochemical methods usually take at least 24–48 h, including conventional culture, to identify those bacteria or fungi and can lead to delayed treatment. Matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI–TOF MS), developed by Koichi Tanaka in 1988 [2], has become the reference-standard in many high-income laboratories [3]. MALDI–TOF MS has a high level of accuracy and provides a rapid identification (10–15 min) of microbes compared with biochemical methods [4,5,6]. MALDI–TOF MS can differentiate with high accuracy species that are difficult to be identified by biochemical methods such as Haemophilus, Aggregatibacter, Cardiobacterium, Eikenella and Kingella (HACEK) groups, coagulase-negative Staphylococci or nutritionally variant Streptococci [7,8,9]. MALDI–TOF MS has also been shown to be cost-effective by reducing the length of hospital admission and costs [10,11,12]. Moreover, MALDI–TOF MS has been shown to be able to predict antimicrobial resistance in bacteria [13].
Most studies comparing MALDI–OF MS and biochemical methods are from high-income countries [4,5,6], with few reports from low–middle-income countries or countries (LMIC) with tropical climates [4,14,15]. In 2015, a MALDI–TOF MS was installed in the San Lazaro Hospital (SLH)-Nagasaki Collaborative Research Laboratory and analyzed over 13,000 bacterial and fungal isolates in 5 years. The aim of this study was to assess the performance of MALDI–TOF MS compared to conventional methods in the Philippines.
2. Materials and Methods
This is a retrospective study using secondary data, which were collected from microbiological specimens in the San Lazaro Hospital (SLH), Metro Manila, the Philippines between 1 January 2018 and 15 January 2020. All data were de-identified to respect patient confidentiality and assigned a new code by the laboratory staff in SLH prior to being provided to the investigators.
2.1. Identification of Bacteria by Conventional Biochemical Methods
Bacteria or fungi cultured from clinical samples were sub-cultured for purity where necessary and examined by Gram staining and colonial morphology. Further identification was conducted using the VITEK2 compact system (version 8.01 bioMe’rieux, Marcy l’Etoile, France). In cases where the isolates could not be identified, biochemical tests that help differentiate bacteria through the characterization of their abilities in enzyme production, carbohydrate, protein, and lipid metabolism and compound utilization were performed according to standardized microbiology protocols [16].
2.2. Identification of Bacteria by MALDI–TOF MS
All the isolates were identified by the MALDI Biotyper 3.1 MSP database 5627 (Bruker Daltonik GmbH, Bremen, Germany). The detected spectrum was compared with reference data and evaluated by calculating a score by a unique algorithm. If the score was 2.0 or more, it was considered highly reliable at the species level, if the score was 1.7 or more and less than 2.0, it was highly reliable at the genus level, if it was less than 1.7, it was considered less reliable, and if it could not be identified, the result was ‘No identification’ returned.
3. Results
In total, 3530 sample isolates were tested with conventional biochemical methods. Of these, 1809 samples were tested by VITEK2, and 1721 were tested by manual methods. The concordance was calculated at the species and genus levels. Figure 1 shows the result of concordance between MALDI–TOF MS and VITEK2 or manual tests. Table 1 shows the concordance between MALDI–TOF MS and biochemical methods. The concordance was 95.8% (species level) and 99.8% (genus level) compared with VITEK2, and 96.6% (species level) and 99.9% (genus level) compared with manual biochemical testing. The total concordance was 96.2% (species) and 99.9% (genus).
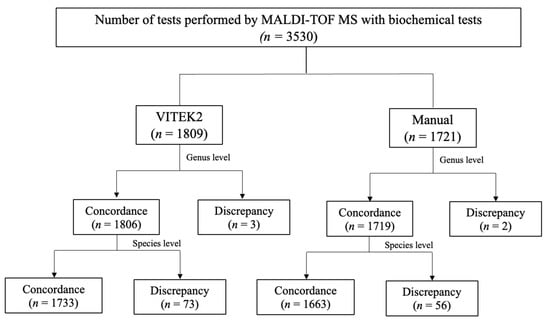
Figure 1.
Flow chart of concordance between MALDI–TOF MS and VITEK2 or manual biochemical testing.

Table 1.
Performance of the MALDI Biotyper in comparison to conventional methods.
