Polymerase Chain Reaction (PCR) as a Potential Point of Care Laboratory Test for Leprosy Diagnosis—A Systematic Review
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
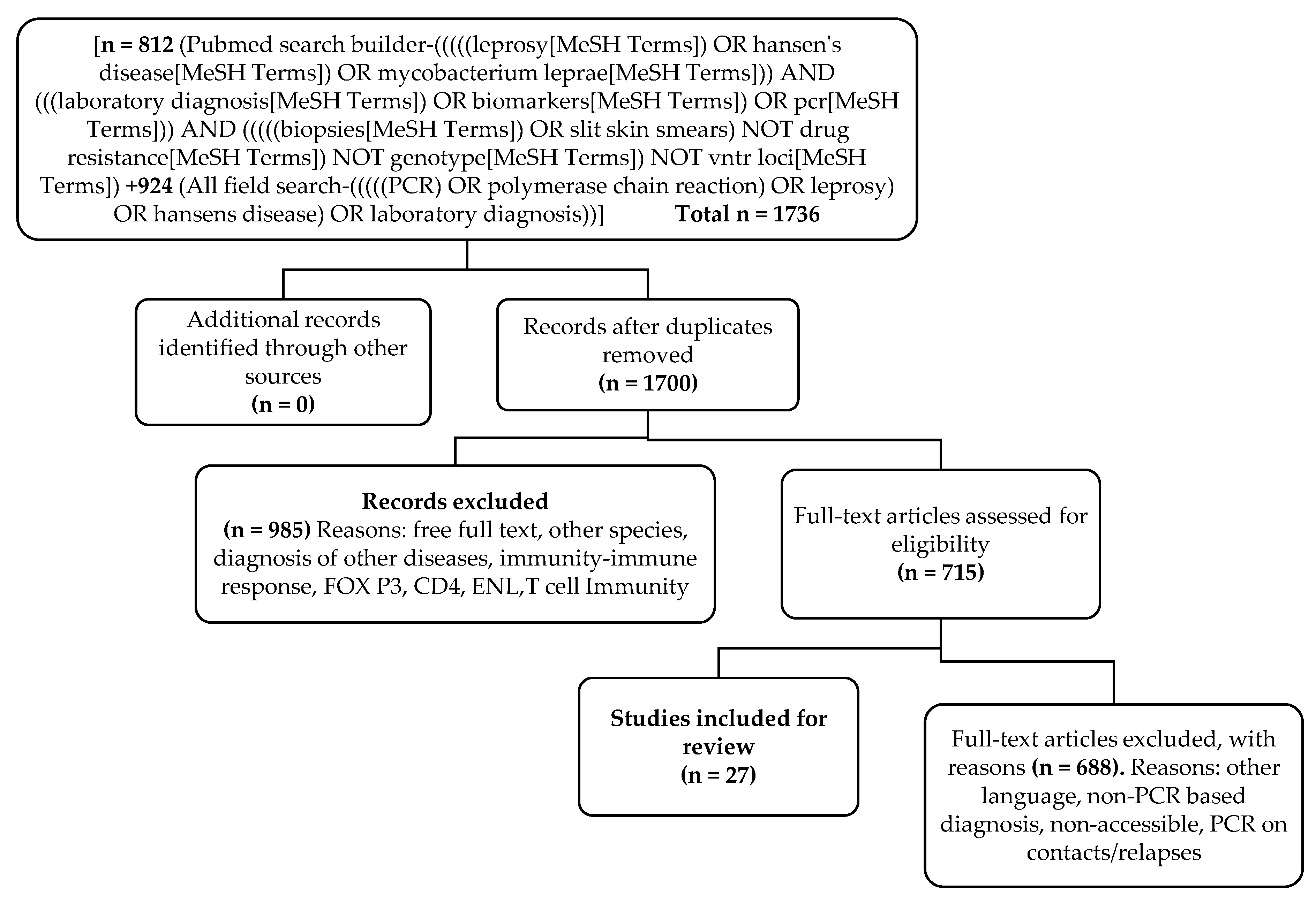
2.2. Data Search
2.3. Data Extraction
2.4. Definitions of Some of the Data Terms Used in the Review
2.5. Data Analysis
3. Results
3.1. Basic Clinical and Geographical Information
3.2. Analysis
3.3. Clinical Classification vs. Sensitivity of AFB and PCR
3.4. Clinical Specimens and PCR
3.5. Gene Markers—PCR Sensitivity
3.6. Method of PCR vs. PCR Sensitivity
4. Conclusions
Limitations of This Systematic Review Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Noordeen, S.K. The epidemiology of leprosy. In Leprosy; Hastings, R.C., Ed.; Churchill Livingstone, Produced by Longman Group Ltd.: Hong Kong, China, 1985; pp. 15–30. [Google Scholar]
- Dockrell, H.M. Leprosy, 3rd ed.; Bryceson, A.D.M., Pfaltzgraff, R.E., Eds.; Churchill Livingstone: Edinburgh, UK, 1990; p. 240. ISBN 0-443-03373-0. [Google Scholar]
- Lockwood, D.N.J. Leprosy. Medicine 2005, 33, 26–29. [Google Scholar] [CrossRef]
- Van Brakel, W.H.; Sihombing, B.; Djarir, H.; Beise, K.; Kusumawardhani, L.; Yulihane, R.; Kurniasari, I.; Kasim, M.; Kesumaningsih, K.I.; Wilder-Smith, A. Disability in people affected by leprosy: The role of impairment, activity, social participation, stigma and discrimination. Glob. Health Action. 2012, 5. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Target Attained, Remaining Endemic Countries Pose Greatest Challenge; Press Release, WHA/2; WHO: Geneva, Switzerland, 2002. [Google Scholar]
- World Health Organization. The Global Leprosy Strategy 2016–2020: Accelerating towards a Leprosy-Free World; WHO: Geneva, Switzerland, 2016. [Google Scholar]
- World Health Organization. Global leprosy update, 2016: Accelerating reduction of disease burden. Wkly. Epidemiol. Rec. 2017, 92, 501–520. [Google Scholar]
- Sermrittirong, S.; van Brakel, W.H. Stigma in leprosy: Concepts, causes and determinants. Lepr. Rev. 2014, 85, 36–47. [Google Scholar] [PubMed]
- Duthie, M.S.; Truman, R.W.; Goto, W.; O’Donnell, J.; Hay, M.N.; Spencer, J.S.; Carter, D.; Reed, S.G. Insight toward early diagnosis of leprosy through analysis of the developing antibody responses of Mycobacterium leprae-infected armadillos. Clin. Vaccine Immunol. 2011, 18, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Veena, S.; Kumar, P.; Shashikala, P.; Gurubasavaraj, H.; Chandrasekhar, H.R. Significance of histopathology in leprosy patients with 1–5 skin lesions with relevance to therapy. J. Lab. Physicians 2011, 3, 21–24. [Google Scholar] [CrossRef] [PubMed]
