Detergent-Based Decellularization for Anisotropic Cardiac-Specific Extracellular Matrix Scaffold Generation
Abstract
:1. Introduction
2. Materials and Methods
2.1. hiPSC-CFs Differentiation and Culture
2.2. Fabrication of Anisotropic Cardiac Fibroblast Cell Sheets
2.3. Decellularization Methods to Fabricate hiPSC-CF-ECM
2.4. Morphological Analysis of ECM Key Structural Proteins
2.5. ECM Components Characterization
2.6. hiPSC-CMs Differentiation and Culture
2.7. hiPSC-CMs Immunofluorescence Staining and Imaging
2.8. hiPSC-CMs Beating Analysis
2.9. Statistical Analysis
3. Results
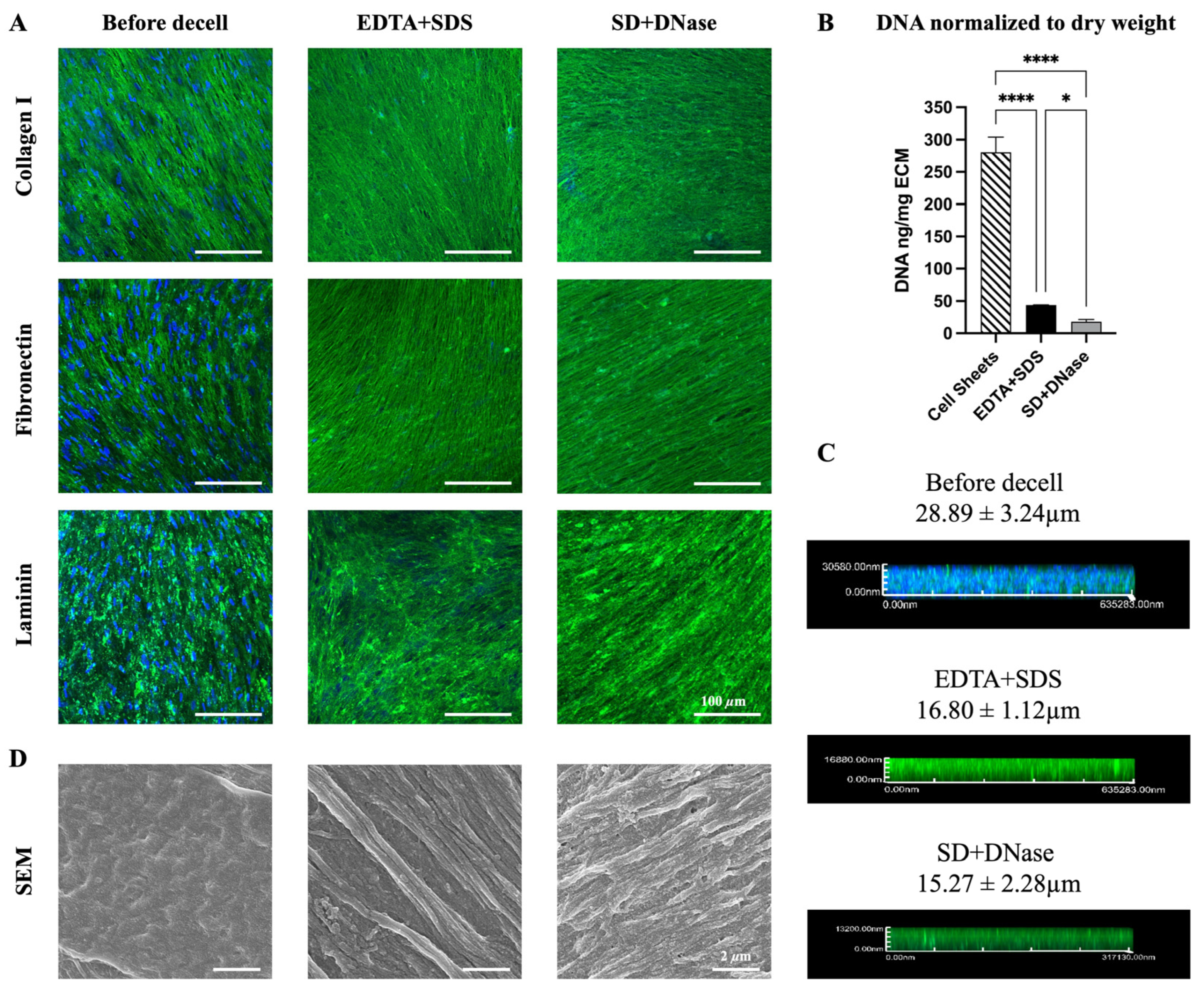
3.1. Characterization of ECM before and after Decellularization
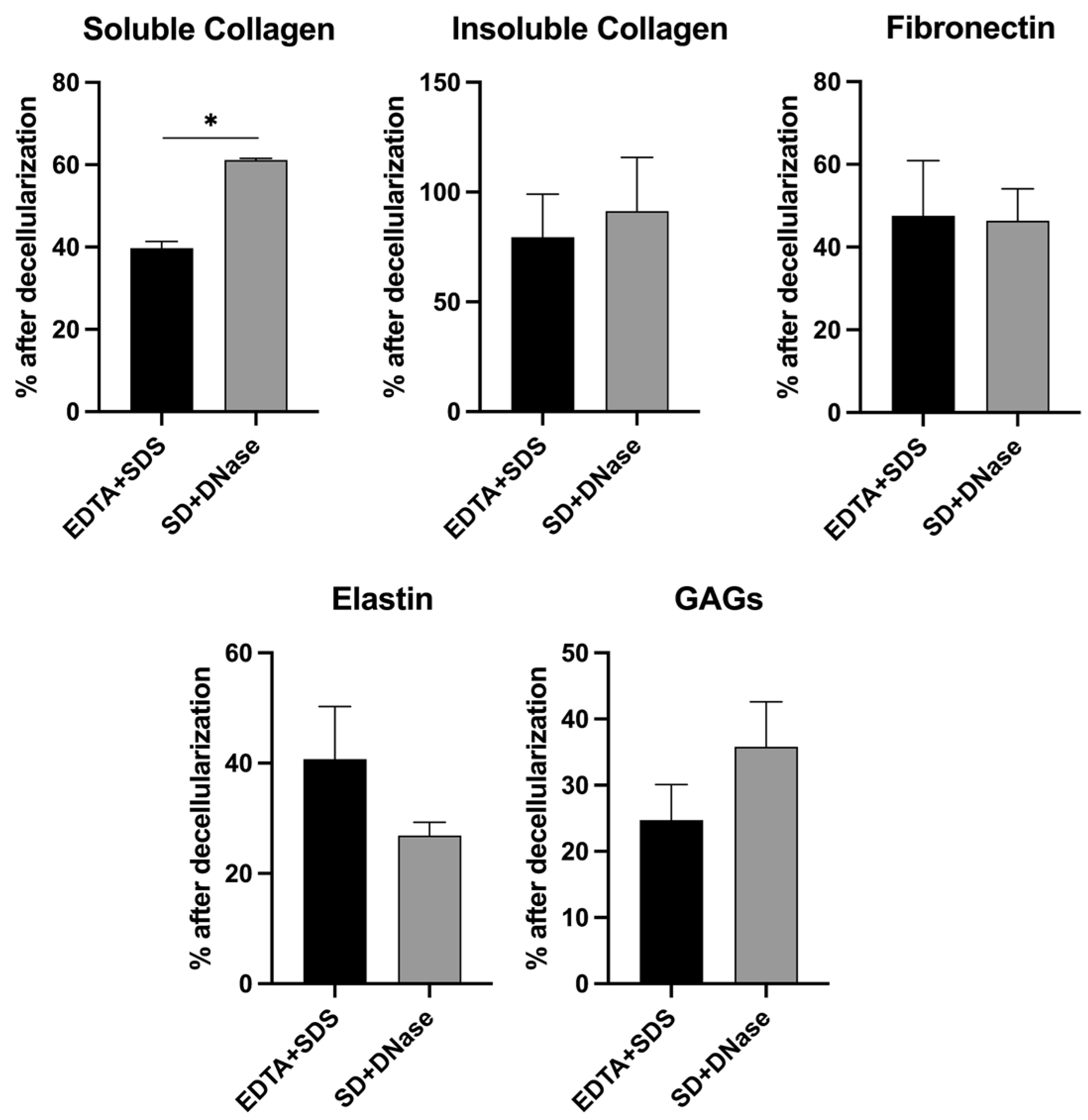
3.2. Quantification of Major ECM Composition
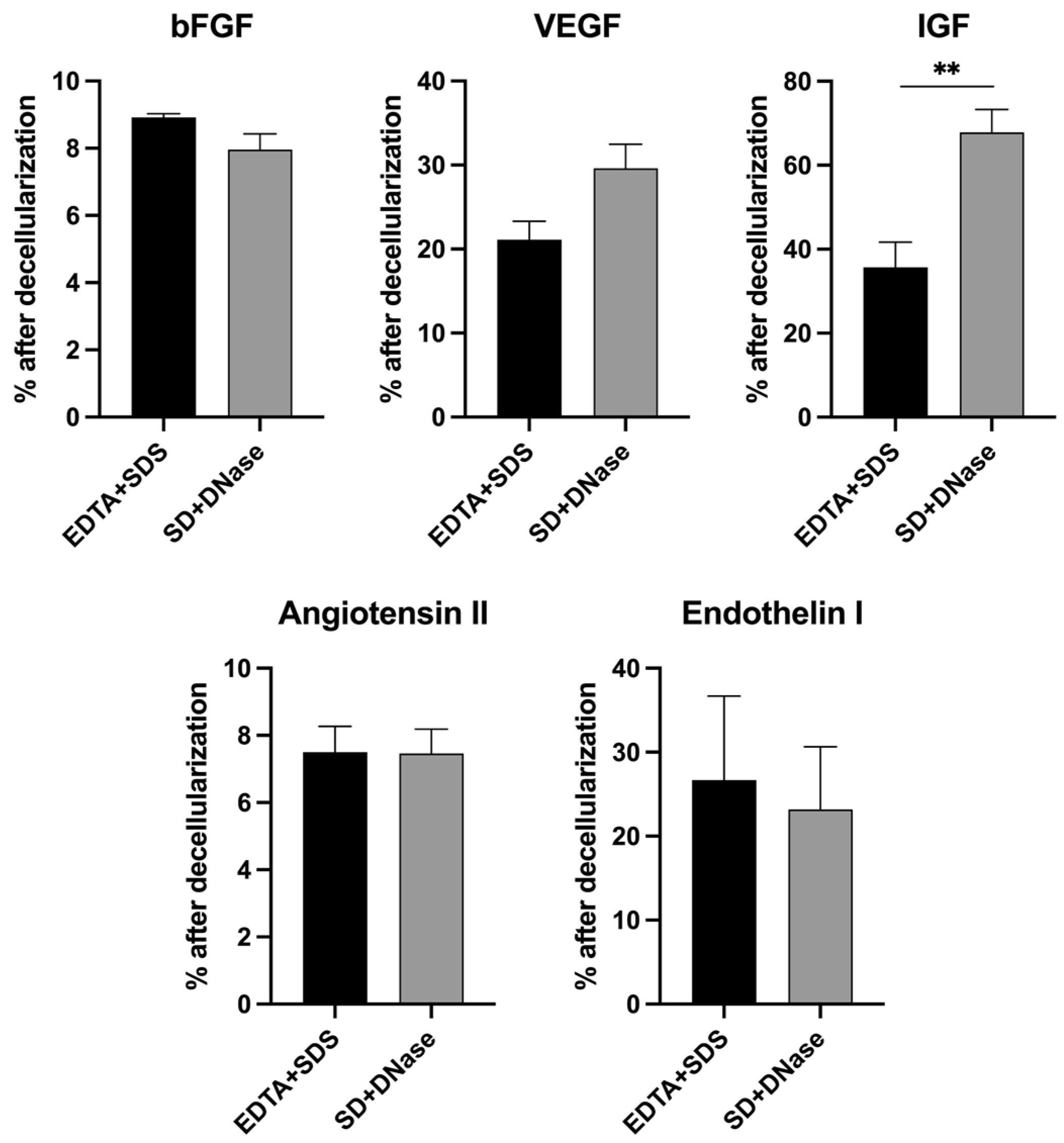
3.3. Quantification of Major ECM Growth Factors
3.4. The Structural and Functional Maturation of hiPSC-CMs on ECM Scaffold
3.5. Beating Analysis of hiPSC-CMs on ECM Scaffold
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qian, Z.; Radke, D.; Jia, W.; Tahtinen, M.; Wang, G.; Zhao, F. Bioengineering scaffolds for regenerative engineering. Encycl. Biomed. Eng. 2018, 1, 444. [Google Scholar]
- Guan, X.; Xu, W.; Zhang, H.; Wang, Q.; Yu, J.; Zhang, R.; Chen, Y.; Xia, Y.; Wang, J.; Wang, D. Transplantation of human induced pluripotent stem cell-derived cardiomyocytes improves myocardial function and reverses ventricular remodeling in infarcted rat hearts. Stem Cell Res. Ther. 2020, 11, 73. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. Engineered tissue patch for cardiac cell therapy. Curr. Treat. Options Cardiovasc. Med. 2015, 17, 399. [Google Scholar] [CrossRef] [PubMed]
- Shadrin, I.Y.; Allen, B.W.; Qian, Y.; Jackman, C.P.; Carlson, A.L.; Juhas, M.E.; Bursac, N. Cardiopatch platform enables maturation and scale-up of human pluripotent stem cell-derived engineered heart tissues. Nat. Commun. 2017, 8, 1825. [Google Scholar] [CrossRef]
- Gao, L.; Gregorich, Z.R.; Zhu, W.; Mattapally, S.; Oduk, Y.; Lou, X.; Kannappan, R.; Borovjagin, A.V.; Walcott, G.P.; Pollard, A.E. Large cardiac muscle patches engineered from human induced-pluripotent stem cell–derived cardiac cells improve recovery from myocardial infarction in swine. Circulation 2018, 137, 1712–1730. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Min, S.; Choi, Y.S.; Jo, S.-H.; Jung, J.H.; Han, K.; Kim, J.; An, S.; Ji, Y.W.; Kim, Y.-G. Tissue extracellular matrix hydrogels as alternatives to Matrigel for culturing gastrointestinal organoids. Nat. Commun. 2022, 13, 1692. [Google Scholar] [CrossRef]
