A User’s Guide to Cell-Free Protein Synthesis
Abstract
:1. Introduction
2. CFPS Reaction Formats
2.1. Commercial Systems
2.2. Coupled and Uncoupled Formats
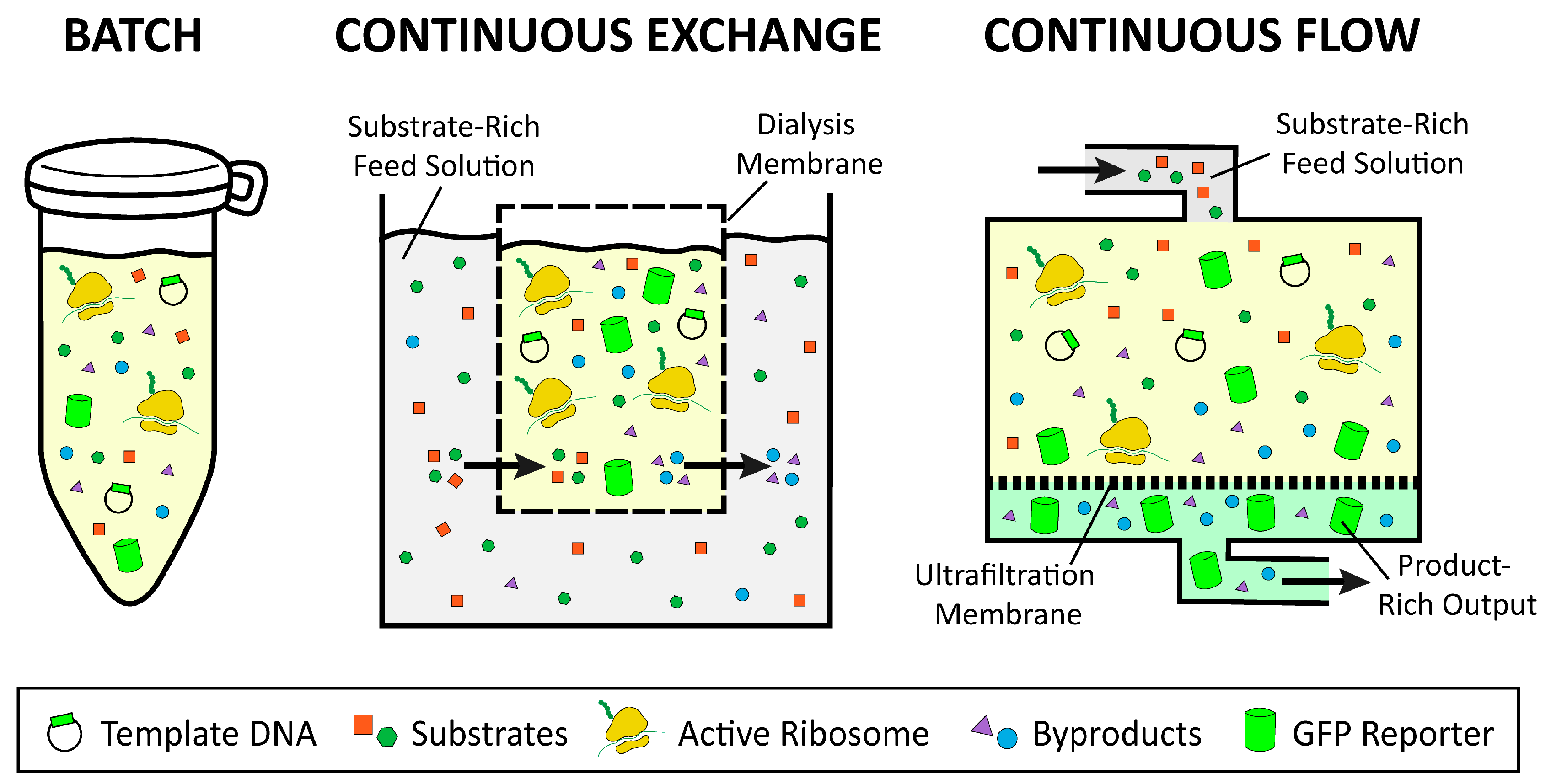
2.3. Batch, Continuous Flow, and Continuous Exchange Formats
2.4. Lyophilization
2.5. Microfluidics Format
3. Applications of Cell-Free Protein Synthesis
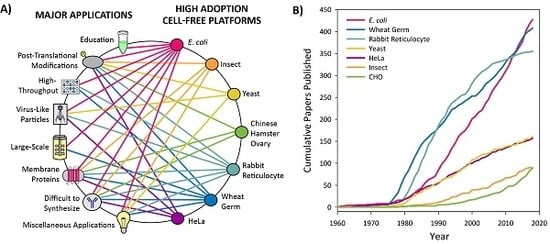
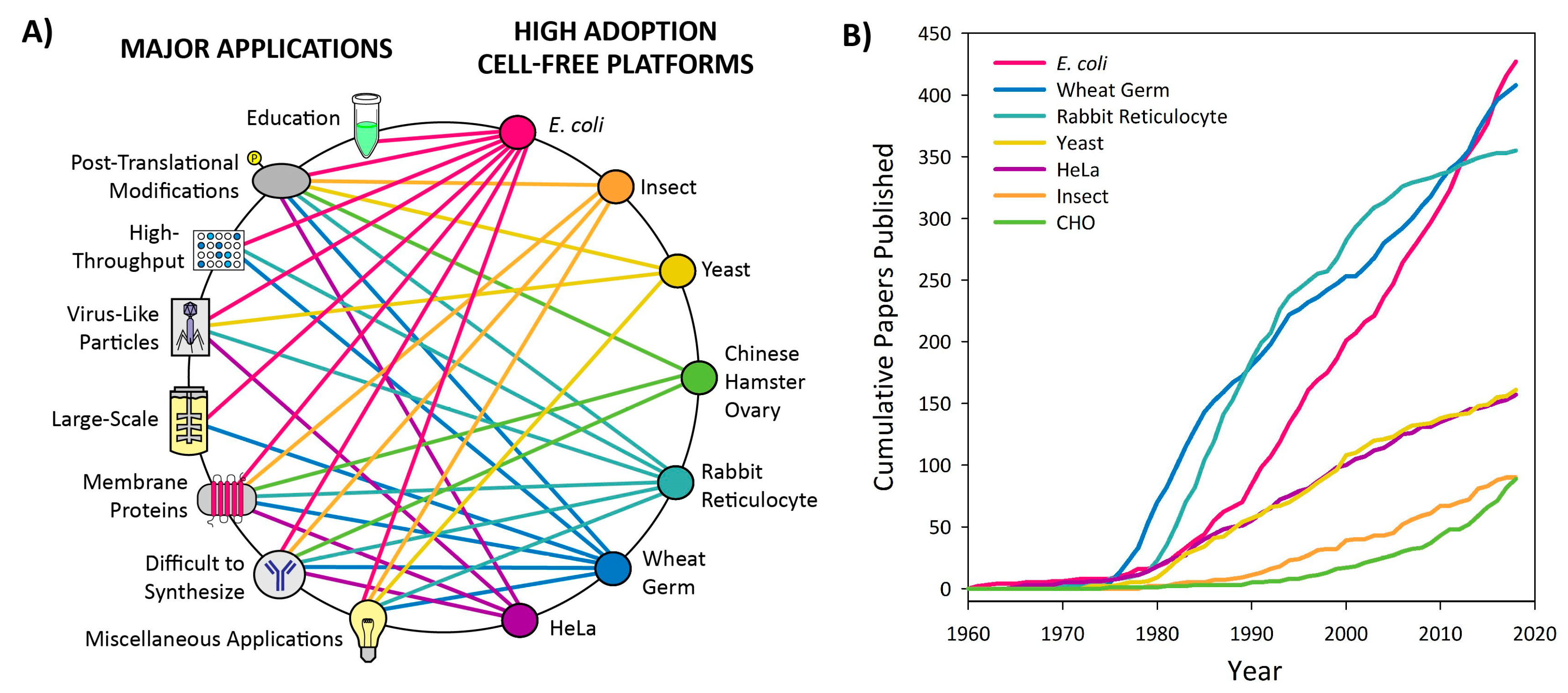
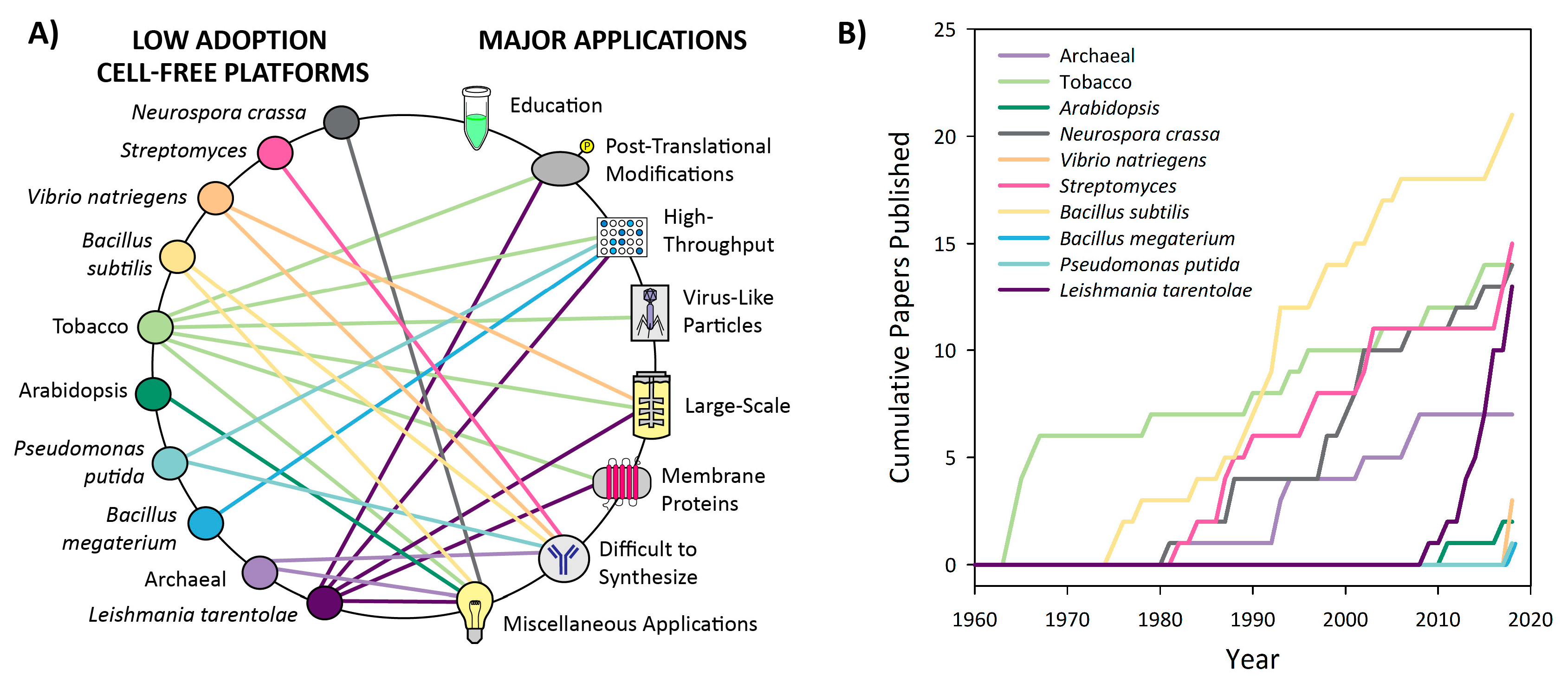
3.1. Introduction to Platform Categorization
3.2. High Adoption Platforms
3.2.1. Education
3.2.2. Post-Translational Modifications (PTMs)
3.2.3. High-Throughput Screening
3.2.4. Virus-Like Particles
3.2.5. Large-Scale
3.2.6. Membrane Proteins
3.2.7. Difficult to Synthesize Proteins
3.2.7.1. Antibodies
3.2.7.2. Large Proteins
3.2.7.3. Ice Structuring Proteins
3.2.7.4. Metalloproteins
3.2.8. Miscellaneous Applications
3.2.8.1. Studies of Translational Machinery
3.2.8.2. Genetic Circuits
3.2.8.3. Metabolic Engineering
3.2.8.4. Genetic Code Expansion
3.3. Low Adoption Platforms
3.3.1. Neurospora crassa
3.3.2. Streptomyces
3.3.3. Vibrio natriegens
3.3.4. Bacillus subtilis
3.3.5. Tobacco
3.3.6. Arabidopsis
3.3.7. Pseudomonas putida
3.3.8. Bacillus megaterium
3.3.9. Archaeal
3.3.10. Leishmania tarentolae
3.4. Recent and Future Applications
4. Methodological Differences between Platforms
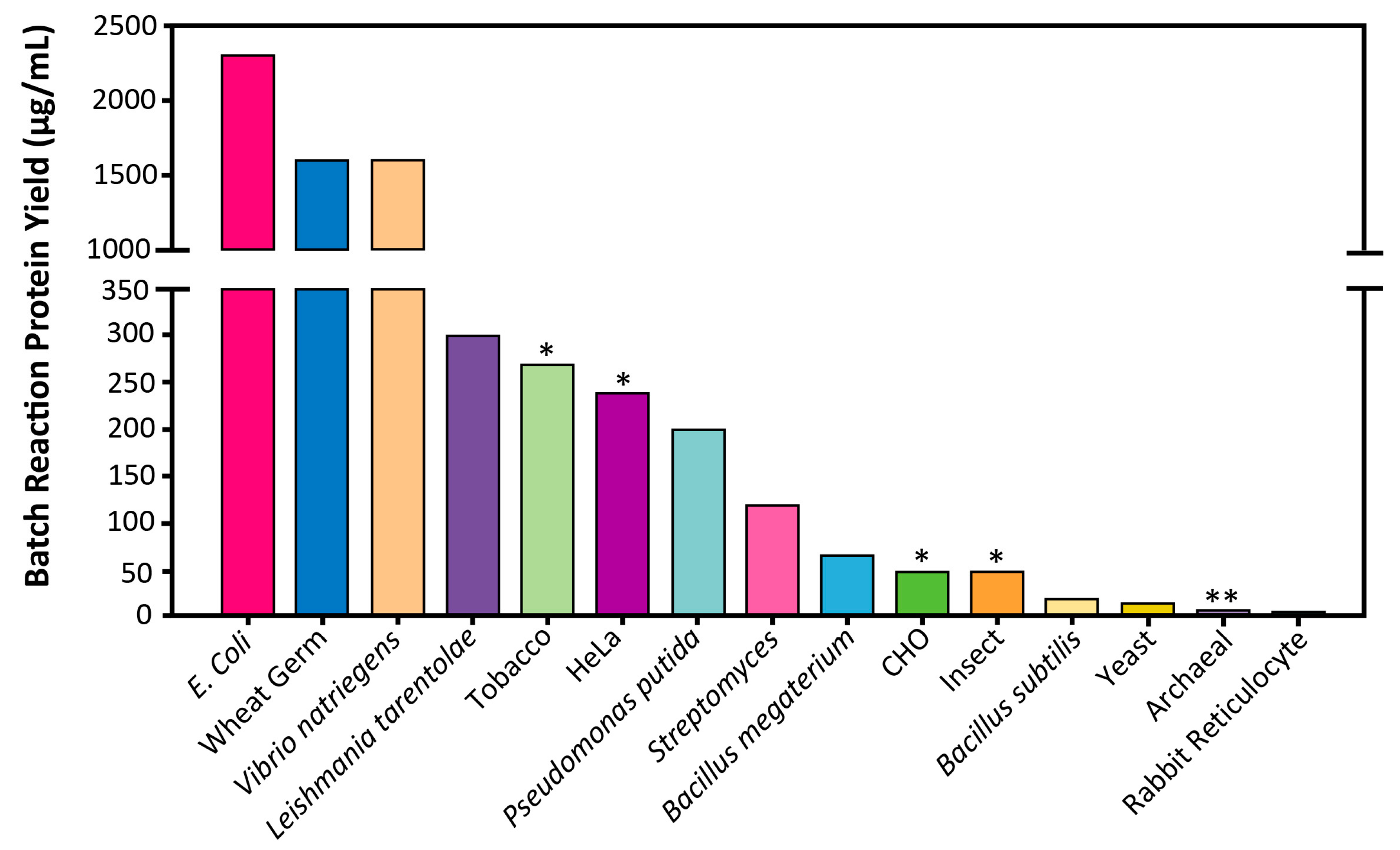
4.1. Productivity
4.2. Growth
4.3. Extract Preparation
4.4. CFPS Reaction Setup
4.5. Time
5. Standard Optimizations
6. Conclusions
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
References
- Nirenberg, M.W.; Matthaei, J.H. The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc. Natl. Acad. Sci. USA 1961, 47, 1588–1602. [Google Scholar] [CrossRef] [PubMed]
- Yin, G.; Garces, E.D.; Yang, J.; Zhang, J.; Tran, C.; Steiner, A.R.; Roos, C.; Bajad, S.; Hudak, S.; Penta, K.; et al. Aglycosylated antibodies and antibody fragments produced in a scalable in vitro transcription-translation system. MAbs 2012, 4, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Woodrow, K.A.; Swartz, J.R. A sequential expression system for high-throughput functional genomic analysis. Proteomics 2007, 7, 3870–3879. [Google Scholar] [CrossRef] [PubMed]
- Carlson, E.D.; Gan, R.; Hodgman, E.C.; Jewet, M.C. Cell-Free Protein Synthesis: Applications Come of Age. Biotechnol. Adv. 2014, 30, 1185–1194. [Google Scholar] [CrossRef] [PubMed]
- Focke, P.J.; Hein, C.; Hoffmann, B.; Matulef, K.; Bernhard, F.; Dötsch, V.; Valiyaveetil, F.I. Combining in Vitro Folding with Cell Free Protein Synthesis for Membrane Protein Expression. Biochemistry 2016, 55, 4212–4219. [Google Scholar] [CrossRef]
- Rosenblum, G.; Cooperman, B.S. Engine out of the chassis: Cell-free protein synthesis and its uses. FEBS Lett. 2014, 588, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, P.; Holland, T.M.; Bundy, B.C. Streamlined extract preparation for Escherichia coli-based cell-free protein synthesis by sonication or bead vortex mixing. Biotechniques 2012, 53, 163–174. [Google Scholar]
- Caschera, F.; Noireaux, V. Synthesis of 2.3 mg/ml of protein with an all Escherichia coli cell-free transcription-translation system. Biochimie 2014, 99, 162–168. [Google Scholar] [CrossRef]
- Harbers, M. Wheat germ systems for cell-free protein expression. FEBS Lett. 2014, 588, 2762–2773. [Google Scholar] [CrossRef]
- Des Soye, B.J.; Davidson, S.R.; Weinstock, M.T.; Gibson, D.G.; Jewett, M.C. Establishing a High-Yielding Cell-Free Protein Synthesis Platform Derived from Vibrio natriegens. ACS Synth. Biol. 2018, 7, 2245–2255. [Google Scholar] [CrossRef]
- Mureev, S.; Kovtun, O.; Nguyen, U.T.T.; Alexandrov, K. Species-independent translational leaders facilitate cell-free expression. Nat. Biotechnol. 2009, 27, 747–752. [Google Scholar] [CrossRef]
- Buntru, M.; Vogel, S.; Stoff, K.; Spiegel, H.; Schillberg, S. A versatile coupled cell-free transcription-translation system based on tobacco BY-2 cell lysates. Biotechnol. Bioeng. 2015, 112, 867–878. [Google Scholar] [CrossRef]
- Mikami, S.; Masutani, M.; Sonenberg, N.; Yokoyama, S.; Imataka, H. An efficient mammalian cell-free translation system supplemented with translation factors. Protein Expr. Purif. 2006, 46, 348–357. [Google Scholar] [CrossRef]
- Wang, H.; Li, J.; Jewett, M.C. Development of a Pseudomonas putida cell-free protein synthesis platform for rapid screening of gene regulatory elements. Synth. Biol. 2018, 3. [Google Scholar] [CrossRef]
- Li, J.; Wang, H.; Jewett, M.C. Expanding the palette of Streptomyces-based cell-free protein synthesis systems with enhanced yields. Biochem. Eng. J. 2018, 130, 29–33. [Google Scholar] [CrossRef]
- Kelwick, R.; Webb, A.J.; MacDonald, J.T.; Freemont, P.S. Development of a Bacillus subtilis cell-free transcription-translation system for prototyping regulatory elements. Metab. Eng. 2016, 38, 370–381. [Google Scholar] [CrossRef]
- Brödel, A.K.; Sonnabend, A.; Kubick, S. Cell-free protein expression based on extracts from CHO cells. Biotechnol. Bioeng. 2014, 111, 25–36. [Google Scholar] [CrossRef]
- Ezure, T.; Suzuki, T.; Higashide, S.; Shintani, E.; Endo, K.; Kobayashi, S.; Shikata, M.; Ito, M.; Tanimizu, K. Cell-Free Protein Synthesis System Prepared from Hi5 Insect Cells by Freeze-Thawing. Biotechnol. Prog 2006, 22, 1570–1577. [Google Scholar] [CrossRef]
- Gan, R.; Jewett, M.C. A combined cell-free transcription-translation system from Saccharomyces cerevisiae for rapid and robust protein synthesis. Biotechnol. J. 2014, 9, 641–651. [Google Scholar] [CrossRef]
- Endoh, T.; Kanai, T.; Sato, Y.T.; Liu, D.V.; Yoshikawa, K.; Atomi, H.; Imanaka, T. Cell-free protein synthesis at high temperatures using the lysate of a hyperthermophile. J. Biotechnol. 2006, 126, 186–195. [Google Scholar] [CrossRef]
- Kobs, G. Selecting the Cell-Free Protein Expression System That Meets Your Experimental Goals; Promega Corporation: Madison, WI, USA, 2008. [Google Scholar]
- Levine, M.Z.; Gregorio, N.E.; Jewett, M.C.; Watts, K.R.; Oza, J.P. Escherichia coli-based cell-free protein synthesis: Protocols for a robust, flexible, and accessible platform technology. J. Vis. Exp. 2019, in press. [Google Scholar] [CrossRef]
- Ohashi, H.; Kanamori, T.; Shimizu, Y.; Ueda, T. A Highly Controllable Reconstituted Cell-Free System -a Breakthrough in Protein Synthesis Research. Curr. Pharm. Biotechnol. 2010, 11, 267–271. [Google Scholar] [CrossRef]
- Shimizu, Y.; Kanamori, T.; Ueda, T. Protein synthesis by pure translation systems. Methods 2005, 36, 299–304. [Google Scholar] [CrossRef]
- Shimizu, Y.; Inoue, A.; Tomari, Y.; Suzuki, T.; Yokogawa, T.; Nishikawa, K.; Ueda, T. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001, 19, 751–755. [Google Scholar] [CrossRef]
- Hillebrecht, J.R.; Chong, S. A comparative study of protein synthesis in in vitro systems: From the prokaryotic reconstituted to the eukaryotic extract-based. BMC Biotechnol. 2008, 8, 1–9. [Google Scholar] [CrossRef]
- Karikó, K.; Muramatsu, H.; Welsh, F.A.; Ludwig, J.; Kato, H.; Akira, S.; Weissman, D. Incorporation of Pseudouridine Into mRNA Yields Superior Nonimmunogenic Vector With Increased Translational Capacity and Biological Stability. Mol. Ther. 2008, 16, 1833–1840. [Google Scholar] [CrossRef]
- Hansen, M.M.K.; Ventosa Rosquelles, M.; Yelleswarapu, M.; Maas, R.J.M.; Van Vugt-Jonker, A.J.; Heus, H.A.; Huck, W.T.S. Protein Synthesis in Coupled and Uncoupled Cell-Free Prokaryotic Gene Expression Systems. ACS Synth. Biol. 2016, 5, 1433–1440. [Google Scholar] [CrossRef]
