A Combined Region- and Pixel-Based Deep Learning Approach for Quantifying Abdominal Adipose Tissue in Adolescents Using Dixon Magnetic Resonance Imaging
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. MRI Image Acquisition
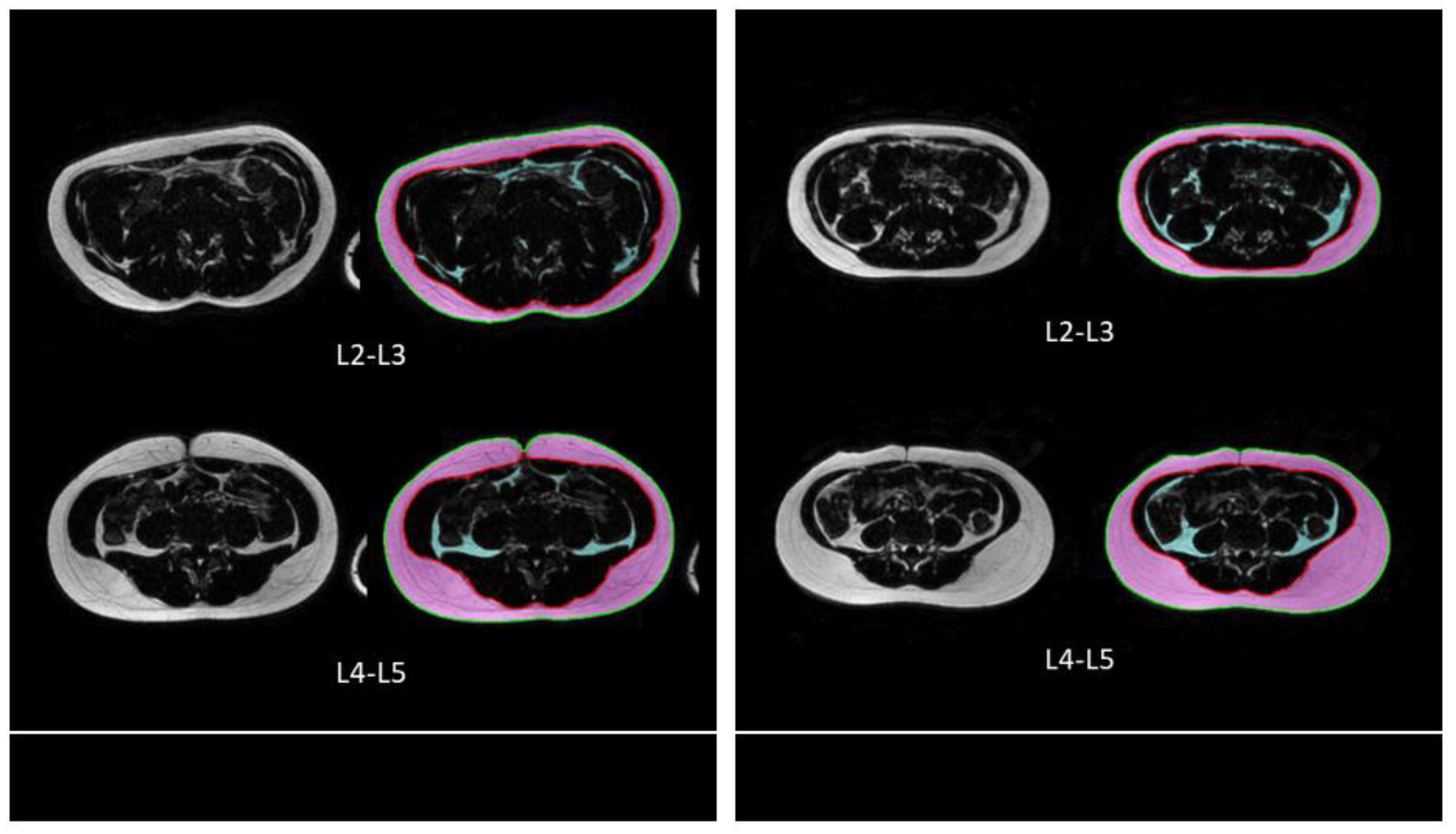
2.3. Image Analysis
2.4. CNN Approach
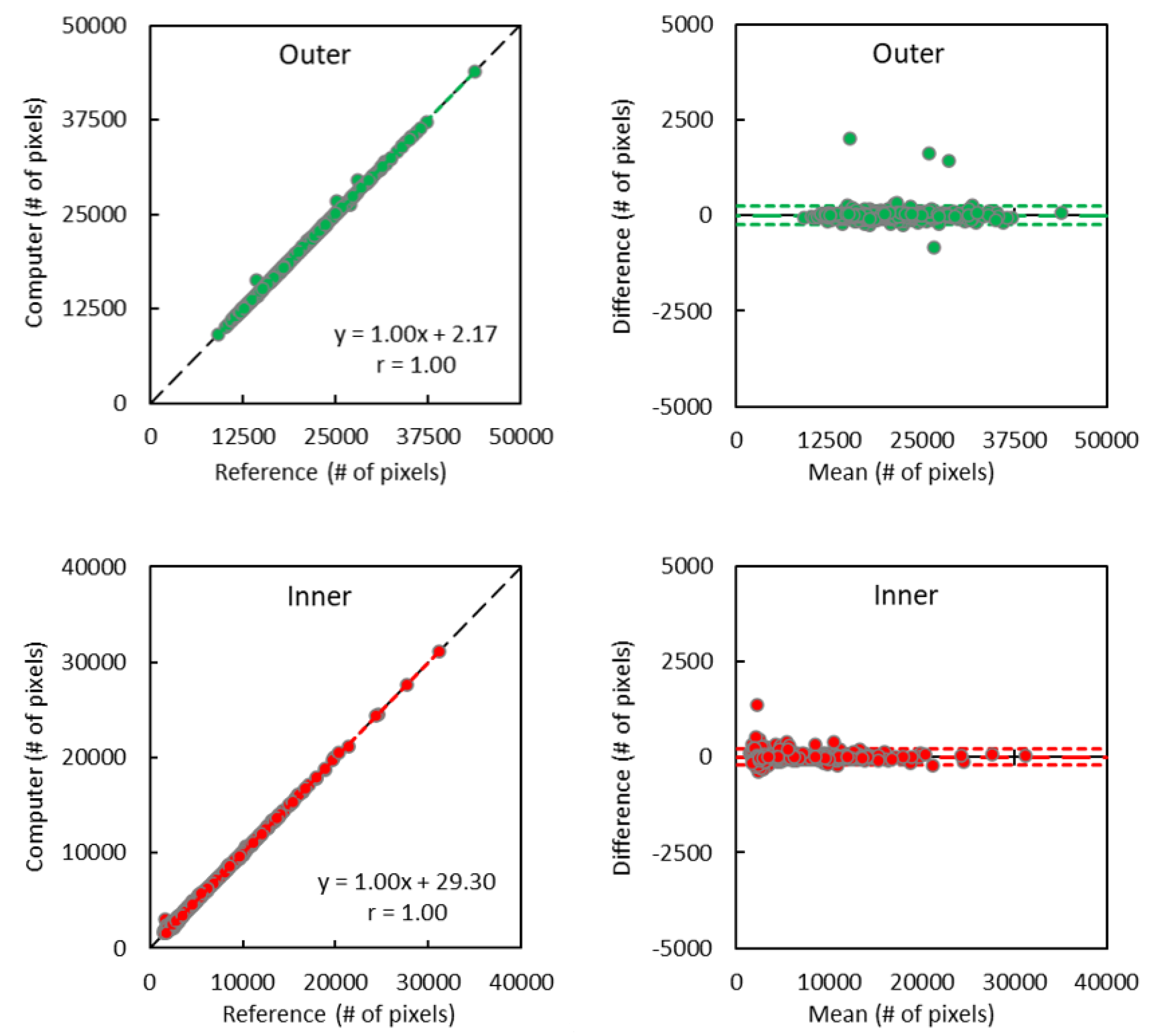
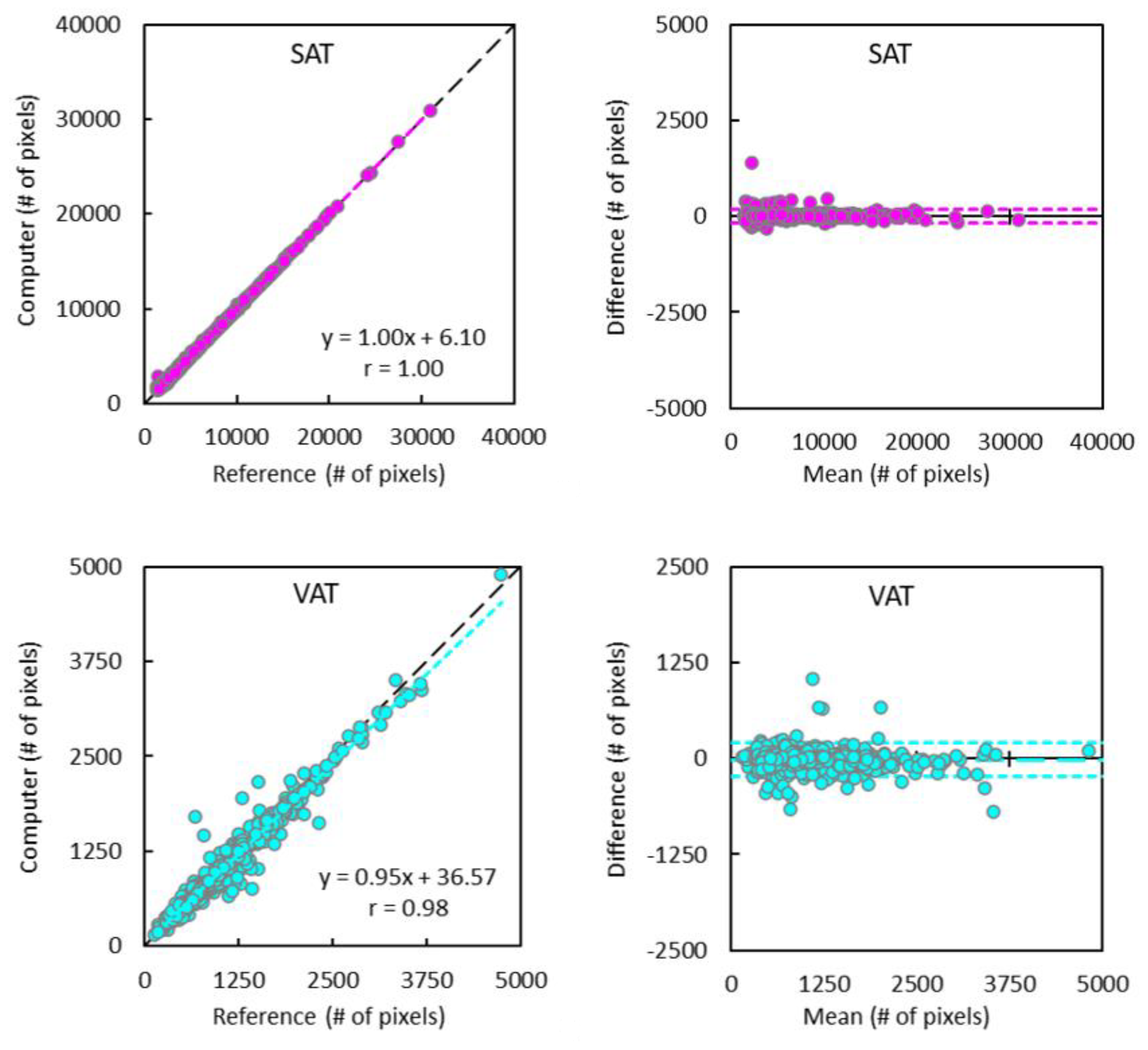
2.5. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 2D | two-dimensional |
| 3D | three-dimensional |
| BMI | body mass index |
| CNN | convolutional neural network |
| CT | computed tomography |
| DEXA | dual-energy X-ray absorptiometry |
| DSC | dice similarity coefficient |
| MRI | magnetic resonance imaging |
| NS | not statistically significant |
| SAT | subcutaneous adipose tissue |
| SD | standard deviation |
| SEM | standard error of the mean |
| VAT | visceral adipose tissue |
References
- Greenberg, A.S.; Obin, M.S. Obesity and the role of adipose tissue in inflammation and metabolism. Am. J. Clin. Nutr. 2006, 83, 461S–465S. [Google Scholar] [CrossRef]
- Peng, Q.; McColl, R.W.; Ding, Y.; Wang, J.; Chia, J.M.; Weatherall, P.T. Automated method for accurate abdominal fat quantification on water-saturated magnetic resonance images. J. Magn. Reson. Imaging 2007, 26, 738–746. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G. Quantitative Imaging of Body Fat Distribution in the Era of Deep Learning. Acad. Radiol. 2021, 28, 1488–1490. [Google Scholar] [CrossRef] [PubMed]
- Ng, M.; Fleming, T.; Robinson, M.; Thomson, B.; Graetz, N.; Margono, C.; Mullany, E.C.; Biryukov, S.; Abbafati, C.; Abera, S.F.; et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet 2014, 384, 766–781. [Google Scholar] [CrossRef] [PubMed]