- Lockwood, D.N.; Nicholls, P.; Smith, W.C.; Das, L.; Barkataki, P.; van Brakel, W.; Suneetha, S. Comparing the clinical and histological diagnosis of leprosy and leprosy reactions in the INFIR cohort of Indian patients with multibacillary leprosy. PLoS Negl. Trop. Dis. 2012, 6, e1702. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M. Leprosy—An overview of clinical features, diagnosis, and treatment. J. Dtsch. Dermatol. Ges. 2017, 15, 801–827. [Google Scholar] [CrossRef] [PubMed]
- Lastória, J.C.; Abreu, M.A. Leprosy: A review of laboratory and therapeutic aspects—Part 2. An. Bras. Dermatol. 2014, 89, 389–401. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, M.C.; Ramuno, N.M.; Fachin, L.R.; Tassa, M.; Rosa, P.S.; Belone, A.F.; Diorio, S.M.; Soares, C.T.; Garlet, G.P.; Trombone, A.P. qPCR detection of Mycobacterium leprae in biopsies and slit skin smear of different leprosy clinical forms. Braz. J. Infect. Dis. 2017, 21, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Speers, D.J. Clinical applications of molecular biology for infectious diseases. Clin. Biochem. Rev. 2006, 27, 39–51. [Google Scholar] [PubMed]
- Niemz, A.; Boyle, D.S. Nucleic acid testing for tuberculosis at the point-of-care in high-burden countries. Expert Rev. Mol. Diagn. 2012, 12, 687–701. [Google Scholar] [CrossRef] [PubMed]
- Martinez, A.N.; Talhari, C.; Moraes, M.O.; Talhari, S. PCR-based techniques for leprosy diagnosis: From the laboratory to the clinic. PLoS Negl. Trop. Dis. 2014, 8, e2655. [Google Scholar] [CrossRef] [PubMed]
- Male, M.M.; Rao, B.G.; Chokkakula, S.; Kasetty, S.; Rao, P.V.R.; Jonnalagada, S.; Reddy, A.M.; Srikantam, A. Molecular screening for primary drug resistance in M. leprae from newly diagnosed leprosy cases from India. Lepr. Rev. 2016, 87, 322–331. [Google Scholar]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; Group, P. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. Ann. Intern. Med. 2009, 151, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Canese, K.; Weis, S. PubMed: The Bibliographic Database. In The NCBI Handbook [Internet], 2nd ed.; National Center for Biotechnology Information (US): Bethesda, MD, USA, 2013. [Google Scholar]
- Chaitanya, V.S.; Cuello, L.; Das, M.; Sudharsan, A.; Ganesan, P.; Kanmani, K.; Rajan, L.; Ebenezer, M. Analysis of a novel multiplex polymerase chain reaction assay as a sensitive tool for the diagnosis of indeterminate and tuberculoid forms of leprosy. Int. J. Mycobacteriol. 2017, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- De Wit, M.Y.; Douglas, J.T.; McFadden, J.; Klatser, P.R. Polymerase chain reaction for detection of Mycobacterium leprae in nasal swab specimens. J. Clin. Microbiol. 1993, 31, 502–506. [Google Scholar] [PubMed]