The concordance of Gram-positive cocci was 100% (genus) and 97.8% (species). The concordance of Gram-negative rods was 99.8% (genus) and 95.1% (species). Among Gram-positive rods, only Corynebacterium diphtheriae was identified by biochemical methods. For Corynebacterium diphtheriae, MALDI–TOF MS had 100% concordance with the biochemical methods. Concordance of Gram-negative cocci was 100% for both genus and species. The concordance of fungi was 100% (genus) and 94.7% (species).
There were 23 isolates that could not be identified by MALDI–TOF MS. Of these, six were regarded as contaminants based on colony morphology and were not tested by biochemical methods. The remaining 17 were identified by biochemical methods: 1 was Streptococcus pneumoniae, three were Pseudomonas (1 aeruginosa, 1 alcaligenes and 1 putida), 1 was Klebsiella pneumoniae, 4 were Cryptococcus (3 neoformans and 1 laurentii), 7 were Candida (3 albicans, 1 krusei, 1 lipolytica, 1 parapsilosis and 1 tropicalis) and 1 was an unidentified fungus.
4. Discussion
This study is the first to compare the performance of MALDI–TOF MS with that of standard biochemical methods in the Philippines. MALDI–TOF MS had high concordance with biochemical methods in the identification of microorganisms at both the species and the genus levels (96.2%, 99.9%). Several studies have shown that MALDI–TOF MS is an accurate and rapid diagnostic test for not only bacteria but also fungi and acid-fast bacilli in high-income countries [17,18,19]. The finding that MALDI–TOF MS is almost equivalent to biochemical methods strongly supports its role in identifying pathogenic microorganisms in this setting. Additional methods are still sometimes needed, including biochemical methods, because MALDI–TOF MS is not good at differentiating and sometimes misidentifies closely related species [20,21,22,23]. For example, Burkholderia pseudomallei, mallei, and thailandensis cannot be differentiated [21], and the same is true for Streptococcus pneumoniae and mitis [16], and Neisseria meningitidis [23]. It may be necessary to modify the sample preparation protocol in such instances [24].
Another limitation of this method is that the accuracy of the results is highly dependent on the spectrum of the database. If an organism is not included in the database, then MALDI–TOF MS cannot identify it or sometimes misidentifies it. It is necessary that the database is up to date. Unfortunately, MALDI–TOF MS databases are proprietary, and regular database updating may not be sustainable for many laboratories, especially in LMICs. One potential solution to this is the creation of a publicly available online platform with a universal database of reference mass spectra [25]. Refining criteria, such as lowering cutoff values, for distinguishing closely related species may also be a workaround to this problem. Supplemental nucleic acid sequencing of the 16S rRNA gene may also help resolve unidentifiable or undifferentiable isolates [26].
In our study, 23 bacteria were not detected by MALDI–TOF MS. These results would be caused by different reasons, for example, the culture not being fresh, the colony being too small, an inadequate sample inoculated into the target plate, or the sample in the target plate being contaminated with culture media/agar.
The other disadvantage of MALDI–TOF MS is its relatively high initial cost. It may be difficult for institutions in LMIC to install this equipment, even in referral hospitals. MALDI–TOF could potentially reduce healthcare-associated costs and reduce the turnaround time for culture results, thereby allowing clinicians to initiate early targeted therapy. A previous study reported that MALDI–TOF MS is cost-effective for the identification of bacteria in an LMIC setting [14].
Even though MALDI–TOF MS displayed high accuracy for the identification of bacteria and fungi, it does not provide an answer in all circumstances. It is important to retain skills in traditional microbiological methods, and, for some microorganisms, molecular methods, such as nucleic acid sequencing, may be the best route to their identification. Furthermore, at present, MALDI–TOF MS does not provide antimicrobial sensitivity test results, and other methods to determine this property will continue to be required.
5. Conclusions
MALDI–TOF MS appears to be an accurate and rapid diagnostic method compared with biochemical methods at not only genus level but also species level. Additionally, with a result available in 10–15 min, MALDI–TOF MS can improve the turnaround time of results. Those advantages could lead to improved infection management and infection control in low- and middle-income countries.