- Després, J.P.; Lemieux, I. Abdominal obesity and metabolic syndrome. Nature 2006, 444, 881–887. [Google Scholar] [CrossRef] [PubMed]
- Magudia, K.; Bridge, C.P.; Bay, C.P.; Farah, S.; Babic, A.; Fintelmann, F.J.; Brais, L.K.; Andriole, K.P.; Wolpin, B.M.; Rosenthal, M.H. Utility of normalized body composition areas, derived from outpatient abdominal CT using a fully automated deep learning method, for predicting subsequent cardiovascular Events. Am. J. Roentgenol. 2022, 1–9. [Google Scholar] [CrossRef]
- Raj, M.; Kumar, R.K. Obesity in children & adolescents. Indian J. Med. Res. 2010, 132, 598. [Google Scholar]
- Cali, A.M.; Caprio, S. Obesity in children and adolescents. J. Clin. Endocrinol. Metab. 2008, 93, s31–s36. [Google Scholar] [CrossRef]
- Whitaker, R.C.; Wright, J.A.; Pepe, M.S.; Seidel, K.D.; Dietz, W.H. Predicting obesity in young adulthood from childhood and parental obesity. N. Engl. J. Med. 1997, 337, 869–873. [Google Scholar] [CrossRef]
- Kway, Y.M.; Thirumurugan, K.; Tint, M.T.; Michael, N.; Shek, L.P.; Yap, F.K.; Tan, K.H.; Godfrey, K.M.; Chong, Y.S.; Fortier, M.V.; et al. Automated Segmentation of Visceral, Deep Subcutaneous, and Superficial Subcutaneous Adipose Tissue Volumes in MRI of Neonates and Young Children. Radiol. Artif. Intell. 2021, 3, e200304. [Google Scholar] [CrossRef]
- Marunowski, K.; Świętoń, D.; Bzyl, W.; Grzywińska, M.; Kaszubowski, M.; Bandosz, P.; Khrichenko, D.; Piskunowicz, M. MRI-Derived Subcutaneous and Visceral Adipose Tissue Reference Values for Children Aged 6 to Under 18 Years. Front. Nutr. 2021, 8, 757274. [Google Scholar] [CrossRef] [PubMed]
- Greco, F.; Mallio, C.A. Artificial intelligence and abdominal adipose tissue analysis: A literature review. Quant. Imaging Med. Surg. 2021, 11, 4461. [Google Scholar] [CrossRef] [PubMed]