- Schaefer, J.A.; Guzman, P.A.; Riemenschneider, S.B.; Kamp, T.J.; Tranquillo, R.T. A cardiac patch from aligned microvessel and cardiomyocyte patches. J. Tissue Eng. Regen. Med. 2018, 12, 546–556. [Google Scholar] [CrossRef]
- Sharma, D.; Ferguson, M.; Kamp, T.J.; Zhao, F. Constructing biomimetic cardiac tissues: A review of scaffold materials for engineering cardiac patches. Emergent Mater. 2019, 2, 181–191. [Google Scholar] [CrossRef]
- Watt, F.M.; Huck, W.T. Role of the extracellular matrix in regulating stem cell fate. Nat. Rev. Mol. Cell Biol. 2013, 14, 467–473. [Google Scholar] [CrossRef]
- Assunção, M.; Dehghan-Baniani, D.; Yiu, C.H.K.; Später, T.; Beyer, S.; Blocki, A. Cell-derived extracellular matrix for tissue engineering and regenerative medicine. Front. Bioeng. Biotechnol. 2020, 8, 602009. [Google Scholar] [CrossRef]
- Beachley, V.Z.; Wolf, M.T.; Sadtler, K.; Manda, S.S.; Jacobs, H.; Blatchley, M.R.; Bader, J.S.; Pandey, A.; Pardoll, D.; Elisseeff, J.H. Tissue matrix arrays for high-throughput screening and systems analysis of cell function. Nat. Methods 2015, 12, 1197–1204. [Google Scholar] [CrossRef] [PubMed]
- Frangogiannis, N.G. The extracellular matrix in ischemic and nonischemic heart failure. Circ. Res. 2019, 125, 117–146. [Google Scholar] [CrossRef] [PubMed]
- Ariyasinghe, N.R.; Lyra-Leite, D.M.; McCain, M.L. Engineering cardiac microphysiological systems to model pathological extracellular matrix remodeling. Am. J. Physiol.-Heart Circ. Physiol. 2018, 315, H771–H789. [Google Scholar] [CrossRef] [PubMed]
- Walker, C.A.; Spinale, F.G. The structure and function of the cardiac myocyte: A review of fundamental concepts. J. Thorac. Cardiovasc. Surg. 1999, 118, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Tao, R.; Campbell, K.F.; Carvalho, J.L.; Ruiz, E.C.; Kim, G.C.; Schmuck, E.G.; Raval, A.N.; da Rocha, A.M.; Herron, T.J. Functional cardiac fibroblasts derived from human pluripotent stem cells via second heart field progenitors. Nat. Commun. 2019, 10, 2238. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.-H.; Wang, Y.-C.; Liu, S.-P.; Shih, T.-R.; Lin, H.-L.; Chen, Y.-M.; Sung, J.-H.; Lu, C.-H.; Wei, J.-R.; Wang, Z.-W. Decellularization and recellularization technologies in tissue engineering. Cell Transplant. 2014, 23, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Mertsch, S.; Hasenzahl, M.; Reichl, S.; Geerling, G.; Schrader, S. Decellularized human corneal stromal cell sheet as a novel matrix for ocular surface reconstruction. J. Tissue Eng. Regen. Med. 2020, 14, 1318–1332. [Google Scholar] [CrossRef] [PubMed]