- Spirin, A.S. High-throughput cell-free systems for synthesis of functionally active proteins. Trends Biotechnol. 2004, 22, 538–545. [Google Scholar] [CrossRef]
- Stech, M.; Quast, R.B.; Sachse, R.; Schulze, C.; Wüstenhagen, D.A.; Kubick, S. A Continuous-Exchange Cell-Free Protein Synthesis System Based on Extracts from Cultured Insect Cells. PLoS ONE 2014, 9, e96635. [Google Scholar] [CrossRef]
- Hong, S.H.; Kwon, Y.-C.; Martin, R.W.; Des Soye, B.J.; de Paz, A.M.; Swonger, K.N.; Ntai, I.; Kelleher, N.L.; Jewett, M.C. Improving cell-free protein synthesis through genome engineering of Escherichia coli lacking release factor 1. Chembiochem 2015, 16, 844–853. [Google Scholar] [CrossRef]
- Endo, Y.; Otsuzuki, S.; Ito, K.; Miura, K. Production of an enzymatic active protein using a continuous flow cell-free translation system. J. Biotechnol. 1992, 25, 221–230. [Google Scholar] [CrossRef]
- Volyanik, E.V.; Dalley, A.; Mckay, I.A.; Leigh, I.; Williams, N.S.; Bustin, S.A. Synthesis of Preparative Amounts of Biologically Active Interleukin-6 Using a Continuous-Flow Cell-Free Translation System. Anal. Biochem. 1993, 214, 289–294. [Google Scholar] [CrossRef]
- Thoring, L.; Kubick, S. Versatile cell-free protein synthesis systems based on chinese hamster ovary cells. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2018; Volume 1850, pp. 289–308. ISBN 978-1-4939-8729-0. [Google Scholar]
- Schoborg, J.A.; Hodgman, C.E.; Anderson, M.J.; Jewett, M.C. Substrate replenishment and byproduct removal improve yeast cell-free protein synthesis. Biotechnol. J. 2014, 9, 630–640. [Google Scholar] [CrossRef]
- Schwarz, D.; Junge, F.; Durst, F.; Frölich, N.; Schneider, B.; Reckel, S.; Sobhanifar, S.; Dötsch, V.; Bernhard, F. Preparative scale expression of membrane proteins in Escherichia coli-based continuous exchange cell-free systems. Nat. Protoc. 2007, 2, 2945–2957. [Google Scholar] [CrossRef]
- Chekulayeva, M.N.; Kurnasov, O.V.; Shirokov, V.A.; Spirin, A.S. Continuous-Exchange Cell-Free Protein-Synthesizing System: Synthesis of HIV-1 Antigen Nef. Biochem. Biophys. Res. Commun. 2001, 280, 914–917. [Google Scholar] [CrossRef]
- Quast, R.B.; Sonnabend, A.; Stech, M.; Wustenhagen, D.A.; Kubick, S. High-yield cell-free synthesis of human EGFR by IRES-mediated protein translation in a continuous exchange cell-free reaction format. Sci. Rep. 2016, 6, 30399. [Google Scholar] [CrossRef]
- Thoring, L.; Dondapati, S.K.; Stech, M.; Wustenhagen, D.A.; Kubick, S. High-yield production of difficult-to-express proteins in a continuous exchange cell-free system based on CHO cell lysates. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef]
- Hunt, J.P.; Yang, S.O.; Wilding, K.M.; Bundy, B.C. The growing impact of lyophilized cell-free protein expression systems. Bioengineered 2017, 8, 325–330. [Google Scholar] [CrossRef]
- Madono, M.; Sawasaki, T.; Morishita, R.; Endo, Y. Wheat germ cell-free protein production system for post-genomic research. N. Biotechnol. 2011, 28, 211–217. [Google Scholar] [CrossRef]
- Smith, M.T.; Berkheimer, S.D.; Werner, C.J.; Bundy, B.C. Lyophilized Escherichia coli-based cell-free systems for robust, high-density, long-term storage. Biotechniques 2014, 56, 186–193. [Google Scholar] [CrossRef]
- Dopp, J.L.; Reuel, N.F. Process optimization for scalable E. coli extract preparation for cell-free protein synthesis. Biochem. Eng. J. 2018, 138, 21–28. [Google Scholar] [CrossRef]
- Pardee, K.; Green, A.A.; Ferrante, T.; Cameron, D.E.; DaleyKeyser, A.; Yin, P.; Collins, J.J. Paper-based synthetic gene networks. Cell 2014, 159, 940–954. [Google Scholar] [CrossRef]
- Pardee, K.; Green, A.A.; Takahashi, M.K.; Braff, D.; Lambert, G.; Lee, J.W.; Ferrante, T.; Ma, D.; Donghia, N.; Fan, M.; et al. Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell 2016, 165, 1255–1266. [Google Scholar] [CrossRef]
- Stark, J.C.; Huang, A.; Nguyen, P.Q.; Dubner, R.S.; Hsu, K.J.; Ferrante, T.C.; Anderson, M.; Kanapskyte, A.; Mucha, Q.; Packett, J.S.; et al. BioBitsTM Bright: A fluorescent synthetic biology education kit. Sci. Adv. 2018, 4, 1–11. [Google Scholar] [CrossRef]
- Pardee, K.; Slomovic, S.; Nguyen, P.Q.; Lee, J.W.; Donghia, N.; Burrill, D.; Ferrante, T.; McSorley, F.R.; Furuta, Y.; Vernet, A.; et al. Portable, On-Demand Biomolecular Manufacturing. Cell 2016, 167, 248–259. [Google Scholar] [CrossRef]
- Gale, B.; Jafek, A.; Lambert, C.; Goenner, B.; Moghimifam, H.; Nze, U.; Kamarapu, S. A Review of Current Methods in Microfluidic Device Fabrication and Future Commercialization Prospects. Inventions 2018, 3, 60. [Google Scholar] [CrossRef]
- Damiati, S.; Mhanna, R.; Kodzius, R.; Ehmoser, E.K. Cell-free approaches in synthetic biology utilizing microfluidics. Genes (Basel) 2018, 9, 144. [Google Scholar] [CrossRef]
- Gerber, D.; Maerkl, S.J.; Quake, S.R. An in vitro microfluidic approach to generating protein-interaction networks. Nat. Methods 2009, 6, 71–74. [Google Scholar] [CrossRef]
- Jiao, Y.; Liu, Y.; Luo, D.; Huck, W.T.S.; Yang, D. Microfluidic-Assisted Fabrication of Clay Microgels for Cell-Free Protein Synthesis. ACS Appl. Mater. Interfaces 2018, 10, 29308–29313. [Google Scholar] [CrossRef]
- Georgi, V.; Georgi, L.; Blechert, M.; Bergmeister, M.; Zwanzig, M.; Wüstenhagen, D.A.; Bier, F.F.; Jung, E.; Kubick, S. On-chip automation of cell-free protein synthesis: New opportunities due to a novel reaction mode. Lab Chip 2016, 16, 269–281. [Google Scholar] [CrossRef]
- Huang, A.; Nguyen, P.Q.; Stark, J.C.; Takahashi, M.K.; Donghia, N.; Ferrante, T.; Dy, A.J.; Hsu, K.J.; Dubner, R.S.; Pardee, K.; et al. BioBitsTM Explorer: A modular synthetic biology education kit. Sci. Adv. 2018, 4. [Google Scholar] [CrossRef]
- Jaroentomeechai, T.; Stark, J.C.; Natarajan, A.; Glasscock, C.J.; Yates, L.E.; Hsu, K.J.; Mrksich, M.; Jewett, M.C.; Delisa, M.P. Single-pot glycoprotein biosynthesis using a cell-free transcription-translation system enriched with glycosylation machinery. Nat. Commun. 2018, 9. [Google Scholar]
- Sanford, J.; Codina, J.; Birnbaumers, L. Gamma subunits of G Proteins, but not their alpha or beta subunits, are polyisoprenylated. J. Biol. Chem. 1991, 266, 9570–9579. [Google Scholar]
- Dalley, J.A.; Bulleid, N.J. The Endoplasmic Reticulum (ER) Translocon can Differentiate between Hydrophobic Sequences Allowing Signals for Glycosylphosphatidylinositol Anchor Addition to be Fully Translocated into the ER Lumen. J. Biol. Chem. 2003, 278, 51749–51757. [Google Scholar] [CrossRef]
- John, D.C.; Grant, M.E.; Bulleid, N.J. Cell-free synthesis and assembly of prolyl 4-hydroxylase: the role of the beta-subunit (PDI) in preventing misfolding and aggregation of the alpha-subunit. EMBO J. 1993, 12, 1587–1595. [Google Scholar] [CrossRef]
- Gibbs, P.E.M.; Zouzias, D.C.; Freedberg, I.M. Differential post-translational modification of human type I keratins synthesized in a rabbit reticulocyte cell-free system. Biochim. Biophys. Acta - Gene Struct. Expr. 1985, 824, 247–255. [Google Scholar] [CrossRef]
- Dougherty, W.G.; Dawn Parks, T. Post-translational processing of the tobacco etch virus 49-kDa small nuclear inclusion polyprotein: Identification of an internal cleavage site and delimitation of VPg and proteinase domains. Virology 1991, 183, 449–456. [Google Scholar] [CrossRef]
- Starr, C.M.; Hanover, J.A. Glycosylation of nuclear pore protein p62. Reticulocyte lysate catalyzes O-linked N-acetylglucosamine addition in vitro. J. Biol. Chem. 1990, 265, 6868–6873. [Google Scholar]