- De Larochellière, E.; Côté, J.; Gilbert, G.; Bibeau, K.; Ross, M.K.; Dion-Roy, V.; Pibarot, P.; Després, J.P.; Larose, É. Visceral/epicardial adiposity in nonobese and apparently healthy young adults: Association with the cardiometabolic profile. Atherosclerosis 2014, 234, 23–29. [Google Scholar] [CrossRef]
- Ibrahim, M.M. Subcutaneous and visceral adipose tissue: Structural and functional differences. Obes. Rev. 2010, 11, 11–18. [Google Scholar] [CrossRef]
- Fox, C.S.; Massaro, J.M.; Hoffmann, U.; Pou, K.M.; Maurovich-Horvat, P.; Liu, C.Y.; Vasan, R.S.; Murabito, J.M.; Meigs, J.B.; Cupples, L.A.; et al. Abdominal visceral and subcutaneous adipose tissue compartments: Association with metabolic risk factors in the Framingham Heart Study. Circulation 2007, 116, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Bjorntorp, P. Metabolic implications of body fat distribution. Diabetes Care 1991, 14, 1132–1143. [Google Scholar] [CrossRef] [PubMed]
- Sumner, A.E.; Bagheri, M.H. Identifying the waist circumference of risk in people of African descent. Nat. Rev. Endocrinol. 2020, 16, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Brambilla, P.; Bedogni, G.; Moreno, L.A.; Goran, M.I.; Gutin, B.; Fox, K.R.; Peters, D.M.; Barbeau, P.; De Simone, M.; Pietrobelli, A. Crossvalidation of anthropometry against magnetic resonance imaging for the assessment of visceral and subcutaneous adipose tissue in children. Int. J. Obes. 2006, 30, 23–30. [Google Scholar] [CrossRef]
- Borga, M.; West, J.; Bell, J.D.; Harvey, N.C.; Romu, T.; Heymsfield, S.B.; Leinhard, O.D. Advanced body composition assessment: From body mass index to body composition profiling. J. Investig. Med. 2018, 66, 1–9. [Google Scholar] [CrossRef]
- Dixon, W.T. Simple proton spectroscopic imaging. Radiology 1984, 153, 189–194. [Google Scholar] [CrossRef]