- Polisetti, N.; Schmid, A.; Schlötzer-Schrehardt, U.; Maier, P.; Lang, S.J.; Steinberg, T.; Schlunck, G.; Reinhard, T. A decellularized human corneal scaffold for anterior corneal surface reconstruction. Sci. Rep. 2021, 11, 2992. [Google Scholar] [CrossRef] [PubMed]
- Ramos, T.; Parekh, M.; Meleady, P.; O’Sullivan, F.; Stewart, R.M.; Kaye, S.B.; Hamill, K.; Ahmad, S. Specific decellularized extracellular matrix promotes the plasticity of human ocular surface epithelial cells. Front. Med. 2022, 9, 974212. [Google Scholar] [CrossRef]
- Yao, S.; Liang, Z.; Lee, Y.W.; Yung, P.S.H.; Lui, P.P.Y. Bioactive decellularized tendon-derived stem cell sheet for promoting graft healing after anterior cruciate ligament reconstruction. Am. J. Sports Med. 2023, 51, 66–80. [Google Scholar] [CrossRef]
- Nokhbatolfoghahaei, H.; Paknejad, Z.; Bohlouli, M.; Rezai Rad, M.; Aminishakib, P.; Derakhshan, S.; Mohammadi Amirabad, L.; Nadjmi, N.; Khojasteh, A. Fabrication of decellularized engineered extracellular matrix through bioreactor-based environment for bone tissue engineering. ACS Omega 2020, 5, 31943–31956. [Google Scholar] [CrossRef]
- Kim, Y.S.; Majid, M.; Melchiorri, A.J.; Mikos, A.G. Applications of decellularized extracellular matrix in bone and cartilage tissue engineering. Bioeng. Transl. Med. 2019, 4, 83–95. [Google Scholar] [CrossRef]
- Kitahara, H.; Yagi, H.; Tajima, K.; Okamoto, K.; Yoshitake, A.; Aeba, R.; Kudo, M.; Kashima, I.; Kawaguchi, S.; Hirano, A. Heterotopic transplantation of a decellularized and recellularized whole porcine heart. Interact. Cardiovasc. Thorac. Surg. 2016, 22, 571–579. [Google Scholar] [CrossRef]
- Hodgson, M.J.; Knutson, C.C.; Momtahan, N.; Cook, A.D. Extracellular matrix from whole porcine heart decellularization for cardiac tissue engineering. In Decellularized Scaffolds and Organogenesis: Methods and Protocols; Humana Press: New York, NY, USA, 2018; pp. 95–102. [Google Scholar]
- Wang, Q.; Yang, H.; Bai, A.; Jiang, W.; Li, X.; Wang, X.; Mao, Y.; Lu, C.; Qian, R.; Guo, F. Functional engineered human cardiac patches prepared from nature’s platform improve heart function after acute myocardial infarction. Biomaterials 2016, 105, 52–65. [Google Scholar] [CrossRef] [PubMed]
- Gregory, E.; Baek, I.H.; Ala-Kokko, N.; Dugan, R.; Pinzon-Herrera, L.; Almodóvar, J.; Song, Y.H. Peripheral Nerve Decellularization for In Vitro Extracellular Matrix Hydrogel Use: A Comparative Study. ACS Biomater. Sci. Eng. 2022, 8, 2574–2588. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Wang, D.; Wei, Q.; Yang, Y. Nerve decellularized matrix composite scaffold with high antibacterial activity for nerve regeneration. Front. Bioeng. Biotechnol. 2022, 9, 840421. [Google Scholar] [CrossRef] [PubMed]