- Shields, D.; Blobel, G. Efficient Cleavage and Segregation of Nascent Presecretory Proteins in a Reticuloqyte Lysate Supplemented with Microsomal Membranes. J. Biol. Chem. 1978, 253, 3573–3576. [Google Scholar]
- Ezure, T.; Suzuki, T.; Shikata, M.; Ito, M.; Ando, E.; Utsumi, T.; Nishimura, O.; Tsunasawa, S. Development of an insect cell-free system. Curr Pharm Biotechnol 2010, 11, 279–284. [Google Scholar] [CrossRef]
- Tarui, H.; Murata, M.; Tani, I.; Imanishi, S.; Nishikawa, S.; Hara, T. Establishment and characterization of cell-free translation/glycosylation in insect cell ( Spodoptera frugiperda 21) extract prepared with high pressure treatment. Appl. Microbiol. Biotechnol. 2001, 55, 446–453. [Google Scholar] [CrossRef]
- Zemella, A.; Thoring, L.; Hoffmeister, C.; Kubick, S. Cell-Free Protein Synthesis: Pros and Cons of Prokaryotic and Eukaryotic Systems. ChemBioChem 2015, 16, 2420–2431. [Google Scholar] [CrossRef]
- Suzuki, T.; Ito, M.; Ezure, T.; Shikata, M.; Ando, E.; Utsumi, T.; Tsunasawa, S.; Nishimura, O. Protein prenylation in an insect cell-free protein synthesis system and identification of products by mass spectrometry. Proteomics 2007, 7, 1942–1950. [Google Scholar] [CrossRef]
- Suzuki, T.; Ezure, T.; Ando, E.; Nishimura, O.; Utsumi, T.; Tsunasawa, S. Preparation of ubiquitin-conjugated proteins using an insect cell-free protein synthesis system. J. Biotechnol. 2010, 145, 73–78. [Google Scholar] [CrossRef]
- Katzen, F.; Kudlicki, W. Efficient generation of insect-based cell-free translation extracts active in glycosylation and signal sequence processing. J. Biotechnol. 2006, 125, 194–197. [Google Scholar] [CrossRef]
- Zampatis, D.E.; Rutz, C.; Furkert, J.; Schmidt, A.; Wüstenhagen, D.; Kubick, S.; Tsopanoglou, N.E.; Schülein, R. The protease-activated receptor 1 possesses a functional and cleavable signal peptide which is necessary for receptor expression. FEBS Lett. 2012, 586, 2351–2359. [Google Scholar] [CrossRef]
- von Groll, U.; Kubick, S.; Merk, H.; Stiege, W.; F, S. Advances in insect-based cell-free protein expression. In Landes Bioscience; Kudlicki, W., Katzen, F., Bennett, P., Eds.; Taylor & Francis: Austin, TX, USA, 2007; ISBN 1734998075. [Google Scholar]
- Suzuki, T.; Ito, M.; Ezure, T.; Shikata, M.; Ando, E.; Utsumi, T.; Tsunasawa, S.; Nishimura, O. N-Terminal protein modifications in an insect cell-free protein synthesis system and their identification by mass spectrometry. Proteomics 2006, 6, 4486–4495. [Google Scholar] [CrossRef]
- Ezure, T.; Suzuki, T.; Shikata, M.; Ito, M.; Ando, E.; Nishimura, O.; Tsunasawa, S. Expression of proteins containing disulfide bonds in an insect cell-free system and confirmation of their arrangements by MALDI-TOF MS. Proteomics 2007, 7, 4424–4434. [Google Scholar] [CrossRef]
- Suzuki, T.; Moriya, K.; Nagatoshi, K.; Ota, Y.; Ezure, T.; Ando, E.; Tsunasawa, S.; Utsumi, T. Strategy for comprehensive identification of human N-myristoylated proteins using an insect cell-free protein synthesis system. Proteomics 2010, 10, 1780–1793. [Google Scholar] [CrossRef]
- Sachse, R.; Wüstenhagen, D.; Šamalíková, M.; Gerrits, M.; Bier, F.F.; Kubick, S. Synthesis of membrane proteins in eukaryotic cell-free systems. Eng. Life Sci. 2013, 13, 39–48. [Google Scholar] [CrossRef]
- Guarino, C.; DeLisa, M.P. A prokaryote-based cell-free translation system that efficiently synthesizes glycoproteins. Glycobiology 2012, 22, 596–601. [Google Scholar] [CrossRef]
- Schoborg, J.A.; Hershewe, J.M.; Stark, J.C.; Kightlinger, W.; Kath, J.E.; Jaroentomeechai, T.; Natarajan, A.; DeLisa, M.P.; Jewett, M.C. A cell-free platform for rapid synthesis and testing of active oligosaccharyltransferases. Biotechnol. Bioeng. 2018, 115, 739–750. [Google Scholar] [CrossRef]
- Rothblatt, J.A.; Meyer, D.I. Secretion in Yeast: Reconstitution of the Translocation and Glycosylation of a-Factor and lnvertase in a Homologous Cell-Free System. Cell 1986, 44, 619–628. [Google Scholar] [CrossRef]
- Shields, D.; Blobel, G. Cell-free synthesis of fish preproinsulin, and processing by heterologous mammalian microsomal membranes (mRNA from islets of Langerhans/wheat germ system/canine pancreatic microsomal membranes/amino-terminal sequences/ sequence homologies). Cell Biol. 1977, 74, 2059–2063. [Google Scholar]
- Mikami, S.; Kobayashi, T.; Yokoyama, S.; Imataka, H. A hybridoma-based in vitro translation system that efficiently synthesizes glycoproteins. J. Biotechnol. 2006, 127, 65–78. [Google Scholar] [CrossRef]
- Morita, E.H.; Sawasaki, T.; Tanaka, R.; Endo, Y.; Kohno, T. A wheat germ cell-free system is a novel way to screen protein folding and function. Protein Sci. 2003, 12, 1216–1221. [Google Scholar] [CrossRef]
- Goshima, N.; Kawamura, Y.; Fukumoto, A.; Miura, A.; Honma, R.; Satoh, R.; Wakamatsu, A.; Yamamoto, J.I.; Kimura, K.; Nishikawa, T.; et al. Human protein factory for converting the transcriptome into an in vitro-expressed proteome. Nat. Methods 2008, 5, 1011–1017. [Google Scholar] [CrossRef]
- Bryan, T.M.; Goodrich, K.J.; Cech, T.R. Telomerase RNA bound by protein motifs specific to telomerase reverse transcriptase. Mol. Cell 2000, 6, 493–499. [Google Scholar] [CrossRef]
- Shao, J.; Prince, T.; Hartson, S.D.; Matts, R.L. Phosphorylation of Serine 13 Is Required for the Proper Function of the Hsp90 Co-chaperone, Cdc37. J. Biol. Chem. 2003, 278, 38117–38120. [Google Scholar] [CrossRef]
- Keefe, A.D.; Szostak, J.W. Functional proteins from a random-sequence library. Nature 2001, 410, 715–718. [Google Scholar] [CrossRef]
- Roberts, R.W.; Szostak, J.W. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc. Natl. Acad. Sci. USA 1997, 94, 12297–12302. [Google Scholar] [CrossRef]
- Thoring, L.; Wustenhagen, D.A.; Borowiak, M.; Stech, M.; Sonnabend, A.; Kubick, S. Cell-free systems based on CHO cell lysates: Optimization strategies, synthesis of “difficult-to-express” proteins and future perspectives. PLoS ONE 2016, 11, e0163670. [Google Scholar] [CrossRef]
- Bundy, B.C.; Swartz, J.R. Efficient disulfide bond formation in virus-like particles. J. Biotechnol. 2011, 154, 230–239. [Google Scholar] [CrossRef]
- Bundy, B.C.; Franciszkowicz, M.J.; Swartz, J.R. Escherichia coli-based cell-free synthesis of virus-like particles. Biotechnol. Bioeng. 2008, 100, 28–37. [Google Scholar] [CrossRef]
- Smith, M.T.; Varner, C.T.; Bush, D.B.; Bundy, B.C. The incorporation of the A2 protein to produce novel Qβ virus-like particles using cell-free protein synthesis. Biotechnol. Prog. 2012, 28, 549–555. [Google Scholar] [CrossRef]
- Patel, K.G.; Swartz, J.R. Surface Functionalization of Virus-Like Particles by Direct Conjugation Using Azide−Alkyne Click Chemistry. Bioconjug. Chem. 2011, 22, 376–387. [Google Scholar] [CrossRef]
- Rustad, M.; Eastlund, A.; Jardine, P.; Noireaux, V. Cell-free TXTL synthesis of infectious bacteriophage T4 in a single test tube reaction. Synth. Biol. 2018, 3. [Google Scholar] [CrossRef]
- Franco, D.; Pathak, H.B.; Cameron, C.E.; Rombaut, B.; Wimmer, E.; Paul, A.V. Stimulation of Poliovirus Synthesis in a HeLa Cell-Free In Vitro Translation-RNA Replication System by Viral Protein 3CDpro. J. Virol. 2005, 79, 6358–6367. [Google Scholar] [CrossRef]
- Spearman, P.; Ratner, L. Human immunodeficiency virus type 1 capsid formation in reticulocyte lysates. J. Virol. 1996, 70, 8187–8194. [Google Scholar]
- Novelli, A.; Boulanger, P.A. Assembly of adenovirus type 2 fiber synthesized in cell-free translation system. J. Biol. Chem. 1991, 266, 9299–9303. [Google Scholar]