- Ma, J. Dixon techniques for water and fat imaging. J. Magn. Reson. Imaging 2008, 28, 543–558. [Google Scholar] [CrossRef] [PubMed]
- Cole, K.M.; Wei, S.M.; Martinez, P.E.; Nguyen, T.V.; Gregory, M.D.; Kippenhan, J.S.; Kohn, P.D.; Soldin, S.J.; Nieman, L.K.; Yanovski, J.A.; et al. The NIMH Intramural Longitudinal Study of the Endocrine and Neurobiological Events Accompanying Puberty: Protocol and rationale for methods and measures. NeuroImage 2021, 234, 117970. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Fidon, L.; Li, W.; Garcia-Peraza-Herrera, L.C.; Ekanayake, J.; Kitchen, N.; Ourselin, S.; Vercauteren, T. Generalised wasserstein dice score for imbalanced multi-class segmentation using holistic convolutional networks. In Proceedings of the International MICCAI Brainlesion Workshop 2017, Quebec City, QC, Canada, 14 September 2017; Springer: Cham, Switzerland, 2017; pp. 64–76. [Google Scholar]
- Borga, M. MRI adipose tissue and muscle composition analysis—A review of automation techniques. Br. J. Radiol. 2018, 91, 20180252. [Google Scholar] [CrossRef]
- Newman, D.; Kelly-Morland, C.; Leinhard, O.D.; Kasmai, B.; Greenwood, R.; Malcolm, P.N.; Romu, T.; Borga, M.; Toms, A.P. Test–retest reliability of rapid whole body and compartmental fat volume quantification on a widebore 3T MR system in normal-weight, overweight, and obese subjects. J. Magn. Reson. Imaging 2016, 44, 1464–1473. [Google Scholar] [CrossRef]
- Leinhard, O.D.; Johansson, A.; Rydell, J.; Smedby, O.; Nystrom, F.; Lundberg, P.; Borga, M. Quantitative abdominal fat estimation using MRI. In Proceedings of the 19th International Conference on Pattern Recognition, Tampa, FL, USA, 8–11 December 2008; pp. 1–4. [Google Scholar]