- El Soury, M.; García-García, Ó.D.; Moretti, M.; Perroteau, I.; Raimondo, S.; Lovati, A.B.; Carriel, V. Comparison of decellularization protocols to generate peripheral nerve grafts: A study on rat sciatic nerves. Int. J. Mol. Sci. 2021, 22, 2389. [Google Scholar] [CrossRef]
- Lu, H.; Hoshiba, T.; Kawazoe, N.; Chen, G. Comparison of decellularization techniques for preparation of extracellular matrix scaffolds derived from three-dimensional cell culture. J. Biomed. Mater. Res. Part A 2012, 100, 2507–2516. [Google Scholar] [CrossRef]
- Hoshiba, T.; Yunoki, S. Comparison of decellularization protocols for cultured cell-derived extracellular matrix—Effects on decellularization efficacy, extracellular matrix retention, and cell functions. J. Biomed. Mater. Res. Part B Appl. Biomater. 2023, 111, 85–94. [Google Scholar] [CrossRef]
- Xing, Q.; Yates, K.; Tahtinen, M.; Shearier, E.; Qian, Z.; Zhao, F. Decellularization of fibroblast cell sheets for natural extracellular matrix scaffold preparation. Tissue Eng. Part C Methods 2015, 21, 77–87. [Google Scholar] [CrossRef]
- Xu, H.; Xu, B.; Yang, Q.; Li, X.; Ma, X.; Xia, Q.; Zhang, Y.; Zhang, C.; Wu, Y.; Zhang, Y. Comparison of decellularization protocols for preparing a decellularized porcine annulus fibrosus scaffold. PLoS ONE 2014, 9, e86723. [Google Scholar] [CrossRef] [PubMed]
- Narciso, M.; Ulldemolins, A.; Junior, C.; Otero, J.; Navajas, D.; Farre, R.; Gavara, N.; Almendros, I. Novel decellularization method for tissue slices. Front. Bioeng. Biotechnol. 2022, 10, 832178. [Google Scholar] [CrossRef] [PubMed]
- Wüthrich, T.; Lese, I.; Haberthür, D.; Zubler, C.; Hlushchuk, R.; Hewer, E.; Maistriaux, L.; Gianello, P.; Lengelé, B.; Rieben, R. Development of vascularized nerve scaffold using perfusion-decellularization and recellularization. Mater. Sci. Eng. C 2020, 117, 111311. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, Z.Q.; Turner, N.J.; Teng, S.F.; Cheng, W.Y.; Zhou, H.Y.; Zhang, L.; Hu, H.W.; Wang, Q.; Badylak, S.F. Perfusion-decellularized skeletal muscle as a three-dimensional scaffold with a vascular network template. Biomaterials 2016, 89, 114–126. [Google Scholar] [CrossRef]
- Ott, H.C.; Matthiesen, T.S.; Goh, S.-K.; Black, L.D.; Kren, S.M.; Netoff, T.I.; Taylor, D.A. Perfusion-decellularized matrix: Using nature’s platform to engineer a bioartificial heart. Nat. Med. 2008, 14, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Fathi, I.; Imura, T.; Inagaki, A.; Nakamura, Y.; Nabawi, A.; Goto, M. Decellularized whole-organ pre-vascularization: A novel approach for organogenesis. Front. Bioeng. Biotechnol. 2021, 9, 756755. [Google Scholar] [CrossRef]