- Yoon, K.H.; Cho, S.N.; Lee, M.K.; Abalos, R.M.; Cellona, R.V.; Fajardo, T.T., Jr.; Guido, L.S.; Dela Cruz, E.C.; Walsh, G.P.; Kim, J.D. Evaluation of polymerase chain reaction amplification of mycobacterium leprae-specific repetitive sequence in biopsy specimens from leprosy patients. J. Clin. Microbiol. 1993, 31, 895–899. [Google Scholar] [PubMed]
- Kamal, R.; Natrajan, M.; Katoch, K.; Katoch, V.M. Evaluation of diagnostic role of in situ PCR on slit-skin smears in pediatric leprosy. Indian J. Lepr. 2010, 82, 195–200. [Google Scholar] [PubMed]
- Kamal, R.; Dayal, R.; Gaidhankar, K.; Biswas, S.; Gupta, S.B.; Kumar, N.; Kumar, R.; Pengoria, R.; Chauhan, DS.; Katoch, K.; et al. RLEP PCR as a definitive diagnostic test for leprosy from skin smear samples in childhood and adolescent leprosy. Indian J. Lepr. 2016, 88, 193–197. [Google Scholar]
- Banerjee, S.; Sarkar, K.; Gupta, S.; Mahapatra, P.S.; Gupta, S.; Guha, S.; Bandhopadhayay, D.; Ghosal, C.; Paine, S.K.; Dutta, R.N.; et al. Multiplex PCR technique could be an alternative approach for early detection of leprosy among close contacts—A pilot study from India. BMC Infect. Dis. 2010, 10, 252. [Google Scholar] [CrossRef] [PubMed]
- Siwakoti, S.; Rai, K.; Bhattarai, N.R.; Agarwal, S.; Khanal, B. Evaluation of polymerase chain reaction (PCR) with slit skin smear examination (SSS) to confirm clinical diagnosis of leprosy in eastern Nepal. PLoS Negl. Trop. Dis. 2016, 10, e0005220. [Google Scholar] [CrossRef] [PubMed]
- Maltempe, F.G.; Baldin, V.P.; Lopes, M.A.; Siqueira, V.L.D.; de Lima Scodro, R.B.; Cardoso, R.F.; Caleffi-Ferracioli, K.R. Critical analysis: Use of polymerase chain reaction to diagnose leprosy. Braz. J. Pharm. Sci. 2016, 52. [Google Scholar] [CrossRef]
- Wichitwechkarn, J.; Karnjan, S.; Shuntawuttisettee, S.; Sornprasit, C.; Kampirapap, K.; Peerapakorn, S. Detection of Mycobacterium leprae infection by PCR. J. Clin. Microbiol. 1995, 33, 45–49. [Google Scholar] [PubMed]
- Kurabachew, M.; Wondimu, A.; Ryon, J.J. Reverse transcription-PCR detection of Mycobacterium leprae in clinical specimens. J. Clin. Microbiol. 1998, 36, 1352–1356. [Google Scholar] [PubMed]