- Sussman, D.L.; Yao, J.; Summers, R.M. Automated fat measurement and segmentation with intensity inhomogeneity correction. In Medical Imaging 2010: Image Processing; International Society for Optics and Photonics: San Diego, CA, USA, 2010; Volume 7623. [Google Scholar]
- Zhou, A.; Murillo, H.; Peng, Q. Novel segmentation method for abdominal fat quantification by MRI. J. Magn. Reson. Imaging 2011, 34, 852–860. [Google Scholar] [CrossRef]
- Hu, H.H.; Chen, J.; Shen, W. Segmentation and quantification of adipose tissue by magnetic resonance imaging. Magn. Reson. Mater. Phys. Biol. Med. 2016, 29, 259–276. [Google Scholar] [CrossRef]
- Christensen, A.N.; Larsen, C.T.; Mandrup, C.M.; Petersen, M.B.; Larsen, R.; Conradsen, K.; Dahl, V.A. Automatic segmentation of abdominal fat in MRI-Scans, using graph-cuts and image derived energies. In Scandinavian Conference on Image Analysis; Springer: Tromso, Norway, 2017; pp. 109–120. [Google Scholar]
- Chaudry, O.; Grimm, A.; Friedberger, A.; Kemmler, W.; Uder, M.; Jakob, F.; Quick, H.H.; Stengel, S.; Engelke, K. Magnetic Resonance Imaging and Bioelectrical Impedance Analysis to Assess Visceral and Abdominal Adipose Tissue. Obesity 2020, 28, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Estrada, S.; Lu, R.; Conjeti, S.; Orozco-Ruiz, X.; Panos-Willuhn, J.; Breteler, M.M.; Reuter, M. FatSegNet: A fully automated deep learning pipeline for adipose tissue segmentation on abdominal dixon MRI. Magn. Reson. Med. 2020, 83, 1471–1483. [Google Scholar] [CrossRef] [PubMed]
- Langner, T.; Hedström, A.; Mörwald, K.; Weghuber, D.; Forslund, A.; Bergsten, P.; Ahlström, H.; Kullberg, J. Fully convolutional networks for automated segmentation of abdominal adipose tissue depots in multicenter water–fat MRI. Magn. Reson. Med. 2019, 81, 2736–2745. [Google Scholar] [CrossRef]