- Price, A.P.; England, K.A.; Matson, A.M.; Blazar, B.R.; Panoskaltsis-Mortari, A. Development of a decellularized lung bioreactor system for bioengineering the lung: The matrix reloaded. Tissue Eng. Part A 2010, 16, 2581–2591. [Google Scholar] [CrossRef]
- Shupe, T.; Williams, M.; Brown, A.; Willenberg, B.; Petersen, B.E. Method for the decellularization of intact rat liver. Organogenesis 2010, 6, 134–136. [Google Scholar] [CrossRef]
- Baptista, P.M.; Orlando, G.; Mirmalek-Sani, S.-H.; Siddiqui, M.; Atala, A.; Soker, S. Whole organ decellularization-a tool for bioscaffold fabrication and organ bioengineering. In Proceedings of the 2009 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Minneapolis, MN, USA, 3–6 September 2009; pp. 6526–6529. [Google Scholar]
- Zhang, X.; Chen, X.; Hong, H.; Hu, R.; Liu, J.; Liu, C. Decellularized extracellular matrix scaffolds: Recent trends and emerging strategies in tissue engineering. Bioact. Mater. 2022, 10, 15–31. [Google Scholar] [CrossRef]
- Gilpin, A.; Yang, Y. Decellularization strategies for regenerative medicine: From processing techniques to applications. BioMed Res. Int. 2017, 2017, 9831534. [Google Scholar] [CrossRef]
- Hussey, G.S.; Pineda Molina, C.; Cramer, M.C.; Tyurina, Y.Y.; Tyurin, V.A.; Lee, Y.C.; El-Mossier, S.O.; Murdock, M.H.; Timashev, P.S.; Kagan, V.E. Lipidomics and RNA sequencing reveal a novel subpopulation of nanovesicle within extracellular matrix biomaterials. Sci. Adv. 2020, 6, eaay4361. [Google Scholar] [CrossRef] [PubMed]
- Nellinger, S.; Mrsic, I.; Keller, S.; Heine, S.; Southan, A.; Bach, M.; Volz, A.C.; Chassé, T.; Kluger, P.J. Cell-derived and enzyme-based decellularized extracellular matrix exhibit compositional and structural differences that are relevant for its use as a biomaterial. Biotechnol. Bioeng. 2022, 119, 1142–1156. [Google Scholar] [CrossRef] [PubMed]
- Mendibil, U.; Ruiz-Hernandez, R.; Retegi-Carrion, S.; Garcia-Urquia, N.; Olalde-Graells, B.; Abarrategi, A. Tissue-specific decellularization methods: Rationale and strategies to achieve regenerative compounds. Int. J. Mol. Sci. 2020, 21, 5447. [Google Scholar] [CrossRef]
- Neishabouri, A.; Khaboushan, A.S.; Daghigh, F.; Kajbafzadeh, A.-M.; Zolbin, M.M. Decellularization in tissue engineering and regenerative medicine: Evaluation, modification, and application methods. Front. Bioeng. Biotechnol. 2022, 10, 805299. [Google Scholar] [CrossRef]