- Phetsuksiri, B.; Rudeeaneksin, J.; Supapkul, P.; Wachapong, S.; Mahotarn, K.; Brennan, P.J. A simplified reverse transcriptase PCR for rapid detection of Mycobacterium leprae in skin specimens. FEMS Immunol. Med. Microbiol. 2006, 48, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Goulart, I.M.; Cardoso, A.M.; Santos, M.S.; Goncalves, M.A.; Pereira, J.E.; Goulart, L.R. Detection of Mycobacterium leprae DNA in skin lesions of leprosy patients by PCR may be affected by amplicon size. Arch. Dermatol. Res. 2007, 299, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Bang, P.D.; Suzuki, K.; Phuong le, T.; Chu, T.M.; Ishii, N.; Khang, T.H. Evaluation of polymerase chain reaction-based detection of Mycobacterium leprae for the diagnosis of leprosy. J. Dermatol. 2009, 36, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Martinez, A.N.; Ribeiro-Alves, M.; Sarno, E.N.; Moraes, M.O. Evaluation of qPCR-based assays for leprosy diagnosis directly in clinical specimens. PLoS Negl. Trop. Dis. 2011, 5, e1354. [Google Scholar] [CrossRef] [PubMed]
- Reja, A.H.; Biswas, N.; Biswas, S.; Dasgupta, S.; Chowdhury, I.H.; Banerjee, S.; Chakraborty, T.; Dutta, P.K.; Bhattacharya, B. Fite-Faraco staining in combination with multiplex polymerase chain reaction: A new approach to leprosy diagnosis. Indian J. Dermatol. Venereol. Leprol. 2013, 79, 693–700. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Xing, Y.; Yuan, L.C.; De Yang, R.; Tan, F.Y.; Zhang, Y.; Li, H.Y. Application of RLEP real-time PCR for detection of M. leprae DNA in paraffin-embedded skin biopsy specimens for diagnosis of paucibacillary leprosy. Am. J. Trop. Med. Hyg. 2014, 90, 524–529. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.S.; Shamsuzzaman, S.M.; Mamun, K.Z. Demography, clinical presentation and laboratory diagnosis of leprosy by microscopy, histopathology and PCR from Dhaka city in Bangladesh. Lepr. Rev. 2017, 88, 122–130. [Google Scholar]
- Caleffi, K.R.; Hirata, R.D.; Hirata, M.H.; Caleffi, E.R.; Siqueira, V.L.; Cardoso, R.F. Use of the polymerase chain reaction to detect Mycobacterium leprae in urine. Braz. J. Med. Biol. Res. 2012, 45, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, V.; Malhotra, K.; Khan, K.; Maurya, P.K.; Singh, A.K.; Thacker, A.K.; Husain, N.; Kulshreshtha, D. Evaluation of polymerase chain reaction in nerve biopsy specimens of patients with Hansen’s disease. J. Neurol. Sci. 2017, 380, 187–190. [Google Scholar] [CrossRef] [PubMed]
- Gama, R.S.; Gomides, T.A.R.; Gama, C.F.M.; Moreira, S.J.M.; de Neves Manta, F.S.; de Oliveira, L.B.P.; Marcal, P.H.F.; Sarno, E.N.; Moraes, M.O.; Garcia, R.M.G.; et al. High frequency of M. leprae DNA detection in asymptomatic household contacts. BMC Infect. Dis. 2018, 18, 153. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Biswas, N.; Kanti Das, N.; Sil, A.; Ghosh, P.; Hasanoor Raja, A.H.; Dasgupta, S.; Kanti Datta, P.; Bhattacharya, B. Diagnosing leprosy: Revisiting the role of the slit-skin smear with critical analysis of the applicability of polymerase chain reaction in diagnosis. Int. J. Dermatol. 2011, 50, 1522–1527. [Google Scholar] [CrossRef] [PubMed]