- Klein, K.C.; Polyak, S.J.; Lingappa, J.R. Unique Features of Hepatitis C Virus Capsid Formation Revealed by De Novo Cell-Free Assembly. J. Virol. 2004, 78, 9257–9269. [Google Scholar] [CrossRef]
- Wang, X.; Liu, J.; Zheng, Y.; Li, J.; Wang, H.; Zhou, Y.; Qi, M.; Yu, H.; Tang, W.; Zhao, W.M. An optimized yeast cell-free system: Sufficient for translation of human papillomavirus 58 L1 mRNA and assembly of virus-like particles. J. Biosci. Bioeng. 2008, 106, 8–15. [Google Scholar] [CrossRef]
- Wang, X.; Liu, J.; Zhao, W.M.; Zhao, K.N. Translational comparison of HPV58 long and short L1 mRNAs in yeast (Saccharomyces cerevisiae) cell-free system. J. Biosci. Bioeng. 2010, 110, 58–65. [Google Scholar] [CrossRef]
- Zawada, J.F.; Yin, G.; Steiner, A.R.; Yang, J.; Naresh, A.; Roy, S.M.; Gold, D.S.; Heinsohn, H.G.; Murray, C.J. Microscale to manufacturing scale-up of cell-free cytokine production-a new approach for shortening protein production development timelines. Biotechnol. Bioeng. 2011, 108, 1570–1578. [Google Scholar] [CrossRef]
- Jackson, R.J.; Hunt, T. Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol. 1983, 96, 50–74. [Google Scholar]
- Beebe, E.T.; Makino, S.I.; Nozawa, A.; Matsubara, Y.; Frederick, R.O.; Primm, J.G.; Goren, M.A.; Fox, B.G. Robotic large-scale application of wheat cell-free translation to structural studies including membrane proteins. N. Biotechnol. 2011, 28, 239–249. [Google Scholar] [CrossRef]
- Schoborg, J.A.; Jewett, M.C. Cell-free protein synthesis: An emerging technology for understanding, harnessing, and expanding the capabilities of biological systems. In Synthetic Biology: Parts, Devices and Applications; John Wiley & Sons: Hoboken, NJ, USA, 2018; pp. 309–330. ISBN 9783527688104. [Google Scholar]
- Oza, J.P.; Aerni, H.R.; Pirman, N.L.; Barber, K.W.; ter Haar, C.M.; Rogulina, S.; Amrofell, M.B.; Isaacs, F.J.; Rinehart, J.; Jewett, M.C. Robust production of recombinant phosphoproteins using cell-free protein synthesis. Nat. Commun. 2015, 6, 8168. [Google Scholar] [CrossRef]
- Voloshin, A.M.; Swartz, J.R. Efficient and scalable method for scaling up cell free protein synthesis in batch mode. Biotechnol. Bioeng. 2005, 91, 516–521. [Google Scholar] [CrossRef]
- Martin, R.W.; Majewska, N.I.; Chen, C.X.; Albanetti, T.E.; Jimenez, R.B.C.; Schmelzer, A.E.; Jewett, M.C.; Roy, V. Development of a CHO-Based Cell-Free Platform for Synthesis of Active Monoclonal Antibodies. ACS Synth. Biol. 2017, 6, 1370–1379. [Google Scholar] [CrossRef]
- Jewett, M.C.; Swartz, J.R. Mimicking the Escherichia coli Cytoplasmic Environment Activates Long-Lived and Efficient Cell-Free Protein Synthesis. Biotechnol. Bioeng. 2004, 86, 19–26. [Google Scholar] [CrossRef]
- Rigaud, J.L. Membrane proteins: Functional and structural studies using reconstituted proteoliposomes and 2-D crystals. Brazilian J. Med. Biol. Res. 2002, 35, 753–766. [Google Scholar] [CrossRef]
- Sachse, R.; Dondapati, S.K.; Fenz, S.F.; Schmidt, T.; Kubick, S. Membrane protein synthesis in cell-free systems: From bio-mimetic systems to bio-membranes. FEBS Lett. 2014, 588, 2774–2781. [Google Scholar] [CrossRef]
- Yadavalli, R.; Sam-Yellowe, T. HeLa Based Cell Free Expression Systems for Expression of Plasmodium Rhoptry Proteins. J. Vis. Exp. 2015, e52772. [Google Scholar] [CrossRef]
- Dondapati, S.K.; Kreir, M.; Quast, R.B.; Wüstenhagen, D.A.; Brüggemann, A.; Fertig, N.; Kubick, S. Membrane assembly of the functional KcsA potassium channel in a vesicle-based eukaryotic cell-free translation system. Biosens. Bioelectron. 2014, 59, 174–183. [Google Scholar] [CrossRef]
- Wilson, C.M.; Farmery, M.R.; Bulleid, N.J. Pivotal role of calnexin and mannose trimming in regulating the endoplasmic reticulum-associated degradation of major histocompatibility complex class I heavy chain. J. Biol. Chem. 2000, 275, 21224–21232. [Google Scholar] [CrossRef]
- Goren, M.A.; Fox, B.G. Wheat germ cell-free translation, purification, and assembly of a functional human stearoyl-CoA desaturase complex. Protein Expr. Purif. 2008, 62, 171–178. [Google Scholar] [CrossRef]
- Nozawa, A.; Nanamiya, H.; Miyata, T.; Linka, N.; Endo, Y.; Weber, A.P.M.; Tozawa, Y. A cell-free translation and proteoliposome reconstitution system for functional analysis of plant solute transporters. Plant Cell Physiol. 2007, 48, 1815–1820. [Google Scholar] [CrossRef]
- Junge, F.; Haberstock, S.; Roos, C.; Stefer, S.; Proverbio, D.; Dötsch, V.; Bernhard, F. Advances in cell-free protein synthesis for the functional and structural analysis of membrane proteins. N. Biotechnol. 2011, 28, 262–271. [Google Scholar] [CrossRef]
- Arduengo, M.; Schenborn, E.; Hurst, R. The Role of Cell-Free Rabbit Reticulocyte Expression Systems in Functional Proteomics. Cell-free Expression; Landes Bioscience: Austin, TX, USA, 2007; pp. 1–18. [Google Scholar]
- Stavnezer, J.; Huang, R.C.C. Synthesis of a Mouse Immunoglobulin Light Chain in a Rabbit Reticulocyte Cell-free System. Nat. New Biol. 1971, 230, 172–176. [Google Scholar] [CrossRef]
- Nicholls, P.J.; Johnson, V.G.; Andrew, S.M.; Hoogenboom, H.R.; Raus, J.C.; Youle, R.J. Characterization of single-chain antibody (sFv)-toxin fusion proteins produced in vitro in rabbit reticulocyte lysate. J. Biol. Chem. 1993, 268, 5302–5308. [Google Scholar]
- Gusdon, B.Y.J.P.; Stavitsky, A.B.; Armentrout, S.A. Synthesis of gamma G antibody and immunoglobulin on polyribosomes in a cell-free system. Proc. Natl. Acad. Sci. 1967, 58, 1189–1196. [Google Scholar] [CrossRef]
- Ryabova, L.A.; Desplancq, D.; Spirin, A.S.; Plückthun, A. Functional antibody production using cell-free translation: Effects of protein disulfide isomerase and chaperones. Nat. Biotechnol. 1997, 15, 79–84. [Google Scholar] [CrossRef]
- Merk, H.; Stiege, W.; Tsumoto, K.; Kumagai, I.; Erdmann, V.A. Cell-free expression of two single-chain monoclonal antibodies against lysozyme: effect of domain arrangement on the expression. J. Biochem. 1999, 125, 328–333. [Google Scholar] [CrossRef]
- Zimmerman, E.S.; Heibeck, T.H.; Gill, A.; Li, X.; Murray, C.J.; Madlansacay, M.R.; Tran, C.; Uter, N.T.; Yin, G.; Rivers, P.J.; et al. Production of site-specific antibody-drug conjugates using optimized non-natural amino acids in a cell-free expression system. Bioconjug. Chem. 2014, 25, 351–361. [Google Scholar] [CrossRef]
- Galeffi, P.; Lombardi, A.; Pietraforte, I.; Novelli, F.; Di Donato, M.; Sperandei, M.; Tornambé, A.; Fraioli, R.; Martayan, A.; Natali, P.G.; et al. Functional expression of a single-chain antibody to ErbB-2 in plants and cell-free systems. J. Transl. Med. 2006, 4, 39. [Google Scholar] [CrossRef]
- Jiang, X.; Ookubo, Y.; Fujii, I.; Nakano, H.; Yamane, T. Expression of Fab fragment of catalytic antibody 6D9 in an Escherichia coli in vitro coupled transcription/translation system. FEBS Lett. 2002, 514, 290–294. [Google Scholar] [CrossRef]
- Groff, D.; Armstrong, S.; Rivers, P.J.; Zhang, J.; Yang, J.; Green, E.; Rozzelle, J.; Liang, S.; Kittle, J.D.; Steiner, A.R.; et al. Engineering toward a bacterial “endoplasmic reticulum” for the rapid expression of immunoglobulin proteins. MAbs 2014, 6, 671–678. [Google Scholar] [CrossRef]
- Kawasaki, T.; Gouda, M.D.; Sawasaki, T.; Takai, K.; Endo, Y. Efficient synthesis of a disulfide-containing protein through a batch cell-free system from wheat germ. Eur. J. Biochem. 2003, 270, 4780–4786. [Google Scholar] [CrossRef]