- Liu, X.; Cai, Y.; Xia, C.; Wu, H.; Li, Q.; Xu, Z.; Lu, F. An innovative method to obtain porous porcine aorta scaffolds for tissue engineering. Artif. Organs 2019, 43, 1162–1169. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Man, K.; Liu, J.; Meckes, B.; Yang, Y. Dissecting Physical and Biochemical Effects in Nanotopographical Regulation of Cell Behavior. ACS Nano 2023, 17, 2124–2133. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Jia, W.; Long, F.; Pati, S.; Chen, Q.; Qyang, Y.; Lee, B.; Choi, C.K.; Zhao, F. Polydopamine and collagen coated micro-grated polydimethylsiloxane for human mesenchymal stem cell culture. Bioact. Mater. 2019, 4, 142–150. [Google Scholar] [CrossRef]
- Lian, X.; Zhang, J.; Azarin, S.M.; Zhu, K.; Hazeltine, L.B.; Bao, X.; Hsiao, C.; Kamp, T.J.; Palecek, S.P. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat. Protoc. 2013, 8, 162–175. [Google Scholar] [CrossRef]
- Zhang, J.; Wilson, G.F.; Soerens, A.G.; Koonce, C.H.; Yu, J.; Palecek, S.P.; Thomson, J.A.; Kamp, T.J. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ. Res. 2009, 104, e30–e41. [Google Scholar] [CrossRef]
- Feng, Y.; Jiang, Z.; Zhang, Y.; Miao, X.; Yu, Q.; Xie, Z.; Yang, G. Stem-cell-derived ECM sheet–implant complexes for enhancing osseointegration. Biomater. Sci. 2020, 8, 6647–6656. [Google Scholar] [CrossRef]
- Yang, L.; Ge, L.; van Rijn, P. Synergistic effect of cell-derived extracellular matrices and topography on osteogenesis of mesenchymal stem cells. ACS Appl. Mater. Interfaces 2020, 12, 25591–25603. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Ferguson, M.; Zhao, F. A step-by-step protocol for generating human fibroblast cell-derived completely biological extracellular matrix scaffolds. In Methods in Cell Biology; Elsevier: Amsterdam, The Netherlands, 2020; Volume 156, pp. 3–13. [Google Scholar]
- Lovekamp, J.J.; Simionescu, D.T.; Mercuri, J.J.; Zubiate, B.; Sacks, M.S.; Vyavahare, N.R. Stability and function of glycosaminoglycans in porcine bioprosthetic heart valves. Biomaterials 2006, 27, 1507–1518. [Google Scholar] [CrossRef] [PubMed]
- Schmuck, E.G.; Mulligan, J.D.; Ertel, R.L.; Kouris, N.A.; Ogle, B.M.; Raval, A.N.; Saupe, K.W. Cardiac fibroblast-derived 3D extracellular matrix seeded with mesenchymal stem cells as a novel device to transfer cells to the ischemic myocardium. Cardiovasc. Eng. Technol. 2014, 5, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Crapo, P.M.; Gilbert, T.W.; Badylak, S.F. An overview of tissue and whole organ decellularization processes. Biomaterials 2011, 32, 3233–3243. [Google Scholar] [CrossRef]