- Rudeeaneksin, J.; Srisungngam, S.; Sawanpanyalert, P.; Sittiwakin, T.; Likanonsakul, S.; Pasadorn, S.; Palittapongarnpim, P.; Brennan, P.J.; Phetsuksiri, B. LightCycler real-time PCR for rapid detection and quantitation of Mycobacterium leprae in skin specimens. FEMS Immunol. Med. Microbiol. 2008, 54, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Dayal, R.; Singh, S.P.; Mathur, P.P.; Katoch, V.M.; Katoch, K.; Natrajan, M. Diagnostic value of in situ polymerase chain reaction in leprosy. Indian J. Pediatr. 2005, 72, 1043–1046. [Google Scholar] [CrossRef] [PubMed]
- Kampirapap, K.; Singtham, N.; Klatser, P.R.; Wiriyawipart, S. DNA amplification for detection of leprosy and assessment of efficacy of leprosy chemotherapy. Int. J. Lepr. Other Mycobact. Dis. 1998, 66, 16–21. [Google Scholar] [PubMed]
- Sundeep Chaitanya, V.; Das, M.; Eisenbach, T.L.; Amoako, A.; Rajan, L.; Horo, I.; Ebenezer, M. Mycobacterium leprae specific genomic target in the promoter region of probable 4-alpha-glucanotransferase (ML1545) gene with potential sensitivity for polymerase chain reaction based diagnosis of leprosy. Int. J. Mycobacteriol. 2016, 5, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Patrocínio, L.G.; Goulart, I.M.; Goulart, L.R.; Patrocinio, J.A.; Ferreira, F.R.; Fleury, R.N. Detection of Mycobacterium leprae in nasal mucosa biopsies by the polymerase chain reaction. FEMS Immunol. Med. Microbiol. 2005, 44, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Boehme, C.C.; Nabeta, P.; Henostroza, G.; Raqib, R.; Rahim, Z.; Gerhardt, M.; Sanga, E.; Hoelscher, M.; Notomi, T.; Hase, T.; et al. Operational feasibility of using loop-mediated isothermal amplification for diagnosis of pulmonary tuberculosis in microscopy centers of developing countries. J. Clin. Microbiol. 2007, 45, 1936–1940. [Google Scholar] [CrossRef] [PubMed]
- Boehme, C.C.; Nabeta, P.; Hillemann, D.; Nicol, M.P.; Shenai, S.; Krapp, F.; Allen, J.; Tahirli, R.; Blakemore, R.; Rustomjee, R.; et al. Rapid molecular detection of tuberculosis and rifampin resistance. N. Engl. J. Med. 2010, 363, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Mori, Y.; Notomi, T. Loop-mediated isothermal amplification (LAMP): A rapid, accurate, and cost-effective diagnostic method for infectious diseases. J. Infect. Chemother. 2009, 15, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Lawn, S.D.; Nicol, M.P. Xpert® MTB/RIF assay: Development, evaluation and implementation of a new rapid molecular diagnostic for tuberculosis and rifampicin resistance. Future Microbiol. 2011, 6, 1067–1082. [Google Scholar] [CrossRef] [PubMed]