- Freedman, D.S.; Khan, L.K.; Dietz, W.H.; Srinivasan, S.R.; Berenson, G.S. Relationship of childhood obesity to coronary heart disease risk factors in adulthood: The Bogalusa Heart Study. Pediatrics 2001, 108, 712–718. [Google Scholar] [CrossRef]
- Pinhas-Hamiel, O.; Dolan, L.M.; Daniels, S.R.; Standiford, D.; Khoury, P.R.; Zeitler, P. Increased incidence of non-insulin-dependent diabetes mellitus among adolescents. J. Pediatr. 1996, 128, 608–615. [Google Scholar] [CrossRef] [PubMed]
- Demerath, E.W.; Shen, W.; Lee, M.; Choh, A.C.; Czerwinski, S.A.; Siervogel, R.M.; Towne, B. Approximation of total visceral adipose tissue with a single magnetic resonance image. Am. J. Clin. Nutr. 2007, 85, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Maislin, G.; Ahmed, M.M.; Gooneratne, N.; Thorne-Fitzgerald, M.; Kim, C.; Teff, K.; Arnardottir, E.S.; Benediktsdottir, B.; Einarsdottir, H.; Juliusson, S.; et al. Single slice vs. volumetric MR assessment of visceral adipose tissue: Reliability and validity among the overweight and obese. Obesity 2012, 20, 2124–2132. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Rao, S.X.; Chen, C.Z.; Li, R.C.; Zeng, M.S. Usefulness of two-point Dixon fat-water separation technique in gadoxetic acid-enhanced liver magnetic resonance imaging. World J. Gastroenterol. 2015, 21, 5017–5022. [Google Scholar] [CrossRef] [PubMed]

| Participants = 136 | ||||||
| Gender | Boy | Girl | ||||
| 78 | 58 | |||||
| Race/Ethnicity | White | Black | Asian | Hispanic | Mixed | Other |
| 68 | 23 | 12 | 10 | 14 | 9 | |
| MRI Scans = 474 | ||||||
| Range | Mean | SD | SEM | Min | Max | |
| Age (years) | 11 | 3 | 0.12 | 8 | 18 | |
| Height (cm) | 149.98 | 15.43 | 0.71 | 121.10 | 190.00 | |
| Weight (kg) | 42.12 | 13.38 | 0.61 | 20.10 | 89.50 | |
| BMI (kg/m2) | 18.19 | 2.34 | 0.11 | 13.20 | 25.19 | |
| BMI SD Score * | 0.14 | 0.62 | 0.03 | −1.98 | 1.58 | |