- Nagata, S.; Hanayama, R.; Kawane, K. Autoimmunity and the clearance of dead cells. Cell 2010, 140, 619–630. [Google Scholar] [CrossRef]
- Zambon, J.P.; Atala, A.; Yoo, J.J. Methods to generate tissue-derived constructs for regenerative medicine applications. Methods 2020, 171, 3–10. [Google Scholar] [CrossRef]
- Wang, X.; Chan, V.; Corridon, P.R. Decellularized blood vessel development: Current state-of-the-art and future directions. Front. Bioeng. Biotechnol. 2022, 10, 951644. [Google Scholar] [CrossRef]
- Silva, A.C.; Pereira, C.; Fonseca, A.C.R.; Pinto-do-Ó, P.; Nascimento, D.S. Bearing my heart: The role of extracellular matrix on cardiac development, homeostasis, and injury response. Front. Cell Dev. Biol. 2021, 8, 621644. [Google Scholar] [CrossRef]
- Wittig, C.; Szulcek, R. Extracellular matrix protein ratios in the human heart and vessels: How to distinguish pathological from physiological changes? Front. Physiol. 2021, 12, 708656. [Google Scholar] [CrossRef]
- Liu, X.; Li, N.; Gong, D.; Xia, C.; Xu, Z. Comparison of detergent-based decellularization protocols for the removal of antigenic cellular components in porcine aortic valve. Xenotransplantation 2018, 25, e12380. [Google Scholar] [CrossRef]
- Wallis, J.M.; Borg, Z.D.; Daly, A.B.; Deng, B.; Ballif, B.A.; Allen, G.B.; Jaworski, D.M.; Weiss, D.J. Comparative assessment of detergent-based protocols for mouse lung de-cellularization and re-cellularization. Tissue Eng. Part C Methods 2012, 18, 420–432. [Google Scholar] [CrossRef]
- Corda, S.; Jane-Lise, S.; Rappaport, L. Extracellular matrix and growth factors during heart growth. Heart Fail. Rev. 2000, 5, 119. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, M. Structure and function of VEGF/VEGF-receptor system involved in angiogenesis. Cell Struct. Funct. 2001, 26, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Vatner, S.F. FGF induces hypertrophy and angiogenesis in hibernating myocardium. Circ. Res. 2005, 96, 705–707. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.E.; Hsieh, P.C.; Takahashi, T.; Song, Q.; Zhang, S.; Kamm, R.D.; Grodzinsky, A.J.; Anversa, P.; Lee, R.T. Local myocardial insulin-like growth factor 1 (IGF-1) delivery with biotinylated peptide nanofibers improves cell therapy for myocardial infarction. Proc. Natl. Acad. Sci. USA 2006, 103, 8155–8160. [Google Scholar] [CrossRef] [PubMed]
- Vinciguerra, M.; Santini, M.P.; Claycomb, W.C.; Ladurner, A.G.; Rosenthal, N. Local IGF-1 isoform protects cardiomyocytes from hypertrophic and oxidative stresses via SirT1 activity. Aging 2010, 2, 43. [Google Scholar] [CrossRef]