- Arora, D.R.; Maheshwari, M.; Arora, B. Rapid point-of-care testing for detection of HIV and clinical monitoring. ISRN AIDS 2013, 2013, 287269. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Nhem, S.; Dourng, D.; Ménard, D. Malaria rapid diagnostic test as point-of-care test: Study protocol for evaluating the VIKIA® Malaria Ag Pf/Pan. Malar. J. 2015, 14, 114. [Google Scholar] [CrossRef] [PubMed]
| Sl. No. | Type of Sample | PCR Type | Marker/Gene | No. of Patients Studied | Smear Microscopy | PCR | p Value | First Author | Study Location/Country | Reference No. | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. Tested | No. Positive | % | No. Tested | No. Positive | % | |||||||||
| 1 | Skin biopsy | Conventional PCR | RLEP | 102 | 102 | 63 | 61.76 | 102 | 59 | 57.84 | NA | Michelle de Campos Soriani Azevedo | Brazil | [13] |
| 2 | Skin biopsy | Multiplex PCR | RLEP | 220 | 220 | 122 | 55.45 | 220 | 164 | 74.55 | p < 0.05 | V Sundeep Chaitanya | India | [21] |
| 3 | Skin biopsy | Multiplex PCR | M- PCR | 220 | 220 | 122 | 55.45 | 220 | 205 | 93.18 | NA | V Sundeep Chaitanya | India | [21] |
| 4 | Nasal swabs | Conventional PCR | 531 bp fragment | 103 | 0 | 0 | 0 | 103 | 82 | 79.61 | NA | Madeleine Y. L. de Wit | Philippines | [22] |
| 5 | SSS | Conventional PCR | 372 bp fragment | 102 | 102 | 62 | 60.78 | 102 | 95 | 93.14 | NA | Kyeong-Han Yoon | Philippines | [23] |
| 6 | Skin biopsy | Conventional PCR | 372 bp | 102 | 102 | 87 | 85.29 | 102 | 95 | 93.14 | NA | Kyeong-Han Yoon | Philippines | [23] |
| 7 | SSS | in situ PCR | 530 bp fragment | 25 | 25 | 5 | 20 | 25 | 18 | 72 | p = 0.01 | R Kamal | India | [24] |
| 8 | SSS | Conventional PCR | RLEP | 73 | 73 | 17 | 23.29 | 73 | 56 | 76.71 | p < 0.001 | R Kamal | India | [25] |
| 9 | SSS | Multiplex PCR | 372&201 bp | 439 | 439 | 223 | 50.8 | 439 | 371 | 84.51 | NA | Surajita Banerjee | India | [26] |
| 10 | SSS | Conventional PCR | RLEP | 50 | 50 | 9 | 18 | 50 | 36 | 72 | NA | Shraddha Siwakoti | Nepal | [27] |
| 11 | SSS | Conventional PCR | PCR-LP | 91 | 91 | 21 | 23.08 | 91 | 22 | 24.18 | NA | Flaviane Granero Maltempe | Brazil | [28] |
| 12 | SSS | Conventional PCR | PCR-P | 91 | 91 | 21 | 23.08 | 91 | 17 | 18.68 | p > 0.05 | Flaviane Granero Maltempe | Brazil | [28] |
| 13 | SSS | Conventional PCR | pra gene | 53 | 0 | 0 | 0 | 53 | 17 | 32.08 | NA | Jesdawan Wichitwechkaran | Bangkok | [29] |
| 14 | Skin biopsy | Conventional PCR | pra gene | 53 | 0 | 0 | 0 | 53 | 35 | 66.04 | NA | Jesdawan Wichitwechkaran | Bangkok | [29] |