Author Contributions
Conceptualization, M.O., C.S.; methodology, J.M., A.C., M.C., A.M.G.V., C.M.P., N.S.; formal analysis, M.O., C.S.; investigation, M.O., N.S., C.M.P., C.S.; resources, M.C.B., Z.D.M.; data curation, M.C.B.; writing—original draft preparation, M.O.; writing—review and editing, M.O., C.S., C.M.P., A.M.G.V., S.S., S.T., B.G.D.; supervision, C.S., K.A., E.F.O.T., D.V.U.; project administration, S.S., T.U.; funding acquisition, C.S., K.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
All data were organized by laboratory staff in SLH, were de-identified in SLH to remove information such as hospital ID, name, address by laboratory staff prior to being provided to the investigators and were assigned a code to respect patient confidentiality. Ethical approval was obtained from the ethical committee of both School of Tropical Medicine and Global Health, Nagasaki University (Ref. No. 103), and San Lazaro Hospital (protocol number: SLH-RERU-2020-009-E).
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef]
- Tanaka, K.; Waki, H.; Ido, Y.; Akita, S.; Yoshida, Y.; Yoshida, T.; Matsuo, T. Protein and polymer analyses up to m/z 100 000 by laser ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 1988, 2, 151–153. [Google Scholar] [CrossRef]
- Schubert, S.; Kostrzewa, M. MALDI-TOF MS in the Microbiology Laboratory: Current Trends. Curr. Issues Mol. Biol. 2017, 23, 17–20. [Google Scholar] [CrossRef]
- Kassim, A.; Pflüger, V.; Premji, Z.; Daubenberger, C.; Revathi, G. Comparison of biomarker based Matrix Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS) and conventional methods in the identification of clinically relevant bacteria and yeast. BMC Microbiol. 2017, 17, 1–8. [Google Scholar] [CrossRef]
- Zhou, C.; Hu, B.; Zhang, X.; Huang, S.; Shan, Y.; Ye, X. The value of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in identifying clinically relevant bacteria: A comparison with automated microbiology system. J. Thorac. Dis. 2014, 6, 545–552. [Google Scholar] [CrossRef]
- van Veen, S.Q.; Claas, E.C.J.; Kuijper, E.J. High-Throughput Identification of Bacteria and Yeast by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry in Conventional Medical Microbiology Laboratories. J. Clin. Microbiol. 2010, 48, 900–907. [Google Scholar] [CrossRef]
- Powell, E.A.; Blecker-Shelly, D.; Montgomery, S.; Mortensen, J.E. Application of Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry for Identification of the Fastidious Pediatric Pathogens Aggregatibacter, Eikenella, Haemophilus, and Kingella. J. Clin. Microbiol. 2013, 51, 3862–3864. [Google Scholar] [CrossRef][Green Version]
- Martins, K.B.; Ferreira, A.M.; Mondelli, A.L.; Rocchetti, T.T.; Lr de S da Cunha, M.D. Evaluation of MALDI-TOF VITEK ® MS and VITEK® 2 system for the identification of Staphylococcus saprophyticus. Future Microbiol. 2018, 13, 1603–1609. [Google Scholar] [CrossRef]
- Ratcliffe, P.; Fang, H.; Thidholm, E.; Boräng, S.; Westling, K.; Özenci, V. Comparison of MALDI-TOF MS and VITEK 2 system for laboratory diagnosis of Granulicatella and Abiotrophia species causing invasive infections. Diagn. Microbiol. Infect. Dis. 2013, 77, 216–219. [Google Scholar] [CrossRef] [PubMed]
- Cavalieri, S.J.; Kwon, S.; Vivekanandan, R.; Ased, S.; Carroll, C.; Anthone, J.; Schmidt, D.; Baysden, M.; Destache, C.J. Effect of antimicrobial stewardship with rapid MALDI-TOF identification and Vitek 2 antimicrobial susceptibility testing on hospitalization outcome. Diagn. Microbiol. Infect. Dis. 2019, 95, 208–211. [Google Scholar] [CrossRef] [PubMed]
- Perez, K.K.; Olsen, R.J.; Musick, W.L.; Cernoch, P.L.; Davis, J.R.; Land, G.A.; Peterson, L.E.; Musser, J.M. Integrating Rapid Pathogen Identification and Antimicrobial Stewardship Significantly Decreases Hospital Costs. Arch. Pathol. Lab. Med. 2012, 137, 1247–1254. [Google Scholar] [CrossRef]