| BMI Percentile * (%) | 54.77 | 21.48 | 0.99 | 2.37 | 94.33 | |
| Body Fat Percentage † (%) | 26.01 | 5.87 | 0.28 | 13.67 | 46.61 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ogunleye, O.A.; Raviprakash, H.; Simmons, A.M.; Bovell, R.T.M.; Martinez, P.E.; Yanovski, J.A.; Berman, K.F.; Schmidt, P.J.; Jones, E.C.; Bagheri, H.; et al. A Combined Region- and Pixel-Based Deep Learning Approach for Quantifying Abdominal Adipose Tissue in Adolescents Using Dixon Magnetic Resonance Imaging. Tomography 2023, 9, 139-149. https://doi.org/10.3390/tomography9010012
Ogunleye OA, Raviprakash H, Simmons AM, Bovell RTM, Martinez PE, Yanovski JA, Berman KF, Schmidt PJ, Jones EC, Bagheri H, et al. A Combined Region- and Pixel-Based Deep Learning Approach for Quantifying Abdominal Adipose Tissue in Adolescents Using Dixon Magnetic Resonance Imaging. Tomography. 2023; 9(1):139-149. https://doi.org/10.3390/tomography9010012
Chicago/Turabian StyleOgunleye, Olanrewaju A., Harish Raviprakash, Ashlee M. Simmons, Rhasaan T.M. Bovell, Pedro E. Martinez, Jack A. Yanovski, Karen F. Berman, Peter J. Schmidt, Elizabeth C. Jones, Hadi Bagheri, and et al. 2023. "A Combined Region- and Pixel-Based Deep Learning Approach for Quantifying Abdominal Adipose Tissue in Adolescents Using Dixon Magnetic Resonance Imaging" Tomography 9, no. 1: 139-149. https://doi.org/10.3390/tomography9010012
APA StyleOgunleye, O. A., Raviprakash, H., Simmons, A. M., Bovell, R. T. M., Martinez, P. E., Yanovski, J. A., Berman, K. F., Schmidt, P. J., Jones, E. C., Bagheri, H., Biassou, N. M., & Hsu, L.-Y. (2023). A Combined Region- and Pixel-Based Deep Learning Approach for Quantifying Abdominal Adipose Tissue in Adolescents Using Dixon Magnetic Resonance Imaging. Tomography, 9(1), 139-149. https://doi.org/10.3390/tomography9010012