| 15 | Skin biopsy | RT-PCR | 16S rRNA | 50 | 50 | 33 | 66 | 50 | 41 | 82 | NA | Mekonnen Kurabachew | Ethiopia | [30] |
| 16 | Nasal mucosal biopsies | RT-PCR | 16S rRNA | 60 | 60 | 24 | 40 | 60 | 47 | 78.33 | NA | Benjawan Phetsuksiri | Bangkok | [31] |
| 17 | Skin biopsy | Conventional PCR | RLEP | 110 | 110 | 43 | 39.09 | 110 | 81 | 73.64 | NA | Isabela Maria Bernardes Goulart | Brazil | [32] |
| 18 | Skin biopsy | Conventional PCR | 372 bp | 110 | 110 | 43 | 39.09 | 110 | 58 | 52.73 | NA | Isabela Maria Bernardes Goulart | Brazil | [32] |
| 19 | Skin biopsy | qPCR | 16S rRNA | 69 | 69 | 0 | 0 | 69 | 53 | 76.81 | NA | Pham Dang Bang | Vietnam | [33] |
| 20 | Skin biopsy | qPCR | RLEP | 47 | 0 | 0 | 0 | 47 | 38 | 80.85 | NA | Alejandra Nóbrega Martinez | Brazil | [34] |
| 21 | Skin biopsy | qPCR | 16S rRNA | 47 | 0 | 0 | 0 | 47 | 24 | 51.06 | NA | Alejandra Nóbrega Martinez | Brazil | [34] |
| 22 | Skin biopsy | qPCR | sodA | 47 | 0 | 0 | 0 | 47 | 22 | 46.81 | NA | Alejandra Nóbrega Martinez | Brazil | [34] |
| 23 | Skin biopsy | qPCR | 85B | 47 | 0 | 0 | 0 | 47 | 26 | 55.32 | NA | Alejandra Nóbrega Martinez | Brazil | [34] |
| 24 | Skin biopsy | Multiplex PCR | 372bp &201 bp | 165 | 165 | 84 | 50.91 | 165 | 111 | 67.27 | NA | Abu Hena Hasanoor Reja | India | [35] |
| 25 | Skin biopsy | qPCR | RLEP & 372 bp fragment | 51 | 51 | 18 | 35.29 | 51 | 38 | 74.51 | p > 0.05 | Wen Yan | China | [36] |
| 26 | Skin biopsy | Nested PCR | RLEP &372 bp fragment | 51 | 51 | 18 | 35.29 | 51 | 37 | 72.55 | NA | Wen Yan | China | [36] |
| 27 | Skin biopsy | Conventional PCR | 530 bp fragment | 55 | 55 | 9 | 16.36 | 55 | 40 | 72.73 | NA | Mohammad Shah Alam | Bangladesh | [37] |
| 28 | Urine | Conventional PCR | pra gene | 73 | 73 | 0 | 0 | 73 | 34 | 46.58 | p > 0.05 | K.R. Caleffi | Brazil | [38] |
| 29 | Nerve biopsy | Conventional PCR | 375 bp fragment | 35 | 35 | 13 | 37.14 | 35 | 22 | 62.86 | NA | Vandana Tiwari | India | [39] |
| 30 | SSS | qPCR | 16S rRNA | 43 | 43 | 13 | 30.23 | 43 | 18 | 41.86 | NA | Rafael Silva Gama | Brazil | [40] |
| 31 | blood | qPCR | 16S rRNA | 43 | 0 | 0 | 0 | 43 | 6 | 13.95 | NA | Rafael Silva Gama | Brazil | [40] |
| 32 | SSS | Multiplex PCR | 372 bp fragment | 164 | 164 | 65 | 39.63 | 164 | 135 | 82.32 | p < 0.0001 | Surajita Banerjee | India | [41] |
| 33 | SSS | qPCR | 16S rRNA | 66 | 66 | 36 | 54.55 | 66 | 52 | 78.79 | NA | Janisara Rudeeaneksin | Bangkok | [42] |
| 34 | Skin biopsy | in situ PCR | 530 bp fragment | 20 | 20 | 2 | 10 | 20 | 12 | 60 | NA | R. Dayal | India | [43] |
| 35 | SSS | Conventional PCR | pra gene | 122 | 122 | 49 | 40.16 | 122 | 86 | 70.49 | p < 0.001 | Kowit Kampirapap | Bangkok | [44] |