- Stech, M.; Merk, H.; Schenk, J.A.; Stöcklein, W.F.M.; Wüstenhagen, D.A.; Micheel, B.; Duschl, C.; Bier, F.F.; Kubick, S. Production of functional antibody fragments in a vesicle-based eukaryotic cell-free translation system. J. Biotechnol. 2012, 164, 220–231. [Google Scholar] [CrossRef]
- Stech, M.; Hust, M.; Schulze, C.; Dübel, S.; Kubick, S. Cell-free eukaryotic systems for the production, engineering, and modification of scFv antibody fragments. Eng. Life Sci. 2014, 14, 387–398. [Google Scholar] [CrossRef]
- Goering, A.W.; Li, J.; McClure, R.A.; Thomson, R.J.; Jewett, M.C.; Kelleher, N.L. In vitro reconstruction of nonribosomal peptide biosynthesis directly from DNA using cell-free protein synthesis. ACS Synth. Biol. 2017, 6, 39–44. [Google Scholar] [CrossRef]
- Mikami, S.; Kobayashi, T.; Masutani, M.; Yokoyama, S.; Imataka, H. A human cell-derived in vitro coupled transcription/translation system optimized for production of recombinant proteins. Protein Expr. Purif. 2008, 62, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, Y.; Rooney, L.; Skach, W.R. In vitro methods for CFTR biogenesis. Methods Mol. Biol. 2011, 741, 233–253. [Google Scholar]
- Brödel, A.K.; Raymond, J.A.; Duman, J.G.; Bier, F.F.; Kubick, S. Functional evaluation of candidate ice structuring proteins using cell-free expression systems. J. Biotechnol. 2013, 163, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lawton, T.J.; Kostecki, J.S.; Nisthal, A.; Fang, J.; Mayo, S.L.; Rosenzweig, A.C.; Jewett, M.C. Cell-free protein synthesis enables high yielding synthesis of an active multicopper oxidase. Biotechnol. J. 2016, 11, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Boyer, M.E.; Stapleton, J.A.; Kuchenreuther, J.M.; Wang, C.; Swartz, J.R. Cell-free synthesis and maturation of [FeFe] hydrogenases. Biotechnol. Bioeng. 2008, 99, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Kuchenreuther, J.M.; Shiigi, S.A.; Swartz, J.R. Cell-Free Synthesis of the H-Cluster: A Model for the In Vitro Assembly of Metalloprotein Metal Centers. In Methods in molecular biology (Clifton, N.J.); Humana Press: Totowa, NJ, USA, 2014; Volume 1122, pp. 49–72. [Google Scholar]
- Ahn, J.-H.; Hwang, M.-Y.; Oh, I.-S.; Park, K.-M.; Hahn, G.-H.; Choi, C.-Y.; Kim, D.-M. Preparation method forEscherichia coli S30 extracts completely dependent upon tRNA addition to catalyze cell-free protein synthesis. Biotechnol. Bioprocess Eng. 2006, 11, 420–424. [Google Scholar] [CrossRef]
- Yokogawa, T.; Kitamura, Y.; Nakamura, D.; Ohno, S.; Nishikawa, K. Optimization of the hybridization-based method for purification of thermostable tRNAs in the presence of tetraalkylammonium salts. Nucleic Acids Res. 2010, 38, e89. [Google Scholar] [CrossRef] [PubMed]
- Panthu, B.; Ohlmann, T.; Perrier, J.; Schlattner, U.; Jalinot, P.; Elena-Herrmann, B.; Rautureau, G.J.P. Cell-Free Protein Synthesis Enhancement from Real-Time NMR Metabolite Kinetics: Redirecting Energy Fluxes in Hybrid RRL Systems. ACS Synth. Biol. 2018, 7, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Jewett, M.C.; Fritz, B.R.; Timmerman, L.E.; Church, G.M. In vitro integration of ribosomal RNA synthesis, ribosome assembly, and translation. Mol. Syst. Biol. 2013, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Timm, A.C.; Shankles, P.G.; Foster, C.M.; Doktycz, M.J.; Retterer, S.T. Toward Microfluidic Reactors for Cell-Free Protein Synthesis at the Point-of-Care. Small 2016, 12, 810–817. [Google Scholar] [CrossRef] [PubMed]
- Brophy, J.A.N.; Voigt, C.A. Principles of genetic circuit design. Nat. Methods 2014, 11, 508–520. [Google Scholar] [CrossRef] [PubMed]
- Noireaux, V.; Bar-Ziv, R.; Libchaber, A. Principles of cell-free genetic circuit assembly. Proc. Natl. Acad. Sci. 2003, 100, 12672–12677. [Google Scholar] [CrossRef] [PubMed]
- Karig, D.K.; Jung, S.-Y.; Srijanto, B.; Collier, C.P.; Simpson, M.L. Probing Cell-Free Gene Expression Noise in Femtoliter Volumes. ACS Synth. Biol. 2013, 2, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Siegal-Gaskins, D.; Noireaux, V.; Murray, R.M. Biomolecular resource utilization in elementary cell-free gene circuits. In Proceedings of the 2013 American Control Conference, Washington, DC, USA, 17–19 June 2013. [Google Scholar]
- Karzbrun, E.; Shin, J.; Bar-Ziv, R.H.; Noireaux, V. Coarse-grained dynamics of protein synthesis in a cell-free system. Phys. Rev. Lett. 2011, 106. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Noireaux, V. An E. coli cell-free expression toolbox: Application to synthetic gene circuits and artificial cells. ACS Synth. Biol. 2012, 1, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Maerkl, S.J.; Murray, R.M.; Sun, Z.Z.; Niederholtmeyer, H.; Hori, Y.; Yeung, E.; Verpoorte, A. Rapid cell-free forward engineering of novel genetic ring oscillators. Elife 2015, 4, 1–18. [Google Scholar]
- Noireaux, V.; Singhal, V.; Sun, Z.Z.; Murray, R.M.; Spring, K.J.; Chappell, J.; Hayes, C.A.; Lucks, J.B.; Fall, C.P.; Al-Khabouri, S.; et al. Rapidly Characterizing the Fast Dynamics of RNA Genetic Circuitry with Cell-Free Transcription–Translation (TX-TL) Systems. ACS Synth. Biol. 2014, 4, 503–515. [Google Scholar]
- Halleran, A.D.; Murray, R.M. Cell-Free and in Vivo Characterization of Lux, Las, and Rpa Quorum Activation Systems in E. coli. ACS Synth. Biol. 2018, 7, 752–755. [Google Scholar] [CrossRef] [PubMed]
- Sen, S.; Apurva, D.; Satija, R.; Siegal, D.; Murray, R.M. Design of a Toolbox of RNA Thermometers. ACS Synth. Biol. 2017, 6, 1461–1470. [Google Scholar] [CrossRef]
- Dudley, Q.M.; Anderson, K.C.; Jewett, M.C. Cell-Free Mixing of Escherichia coli Crude Extracts to Prototype and Rationally Engineer High-Titer Mevalonate Synthesis. ACS Synth. Biol. 2016, 5, 1578–1588. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Zhao, J.; Lian, J.; Xu, Z. Cell-free protein synthesis enabled rapid prototyping for metabolic engineering and synthetic biology. Synth. Syst. Biotechnol. 2018, 3, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Khattak, W.A.; Ul-Islam, M.; Ullah, M.W.; Yu, B.; Khan, S.; Park, J.K. Yeast cell-free enzyme system for bio-ethanol production at elevated temperatures. Process Biochem. 2014, 49, 357–364. [Google Scholar] [CrossRef]
- Kay, J.E.; Jewett, M.C. Lysate of engineered Escherichia coli supports high-level conversion of glucose to 2,3-butanediol. Metab. Eng. 2015, 32, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Quast, R.B.; Mrusek, D.; Hoffmeister, C.; Sonnabend, A.; Kubick, S. Cotranslational incorporation of non-standard amino acids using cell-free protein synthesis. FEBS Lett. 2015, 589, 1703–1712. [Google Scholar] [CrossRef] [PubMed]
- Albayrak, C.; Swartz, J.R. Cell-free co-production of an orthogonal transfer RNA activates efficient site-specific non-natural amino acid incorporation. Nucleic Acids Res. 2013, 41, 5949–5963. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.W.; Des Soye, B.J.; Kwon, Y.-C.; Kay, J.; Davis, R.G.; Thomas, P.M.; Majewska, N.I.; Chen, C.X.; Marcum, R.D.; Weiss, M.G.; et al. Cell-free protein synthesis from genomically recoded bacteria enables multisite incorporation of noncanonical amino acids. Nat. Commun. 2018, 9, 1203. [Google Scholar] [CrossRef] [PubMed]