- Bhavsar, S.M.; Dingle, T.; Hamula, C.L. The impact of blood culture identification by MALDI-TOF MS on the antimicrobial management of pediatric patients. Diagn. Microbiol. Infect. Dis. 2018, 92, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Flores-Treviño, S.; Garza-González, E.; Mendoza-Olazarán, S.; Morfín-Otero, R.; Camacho-Ortiz, A.; Rodríguez-Noriega, E.; Martinez-Melendez, A.; Bocanegra-Ibarias, P. Screening of biomarkers of drug resistance or virulence in ESCAPE pathogens by MALDI-TOF mass spectrometry. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Sow, D.; Fall, B.; Ndiaye, M.; Ba, B.S.; Sylla, K.; Tine, R.; Lô, A.C.; Abiola, A.; Wade, B.; Dieng, T.; et al. Usefulness of MALDI-TOF Mass Spectrometry for Routine Identification of Candida Species in a Resource-Poor Setting. Mycopathologia 2015, 180, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Wattal, C.; Oberoi, J.K.; Goel, N.; Raveendran, R.; Khanna, S. Matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS) for rapid identification of micro-organisms in the routine clinical microbiology laboratory. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 36, 807–812. [Google Scholar] [CrossRef]
- Vandepitte, J.; Verhaegen, J.; Engbaek, K.; Rohner, P.; Piot, P.; Heuck, C.C. Basic Laboratory Procedures in Clinical Bacteriology, 2nd ed.; World Health Organization: Geneva, Switzerland, 2003. [Google Scholar]
- Risch, M.; Radjenovic, D.; Han, J.N.; Wydler, M.; Nydegger, U.; Risch, L. Comparison of MALDI TOF with conventional identification of clinically relevant bacteria. Swiss Med Wkly. 2010, 140. [Google Scholar] [CrossRef]
- TeKippe, E.M.; Burnham, C.-A.D. Evaluation of the Bruker Biotyper and VITEK MS MALDI-TOF MS systems for the identification of unusual and/or difficult-to-identify microorganisms isolated from clinical specimens. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 2163–2171. [Google Scholar] [CrossRef]
- Luo, L.; Cao, W.; Chen, W.; Zhang, R.; Jing, L.; Chen, H.; Yu, F.; Yue, J. Evaluation of the VITEK MS knowledge base version 3.0 for the identification of clinically relevant Mycobacterium species. Emerg Microbes Infect. 2018, 7, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Pauker, V.I.; Thoma, B.R.; Grass, G.; Bleichert, P.; Hanczaruk, M.; Zöller, L.; Zange, S. Improved Discrimination of Bacillus anthracis from Closely Related Species in the Bacillus cereus Sensu Lato Group Based on Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry. J. Clin. Microbiol. 2018, 56, e01900-17. [Google Scholar] [CrossRef]
- Cunningham, S.A.; Patel, R. Importance of Using Bruker’s Security-Relevant Library for Biotyper Identification of Burkholderia pseudomallei, Brucella Species, and Francisella tularensis. J. Clin. Microbiol. 2013, 51, 1639–1640. [Google Scholar] [CrossRef]
- Khot, P.D.; Couturier, M.R.; Wilson, A.R.; Croft, A.; Fisher, M.A. Optimization of Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry Analysis for Bacterial Identification. J. Clin. Microbiol. 2012, 50, 3845–3852. [Google Scholar] [CrossRef] [PubMed]
- Hong, E.; Bakhalek, Y.; Taha, M.-K. Identification of Neisseria meningitidis by MALDI-TOF MS may not be reliable. Clin. Microbiol. Infect. 2019, 25, 717–722. [Google Scholar] [CrossRef] [PubMed]
- Panda, A.; Ghosh, A.K.; Mirdha, B.R.; Xess, I.; Paul, S.; Samantaray, J.C.; Srinivasan, A.; Khalil, S.; Rastogi, N.; Dabas, Y. MALDI-TOF mass spectrometry for rapid identification of clinical fungal isolates based on ribosomal protein biomarkers. J. Microbiol. Methods 2015, 109, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Starostin, K.V.; Demidov, E.A.; Ershov, N.I.; Bryanskaya, A.V.; Efimov, V.M.; Shlyakhtun, V.N.; Peltek, S.E. Creation of an Online Platform for Identification of Microogranisms: Peak-Picking or Full-Spectrum Analysis. Front. Microbiol. 2020, 11, 609033. [Google Scholar] [CrossRef]
- Patel, R. MALDI-TOF MS for the Diagnosis of Infectious Diseases. Clin. Chem. 2015, 61, 100–111. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).