- Zhang, M.; Prosser, B.L.; Bamboye, M.A.; Gondim, A.N.; Santos, C.X.; Martin, D.; Ghigo, A.; Perino, A.; Brewer, A.C.; Ward, C.W. Contractile function during angiotensin-II activation: Increased Nox2 activity modulates cardiac calcium handling via phospholamban phosphorylation. J. Am. Coll. Cardiol. 2015, 66, 261–272. [Google Scholar] [CrossRef]
- Paradis, P.; Dali-Youcef, N.; Paradis, F.W.; Thibault, G.; Nemer, M. Overexpression of angiotensin II type I receptor in cardiomyocytes induces cardiac hypertrophy and remodeling. Proc. Natl. Acad. Sci. USA 2000, 97, 931–936. [Google Scholar] [CrossRef]
- Agapitov, A.V.; Haynes, W.G. Role of endothelin in cardiovascular disease. J. Renin-Angiotensin-Aldosterone Syst. 2002, 3, 1–15. [Google Scholar] [CrossRef]
- Sharma, S.; Jackson, P.; Makan, J. Cardiac troponins. J. Clin. Pathol. 2004, 57, 1025–1026. [Google Scholar] [CrossRef]
- Hsu, C.P.; Moghadaszadeh, B.; Hartwig, J.H.; Beggs, A.H. Sarcomeric and nonmuscle α-actinin isoforms exhibit differential dynamics at skeletal muscle Z-lines. Cytoskeleton 2018, 75, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Beauchamp, P.; Yamada, K.A.; Baertschi, A.J.; Green, K.; Kanter, E.M.; Saffitz, J.E.; Kléber, A.G. Relative contributions of connexins 40 and 43 to atrial impulse propagation in synthetic strands of neonatal and fetal murine cardiomyocytes. Circ. Res. 2006, 99, 1216–1224. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, T.-A.; Sharma, D.; Jia, W.; Ha, D.; Man, K.; Zhang, J.; Yang, Y.; Zhou, Y.; Kamp, T.J.; Zhao, F. Detergent-Based Decellularization for Anisotropic Cardiac-Specific Extracellular Matrix Scaffold Generation. Biomimetics 2023, 8, 551. https://doi.org/10.3390/biomimetics8070551
Chen T-A, Sharma D, Jia W, Ha D, Man K, Zhang J, Yang Y, Zhou Y, Kamp TJ, Zhao F. Detergent-Based Decellularization for Anisotropic Cardiac-Specific Extracellular Matrix Scaffold Generation. Biomimetics. 2023; 8(7):551. https://doi.org/10.3390/biomimetics8070551
Chicago/Turabian StyleChen, Te-An, Dhavan Sharma, Wenkai Jia, Donggi Ha, Kun Man, Jianhua Zhang, Yong Yang, Yuxiao Zhou, Timothy J. Kamp, and Feng Zhao. 2023. "Detergent-Based Decellularization for Anisotropic Cardiac-Specific Extracellular Matrix Scaffold Generation" Biomimetics 8, no. 7: 551. https://doi.org/10.3390/biomimetics8070551
APA StyleChen, T.-A., Sharma, D., Jia, W., Ha, D., Man, K., Zhang, J., Yang, Y., Zhou, Y., Kamp, T. J., & Zhao, F. (2023). Detergent-Based Decellularization for Anisotropic Cardiac-Specific Extracellular Matrix Scaffold Generation. Biomimetics, 8(7), 551. https://doi.org/10.3390/biomimetics8070551