- Ozer, E.; Chemla, Y.; Schlesinger, O.; Aviram, H.Y.; Riven, I.; Haran, G.; Alfonta, L. In vitro suppression of two different stop codons. Biotechnol. Bioeng. 2017, 114, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Benítez-Mateos, A.I.; Llarena, I.; Sánchez-Iglesias, A.; López-Gallego, F. Expanding One-Pot Cell-Free Protein Synthesis and Immobilization for On-Demand Manufacturing of Biomaterials. ACS Synth. Biol. 2018, 7, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Wu, Y.; Mureev, S.; Alexandrov, K. Oligonucleotide-mediated tRNA sequestration enables one-pot sense codon reassignment in vitro. Nucleic Acids Res. 2018, 46, 6387–6400. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Bu, N.; Lu, Y.; Gao, W.; Bu, N.; Lu, Y. Efficient Incorporation of Unnatural Amino Acids into Proteins with a Robust Cell-Free System. Methods Protoc. 2019, 2, 16. [Google Scholar] [CrossRef]
- Quast, R.B.; Kortt, O.; Henkel, J.; Dondapati, S.K.; Wüstenhagen, D.A.; Stech, M.; Kubick, S. Automated production of functional membrane proteins using eukaryotic cell-free translation systems. J. Biotechnol. 2015, 203, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Stech, M.; Brödel, A.K.; Quast, R.B.; Sachse, R.; Kubick, S. Cell-Free Systems: Functional Modules for Synthetic and Chemical Biology. In Advances in Biochemical Engineering/Biotechnology; Springer: Berlin/Heidelberg, Germany, 2013; Volume 137, pp. 67–102. [Google Scholar]
- Wu, C.; Dasgupta, A.; Shen, L.; Bell-Pedersen, D.; Sachs, M.S. The cell free protein synthesis system from the model filamentous fungus Neurospora crassa. Methods 2018, 137, 11–19. [Google Scholar] [CrossRef]
- Szczesna-Skorupa, E.; Filipowicz, W.; Paszewski, A. The cell-free protein synthesis system from the “slime” mutant of Neurospora crassa. Preparation and characterisation of importance of 7-methylguanosine for translation of viral and cellular mRNAs. Eur. J. Biochem. 1981, 121, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Curle, C.A.; Kapoor, M. A Neurospora crassa heat-shocked cell lysate translates homologous and heterologous messenger RNA efficiently, without preference for heat shock messages. Curr. Genet. 1988, 13, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Sachs, M.S.; Wang, Z.; Gaba, A.; Fang, P.; Belk, J.; Ganesan, R.; Amrani, N.; Jacobson, A. Toeprint analysis of the positioning of translation apparatus components at initiation and termination codons of fungal mRNAs. Methods 2002, 26, 105–114. [Google Scholar] [CrossRef]
- Wu, C.; Wei, J.; Lin, P.-J.; Tu, L.; Deutsch, C.; Johnson, A.E.; Sachs, M.S. Arginine Changes the Conformation of the Arginine Attenuator Peptide Relative to the Ribosome Tunnel. J. Mol. Biol. 2012, 416, 518–533. [Google Scholar] [CrossRef]
- Wei, J.; Wu, C.; Sachs, M.S. The Arginine Attenuator Peptide Interferes with the Ribosome Peptidyl Transferase Center. Mol. Cell. Biol. 2012, 32, 2396–2406. [Google Scholar] [CrossRef]
- Yu, C.-H.; Dang, Y.; Zhou, Z.; Wu, C.; Zhao, F.; Sachs, M.S.; Liu, Y. Codon Usage Influences the Local Rate of Translation Elongation to Regulate Co-translational Protein Folding. Mol. Cell 2015, 59, 744–754. [Google Scholar] [CrossRef]
- Li, J.; Wang, H.; Kwon, Y.C.; Jewett, M.C. Establishing a high yielding streptomyces-based cell-free protein synthesis system. Biotechnol. Bioeng. 2017, 114, 1343–1353. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.; Rae, S.; Cundliffe, E. Coupled transcription--translation in extracts of Streptomyces lividans. Mol. Gen. Genet. 1984, 195, 39–43. [Google Scholar] [CrossRef]
- Wiegand, D.J.; Lee, H.H.; Ostrov, N.; Church, G.M. Establishing a Cell-Free Vibrio natriegens Expression System. ACS Synth. Biol. 2018, 7, 2475–2479. [Google Scholar] [CrossRef]
- Failmezger, J.; Scholz, S.; Blombach, B.; Siemann-Herzberg, M. Cell-free protein synthesis from fast-growing Vibrio natriegens. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Tominaga, A.; Kobayashi, Y. Kasugamycin-resistant mutants of Bacillus subtilis. J. Bacteriol. 1978, 135, 1149–1150. [Google Scholar]
- Stallcup, M.R.; Sharrock, W.J.; Rabinowitz, J.C. Specificity of bacterial ribosomes and messenger ribonucleic acids in protein synthesis reactions in vitro. J. Biol. Chem. 1976, 251, 2499–2510. [Google Scholar]
- Villafane, R.; Bechhofer, D.H.; Narayanan, C.S.; Dubnau, D. Replication control genes of plasmid pE194. J. Bacteriol. 1987, 169, 4822–4829. [Google Scholar] [CrossRef]
- Spencer, D.; Wildman, S.G. The Incorporation of Amino Acids into Protein by Cell-free Extracts from Tobacco Leaves. Biochemistry 1964, 3, 954–959. [Google Scholar] [CrossRef]
- Boardman, N.K.; Francki, R.I.B.; Wildman, S.G. Protein synthesis by cell-free extracts of tobacco leaves: III. Comparison of the physical properties and protein synthesizing activities of 70 s chloroplast and 80 s cytoplasmic ribosomes. J. Mol. Biol. 1966, 17, 470–487. [Google Scholar] [CrossRef]
- Phelps, R.H.; Sequeira, L. Synthesis of Indoleacetic Acid via Tryptamine by a Cell-free System from Tobacco Terminal Buds. Plant Physiol. 1967, 42, 1161–1163. [Google Scholar] [CrossRef]
- Guo, Z.H.; Severson, R.F.; Wagner, G.J. Biosynthesis of the Diterpene cis-Abienol in Cell-Free Extracts of Tobacco Trichomes. Arch. Biochem. Biophys. 1994, 308, 103–108. [Google Scholar] [CrossRef]
- Cooke, R.; Penon, P. In vitro transcription from cauliflower mosaic virus promoters by a cell-free extract from tobacco cells. Plant Mol. Biol. 1990, 14, 391–405. [Google Scholar] [CrossRef]
- Chen, C.; Melitz, D.K. Cytokinin biosynthesis in a cell-free system from cytokinin-autotrophic tobacco tissue cultures. FEBS Lett. 1979, 107, 15–20. [Google Scholar] [CrossRef]
- Hao, D.; Yeoman, M.M. Nicotine N-demethylase in cell-free preparations from tobacco cell cultures. Phytochemistry 1996, 42, 325–329. [Google Scholar] [CrossRef]
- Komoda, K.; Naito, S.; Ishikawa, M. Replication of plant RNA virus genomes in a cell-free extract of evacuolated plant protoplasts. Proc. Natl. Acad. Sci. 2004, 101, 1863–1867. [Google Scholar] [CrossRef]
- Gursinsky, T.; Schulz, B.; Behrens, S.E. Replication of Tomato bushy stunt virus RNA in a plant in vitro system. Virology 2009, 390, 250–260. [Google Scholar] [CrossRef]
- Murota, K.; Hagiwara-Komoda, Y.; Komoda, K.; Onouchi, H.; Ishikawa, M.; Naito, S. Arabidopsis cell-free extract, ACE, a new in vitro translation system derived from arabidopsis callus cultures. Plant Cell Physiol. 2011, 52, 1443–1453. [Google Scholar] [CrossRef]
- Moore, S.J.; MacDonald, J.T.; Wienecke, S.; Ishwarbhai, A.; Tsipa, A.; Aw, R.; Kylilis, N.; Bell, D.J.; McClymont, D.W.; Jensen, K.; et al. Rapid acquisition and model-based analysis of cell-free transcription–translation reactions from nonmodel bacteria. Proc. Natl. Acad. Sci. 2018, 201715806. [Google Scholar] [CrossRef]
- Ruggero, D.; Creti, R.; Londei, P. In vitro translation of archaeal natural mRNAs at high temperature. FEMS Microbiol. Lett. 1993, 107, 89–94. [Google Scholar] [CrossRef]
- Elhardt, D.; Böck, A. An in vitro polypeptide synthesizing system from methanogenic bacteria: Sensitivity to antibiotics. MGG Mol. Gen. Genet. 1982, 188, 128–134. [Google Scholar] [CrossRef]
- Uzawa, T.; Yamagishi, A.; Oshima, T. Polypeptide synthesis directed by DNA as a messenger in cell-free polypeptide synthesis by extreme thermophiles, Thermus thermophilus HB27 and sulfolobus tokodaii strain 7. J. Biochem. 2002, 131, 849–853. [Google Scholar] [CrossRef]
- Uzawa, T.; Hamasaki, N.; Oshima, T. Effects of Novel Polyamines on Cell-Free Polypeptide Catalyzed by Thermus thermophilus HB8 Extract. J. Biochem. 1993, 486, 478–486. [Google Scholar] [CrossRef]
- Kovtun, O.; Mureev, S.; Jung, W.R.; Kubala, M.H.; Johnston, W.; Alexandrov, K. Leishmania cell-free protein expression system. Methods 2011, 55, 58–64. [Google Scholar] [CrossRef]