| 36 | Skin biopsy | Conventional PCR | RLEP | 180 | 180 | 122 | 67.78 | 180 | 114 | 63.33 | p < 0.0001 | V Sundeep Chaitanya | India | [45] |
| 37 | Skin biopsy | Conventional PCR | ML1545 | 180 | 180 | 122 | 67.78 | 180 | 164 | 91.11 | NA | V Sundeep Chaitanya | India | [45] |
| 38 | SSS | Conventional PCR | 372 bp fragment | 52 | 52 | 36 | 69.23 | 52 | 36 | 69.23 | NA | Lucas Gomes Patrocínio | Brazil | [46] |
| Classification | No. of Assay (Reports) Studied | Average Number of Patients/Samples Tested (Range) | No. of Assay Reported the AFB Microscopy | %AFB Positivity Mean (Range) | No. of Assay Reported PCR Tests | %PCR Positivity |
|---|---|---|---|---|---|---|
| Bacillary Load | ||||||
| Paucibacillary | 28 | 37.07 (7–234) | 6 | 25.18 (1.75–35.29) | 28 | 48.63 (7.69–81) |
| Multibacillary | 27 | 61.40 (12–205) | 17 | 62.25 (15.38–100) | 27 | 79.65 (17.39–100) |
| Clinical samples | ||||||
| Slit skin samples | 14 | 101 (25–439) | 12 | 37.73 (18–69.23) | 14 | 60.71 (18.68–93.14) |
| Skin biopsy | 20 | 96.3 (20–220) | 14 | 48.96 (10–85.29) | 20 | 70.27 (46.81–93.18) |
| Number of Assays Studied (n = 38) | Highest Positivity (%PCR) | Lowest Positivity (%PCR) | |
|---|---|---|---|
| Gene markers | |||
| RLEP | 9 | 80.85 | 57.84 |
| 16S rRNA | 10 | 82 | 13.95 |
| Method of PCR | |||
| Conventional | 19 | 93.14 | 18.68 |
| Multiplex | 6 | 93.18 | 67.27 |
| Q-PCR | 6 | 80.85 | 13.95 |
| RT-PCR | 5 | 74.5 | 82 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tatipally, S.; Srikantam, A.; Kasetty, S. Polymerase Chain Reaction (PCR) as a Potential Point of Care Laboratory Test for Leprosy Diagnosis—A Systematic Review. Trop. Med. Infect. Dis. 2018, 3, 107. https://doi.org/10.3390/tropicalmed3040107
Tatipally S, Srikantam A, Kasetty S. Polymerase Chain Reaction (PCR) as a Potential Point of Care Laboratory Test for Leprosy Diagnosis—A Systematic Review. Tropical Medicine and Infectious Disease. 2018; 3(4):107. https://doi.org/10.3390/tropicalmed3040107
Chicago/Turabian StyleTatipally, Sushma, Aparna Srikantam, and Sanjay Kasetty. 2018. "Polymerase Chain Reaction (PCR) as a Potential Point of Care Laboratory Test for Leprosy Diagnosis—A Systematic Review" Tropical Medicine and Infectious Disease 3, no. 4: 107. https://doi.org/10.3390/tropicalmed3040107
APA StyleTatipally, S., Srikantam, A., & Kasetty, S. (2018). Polymerase Chain Reaction (PCR) as a Potential Point of Care Laboratory Test for Leprosy Diagnosis—A Systematic Review. Tropical Medicine and Infectious Disease, 3(4), 107. https://doi.org/10.3390/tropicalmed3040107