- Bhide, M.; Natarajan, S.; Hresko, S.; Aguilar, C.; Bencurova, E. Rapid in vitro protein synthesis pipeline: A promising tool for cost-effective protein array design. Mol. Biosyst. 2014, 10, 1236–1245. [Google Scholar] [CrossRef]
- Perez, J.G.; Stark, J.C.; Jewett, M.C. Cell-Free Synthetic Biology: Engineering Beyond the Cell. Cold Spring Harb. Perspect. Biol. 2016, 8, 1–26. [Google Scholar] [CrossRef]
- Ruehrer, S.; Michel, H. Exploiting Leishmania tarentolae cell-free extracts for the synthesis of human solute carriers. Mol. Membr. Biol. 2013, 30, 288–302. [Google Scholar] [CrossRef]
- Madin, K.; Sawasaki, T.; Ogasawara, T.; Endo, Y. A highly efficient and robust cell-free protein synthesis system prepared from wheat embryos: plants apparently contain a suicide system directed at ribosomes. Proc. Natl. Acad. Sci. USA 2000, 97, 559–664. [Google Scholar] [CrossRef]
- Hodgman, C.E.; Jewett, M.C. Optimized extract preparation methods and reaction conditions for improved yeast cell-free protein synthesis. Biotechnol. Bioeng. 2013, 110, 2643–2654. [Google Scholar] [CrossRef]
- Kubick, S.; Schacherl, J.; Fleisher-Notter, H.; Royall, E.; Roberts, L.O.; Steige, W. In Vitro Translation in an Insect-Based Cell-Free System. In Cell-free Protein Expression; Springer: Berlin/Heidelberg, Germany, 2003; ISBN 978-3-642-63939-5. [Google Scholar]
- Kwon, Y.-C.; Jewett, M.C. High-throughput preparation methods of crude extract for robust cell-free protein synthesis. Sci. Rep. 2015, 5, 8663. [Google Scholar] [CrossRef]
- Krinsky, N.; Kaduri, M.; Shainsky-Roitman, J.; Goldfeder, M.; Ivanir, E.; Benhar, I.; Shoham, Y.; Schroeder, A. A Simple and Rapid Method for Preparing a Cell-Free Bacterial Lysate for Protein Synthesis. PLoS ONE 2016, 11, e0165137. [Google Scholar] [CrossRef]
- Shin, J.; Noireaux, V. Efficient cell-free expression with the endogenous E. Coli RNA polymerase and sigma factor 70. J. Biol. Eng. 2010, 4, 8. [Google Scholar] [CrossRef]
- Nishimura, N.; Kitaoka, Y.; Mimura, A.; Takahara, Y. Continuous protein synthesis system with Escherichia coli S30 extract containing endogenous T7 RNA polymerase. Biotechnol. Lett. 1993, 15, 785–790. [Google Scholar] [CrossRef]
- Lee, K.-H.; Kim, D.-M. Recent advances in development of cell-free protein synthesis systems for fast and efficient production of recombinant proteins. FEMS Microbiol. Lett. 2018, 365, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Condò, I.; Ciammaruconi, A.; Benelli, D.; Ruggero, D.; Londei, P. Cis-acting signals controlling translational initiation in the thermophilic archaeon Sulfolobus solfataricus. Mol. Microbiol. 1999, 34, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Brödel, A.K.; Sonnabend, A.; Roberts, L.O.; Stech, M.; Wüstenhagen, D.A.; Kubick, S. IRES-mediated translation of membrane proteins and glycoproteins in eukaryotic cell-free systems. PLoS ONE 2013, 8, e82234. [Google Scholar]
- Anastasina, M.; Terenin, I.; Butcher, S.J.; Kainov, D.E. A technique to increase protein yield in a rabbit reticulocyte lysate translation system. Biotechniques 2014, 56, 36–39. [Google Scholar] [CrossRef] [PubMed]
- Hodgman, C.E.; Jewett, M.C. Characterizing IGR IRES-mediated translation initiation for use in yeast cell-free protein synthesis. N. Biotechnol. 2014, 31, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Dopp, B.J.L.; Tamiev, D.D.; Reuel, N.F. Cell-free supplement mixtures: Elucidating the history and biochemical utility of additives used to support in vitro protein synthesis in E. coli extract. Biotechnol. Adv. 2019, 37, 246–258. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Gu, L.; Aach, J.; Church, G.M. Improved cell-free RNA and protein synthesis system. PLoS ONE 2014, 9, e106232. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Huang, Q.; Deng, Z.; Xu, Y.; Liu, T. Enhancing the efficiency of cell-free protein synthesis system by systematic titration of transcription and translation components. Biochem. Eng. J. 2018, 138, 47–53. [Google Scholar] [CrossRef]

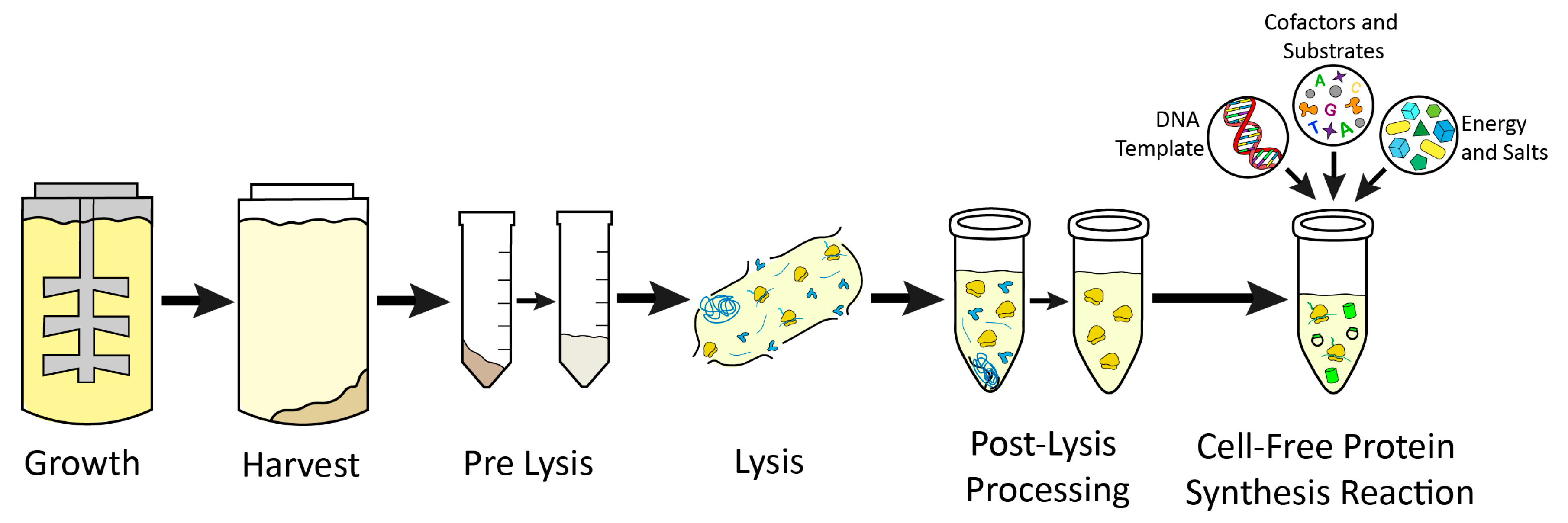
| Growth | |||
|---|---|---|---|
| Platform | Media/Vessel | Harvest | Key Citations |
| E. coli | Media: 2× YPTG (5 g NaCl, 16 g Tryptone, 10 g Yeast extract, 7 g KH2PO4, 3 g KHPO4, pH 7.2/750 mL solution, 18 g Glucose/250 mL solution). Vessel: 2 L Baffled Flask. Conditions: 37 °C, 200 RPM | When OD600 is 3, centrifuge at 5000× g for 10 min at 10 °C. Wash pellet with 30 mL S30 buffer (10 mM Tris OAc, pH 8.2, 14 mM Mg(OAc)2, 60 mM KOAc, 2 mM DTT), then centrifuge at 5000× g for 10 min at 10 °C. Repeat wash 3 times in total. | [22] |
| Wheat Germ | Grind wheat seeds in a mill. | Sieve through 710–850 mm mesh, select embryos via solvent flotation method using a solvent containing 240:600 v/v cyclohexane and carbon tetrachloride. Dry in fume hood overnight. | [194] |
| Yeast | Media: 2% w/v Peptone, 1% w/v Yeast extract, 2% w/v Glucose Vessel: 2.5 L baffled flask Conditions: 30 °C, 250 RPM | When OD600 of 10–12 is reached, centrifuge culture for 10 min at 3000× g. Wash pellet with Buffer A (20 mM HEPES, pH 7.4, 100 mM KOAc, 2 mM Mg(OAc)2). Centrifuge for 10 min at 3000× g and 4 °C. Repeat washing 3 times. | [19] |
| Rabbit Reticulocyte | Make rabbits anemic over 3 days by injections of APH. | Bleed rabbits on day 8. Filter blood through cheesecloth and keep on ice, then centrifuge at 2000 RPM for 10 min. | [98] |
| Insect | Media: Animal component free insect cell medium. Vessel: Fermentor. Conditions: 27 °C | When cell density reaches 4 × 106 cell/mL, centrifuge culture at 200× g for 10 min. Wash once with buffer (40 mM HEPES KOH, pH 7.5, 100 mM KOAc, 4 mM DTT). | [129] |
| HeLa | Media: Minimal essential medium supplemented with 10% heat-inactivated fetal calf serum, 2 mM glutamine, 1 U/mL penicillin, 0.1 mg/mL streptomycin. Vessel: Spinner flask with cell culture controller Conditions: 37 °C, pH 7.2, 67 ppm oxygen, 50 RPM | Harvest when cell density reaches 0.7–0.8 × 106 cells/mL. Wash 3 times with buffer (35 mM HEPES KOH, pH 7.5, 140 mM NaCl, 11 mM glucose). | [13] |
| Chinese Hamster Ovary | Media: Power Chinese hamster ovary-2 chemically defined serum-free media Vessel: Fermentor Conditions: 37 °C | Harvest at 2 × 106 cells/mL cell density by centrifuging at 200× g for 10 min. Wash cells once with buffer (40 mM HEPES KOH, pH 7.5, 100 mM NaOAc, 4 mM DTT). | [17] |
| Extract Prep | |||||
|---|---|---|---|---|---|
| Platform | Pre-Lysis | Lysis | Post-Lysis Processing | Growth and Prep Time | Key Citations |
| E. coli | Resuspend in 1 mL/1 g pellet of S30 buffer by vortexing. | Sonicate on ice for 3 cycles of 45 s on, 59 s off at 50% amplitude. Deliver 800–900 J total for 1.4 mL of resuspended pellet. Supplement with a final concentration of 3 mM DTT. | Centrifuge lysate at 18,000× g and 4 °C for 10 min. Transfer supernatant while avoiding pellet. Perform runoff reaction on supernatant at 37 °C and 250 RPM for 60 min. Centrifuge at 10,000× g and 4 °C for 10 min. Flash freeze supernatant and store at −80 °C. | 1–2 days | [22] |
| Wheat Germ | Wash 3 times with water under vigorous stirring to remove endosperm. | Sonicate for 3 min in 0.5% Nonidet P-40. Wash with sterile water. Grind washed embryos into fine powder in liquid nitrogen and resuspend 5 g in 5mL of 2× Buffer A (40 mM HEPES, pH 7.6, 100 mM KOAc, 5 mM Mg(OAc)2, 2 mM CaCl, 4 mM DTT, 0.3 mM of each of the 20 amino acids). | Centrifuge at 30,000× g for 30 min. Filter supernatant through G-25 column equilibrated with Buffer A. Centrifuge column product at 30,000× g for 10 min. Adjust to 200 A260/mL with Buffer A. Store in liquid nitrogen. | 4–5 days | [64,194] |
| Yeast | Resuspend cells in 1 mL lysis buffer (20 mM HEPES KOH, pH 7.4, 100 mM KOAc, 2 mM Mg(OAc)2, 2 mM DTT, 0.5 mM PMSF) per 1 g cell pellet. | Pass through homogenizer once at 30,000 psig. | Centrifuge homogenate at 30,000× g for 30 min at 4 °C. Then repeat centrifugation with supernatant in a spherical bottom bottle. Desalt supernatant in dialysis tubing with 4 exchanges of 50× volume lysis buffer for 30 min each at 4 °C. Centrifuge at 60,000× g for 20 min at 4 °C. Flash freeze and store at −80 °C. | 1–2 days | [19,195] |
| Rabbit Reticulocyte | Resuspend cells in buffered saline with 5 mM glucose, then centrifuge at 2000 RPM for 10 min. Repeat wash 3 times. | Resuspend cells in 1.5 times the packed cell volume of ice-cold water, mix thoroughly. | Spin lysate at 15,000× g for 20 min at 2 °C. Pour supernatant though 53 μm nylon. Treat with micrococcal nuclease by adding 0.2 mL of 1 mM hemin, 0.1 mL of 5 mg/mL creatine kinase, 0.1 mL of 0.1 M CaCl2, 0.1 mL of micrococcal nuclease. Incubate at 20 °C for 15 min, then add 0.1 mL of 0.2 M EGTA and 60 μL of 10 mg/mL tRNA. Store in liquid nitrogen or at −70 °C. | ~8 days to treat rabbits, 1 day for extract preparation | [98] |
| Insect | Resuspend cells in buffer to final density of 2 × 108 cells/mL. | Mechanically lyse cells by rapidly freezing in liquid nitrogen, then thawing in water bath at 4 °C or use a disruption chamber with 20 kg/cm2 nitrogen gas for 30 min. | Centrifuge lysate at 10,000× g for 10 min. Apply supernatant to G-25 gel filtration column. Pool fractions with highest A260, flash freeze in liquid nitrogen and store at −80 °C. | 1–2 days | [18,129,195,196] |
| HeLa | Resuspend in extraction buffer (20 mM HEPES KOH, pH 7.5, 135 mM KOAc, 30 mM KCl, 1.655 mM Mg(OAc)2) to ~2.3 × 108 cells/mL. | Disrupt cells via 1 MPa nitrogen pressure for 30 min in a cell disruption chamber. | Centrifuge homogenate at 10,000× g for 5 min at 4 °C. Pass supernatant through a PD-10 desalting column equilibrated with extraction buffer. Treat 100 μL of extract with 1 μL of 7500 U/mL nuclease S7 and 1 μL of 100 mM CaCl2 for 5 min at 23 °C, then add 8 μL of 30 mM EGTA. Flash freeze eluted extract in liquid nitrogen and store at −80 °C. | 1–2 days | [13,195] |
| Chinese Hamster Ovary | Resuspend cells in buffer to density of 5 × 108 cells/mL. | Disrupt cells by syringing the pellet through a 20-gauge needle. | Centrifuge lysate at 10,000× g for 10 min. Apply supernatant to G-25 gel filtration column. Pool fractions with an A260 above 100. Treat pooled fractions with 10 U/mL S7 nuclease and 1 mM CaCl2, incubate at room temperature for 2 min, then add 6.7 mM EGTA. Flash freeze in liquid nitrogen and store at −80 °C. | 1–2 days | [17] |
| Cell-Free Protein Synthesis Reaction | ||||
|---|---|---|---|---|
| Platform | Vessel/Conditions | Reaction Composition | Energy Systems | Key Citations |
| E. coli | Vessel: Many vessels can be used, yield increases as the surface area to reaction volume ratio increases Conditions: 30 °C overnight or 37 °C for 4 h | 33% v/v E. coli extract, 16 μg/mL T7RNAP, 16 ng/mL DNA template, Solution A (1.2 mM ATP, 0.85 mM GTP, 0.85 mM UTP, 0.85 mM CTP, 31.50 μg/mL Folinic Acid, 170.60 μg/mL tRNA, 0.40 mM Nicotinamide Adenine Dinucleotide (NAD), 0.27 mM Coenzyme A (CoA), 4 mM Oxalic Acid, 1 mM Putrescine, 1.50 mM Spermidine, 57.33 mM HEPES buffer), Solution B (10 mM Mg(Glu)2, 10 mM NH4(Glu), 130 mM K(Glu), 2 mM of each amino acid, 0.03 M Phosphoenolpyruvate (PEP)) | PEP, glucose + glutamate decarboxylase, or maltodextrin are possible | [22,201] |
| Wheat Germ | Vessel: Not noted Conditions: 26 °C | First, perform an in vitro transcription reaction and isolate mRNA using SP6 RNA polymerase. Set up cell-free translation as follows: 24% v/v wheat germ extract, 4 mM HEPES KOH, pH 7.8, 1.2 mM ATP, 0.25 mM GTP, 16 mM creatine phosphate, 0.45 mg/mL creatine kinase, 2 mM DTT, 0.4 mM spermidine, 0.3 mM of each of the 20 amino acids, 2.5 mM Mg(OAc)2, 100 mM KOAc, 50 μg/mL deacylated tRNA from wheat embryos, 0.05% Nonidet P-40, 1 μM E-64 as proteinase inhibitor, 0.005% NaN3, 0.02 nmol mRNA. | Creatine phosphate + creatine kinase | [194] |
| Yeast | Vessel: 15 μL reactions in 1.5 mL microfuge tubes Conditions: 21 °C | 25 mM HEPES KOH, pH 7.4, 120 mM K(Glu), 6 mM Mg(Glu)2, 1.5 mM ATP, 2 mM GTP, 2 mM CTP, 2 mM UTP, 0.1 mM of each of 20 amino acids, 25 mM creatine phosphate, 2 mM DTT, 0.27 mg/mL creatine phosphokinase, 200 U/mL RNase Inhibitor, 27 μg/mL T7 RNAP, DNA template, and 50% v/v yeast extract | Creatine phosphate + creatine kinase | [19,195] |
| Rabbit Reticulocyte | Vessel: 200 μL reaction performed in an NMR spectrometer Conditions: 30 °C | First, perform an in vitro transcription reaction and isolate mRNA using T7 RNAP. Supplement 1 mL of rabbit reticulocyte lysate with 25 μM hemin, 25 μg creatine kinase, 5 mg phosphocreatine, 50 μg of bovine liver tRNAs, and 2 mM D-glucose. Initiate in vitro translation by combining 27 nM of in vitro transcribed mRNAs, 50% v/v supplemented lysate, 75 mM KCl, 0.75 mM MgCl2, and 20 μM amino acids mix. | Creatine phosphate + creatine kinase | [135] |
| Insect | Vessel: 25 μL reaction, vessel size not noted Conditions: 25 °C | First, perform an in vitro transcription reaction and isolate mRNA using T7 RNAP. Then set up cell-free translation as follows: 1.5 mM Mg(OAc)2, 0.25 mM ATP, 0.1 mM GTP, 0.1 mM EGTA, 40 mM HEPES KOH, pH 7.9, 100 mM KOAc, 20 mM creatine phosphate, 200 μg/mL creatine kinase, 2 mM DTT, 80 μM of each of the 20 amino acids, 0.5 mM PMSF, 1 U/µL RNase inhibitor, 200 μg/mL tRNA, 320 μg/mL mRNA, and 50% v/v insect cell extract. Addition of 20% v/v glycerol to the reaction was also shown to improve yields. | Creatine phosphate + creatine kinase | [18] |
| HeLa | Vessel: 6 μL reaction, vessel not noted Conditions: 32 °C, 1 h | First, perform an in vitro transcription reaction and isolate mRNA using T7 RNAP. Cell-free translation is performed as follows: 75% v/v HeLa cell extract, 30 μM of each of the 20 amino acids, 27 mM HEPES KOH, pH 7.5, 1.2 mM ATP, 0.12 mM GTP, 18 mM creatine phosphate, 0.3 mM spermidine, 44–224 mM KOAc, 16 mM KCl, 1.2 mM Mg(OAc)2, 90 μg/mL calf liver tRNA, 60 μg/mL creatine kinase, and purified mRNA. | Creatine phosphate + creatine kinase | [13] |
| Chinese Hamster Ovary | Vessel: 25 μL reaction, vessel size not noted Conditions: 33 °C, 500 RPM shaking in thermomixer | 25% v/v Chinese hamster ovary cell extract, 100 μM of each of the 20 amino acids, 1.75 mM ATP, 0.30 mM CTP, 0.30 mM GTP, 0.30 mM UTP, 20 nM DNA template, 1 U/μL T7 RNAP, 30 mM HEPES KOH, pH 7.6, 150 mM KOAc, 3.9 mM Mg(OAc)2, 20 mM creatine phosphate, 100 μg/mL creatine kinase, 0.25 mM spermidine, and 2.5 mM DTT. | Creatine phosphate + creatine kinase | [17] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gregorio, N.E.; Levine, M.Z.; Oza, J.P. A User’s Guide to Cell-Free Protein Synthesis. Methods Protoc. 2019, 2, 24. https://doi.org/10.3390/mps2010024
Gregorio NE, Levine MZ, Oza JP. A User’s Guide to Cell-Free Protein Synthesis. Methods and Protocols. 2019; 2(1):24. https://doi.org/10.3390/mps2010024
Chicago/Turabian StyleGregorio, Nicole E., Max Z. Levine, and Javin P. Oza. 2019. "A User’s Guide to Cell-Free Protein Synthesis" Methods and Protocols 2, no. 1: 24. https://doi.org/10.3390/mps2010024
APA StyleGregorio, N. E., Levine, M. Z., & Oza, J. P. (2019). A User’s Guide to Cell-Free Protein Synthesis. Methods and Protocols, 2(1), 24. https://doi.org/10.3390/mps2010024