Chrysanthemum CmHSP90.5 as a Tool to Regulate Heat and Salt Stress Tolerance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Condition
2.2. Cloning and Sequence Analyses of CmHSP90.5
2.3. Subcellular Localization of CmHSP90.5
2.4. Expression Profile of CmHSP90.5
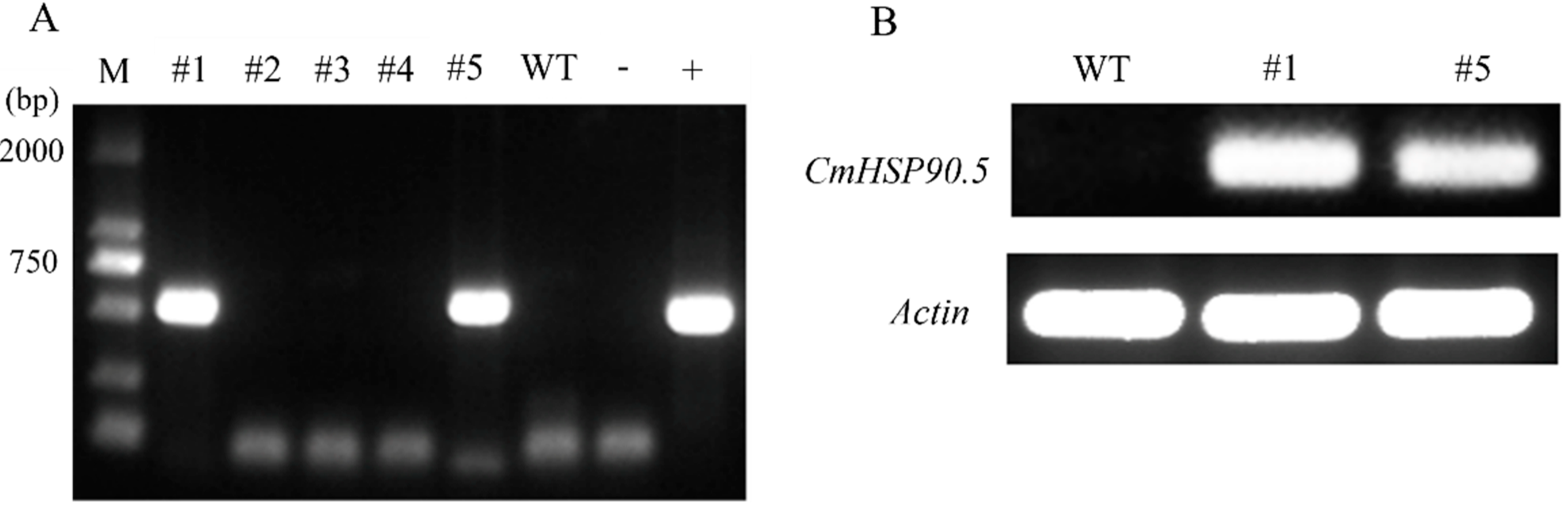
2.5. Generation of CmHSP90.5-Overexpressing Arabidopsis
2.6. Heat Tolerance Analysis of CmHSP90.5-Overexpressing Arabidopsis Plants
2.7. Salt Treatment on Transgenic Arabidopsis Plants
2.8. Determination of the Physiological Indexes of Plants under Stress Conditions
2.9. Expression Profiles of Stress-Related Genes in CmHSP90.5-Overexpressing Arabidopsis Plants
2.10. Statistical Analysis
3. Results
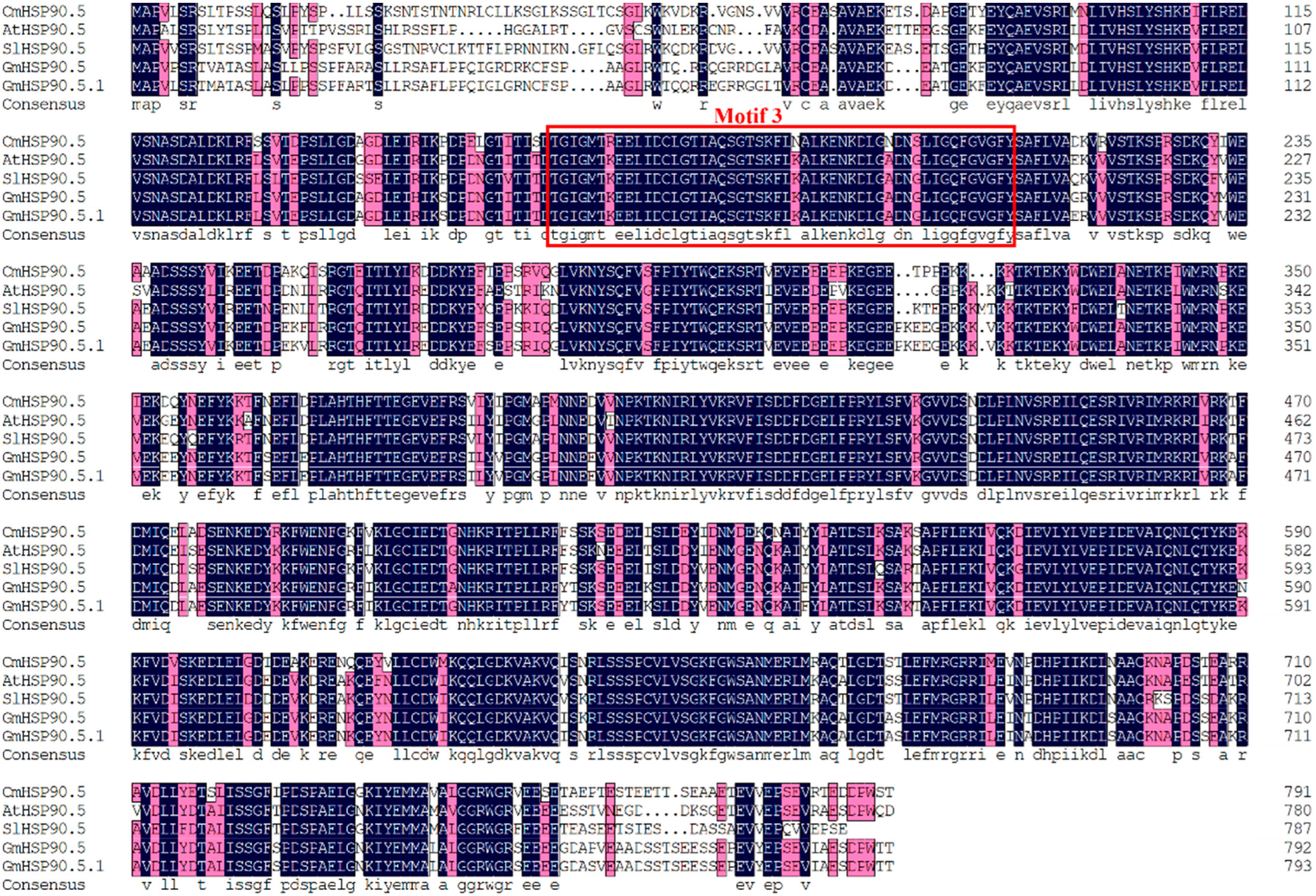
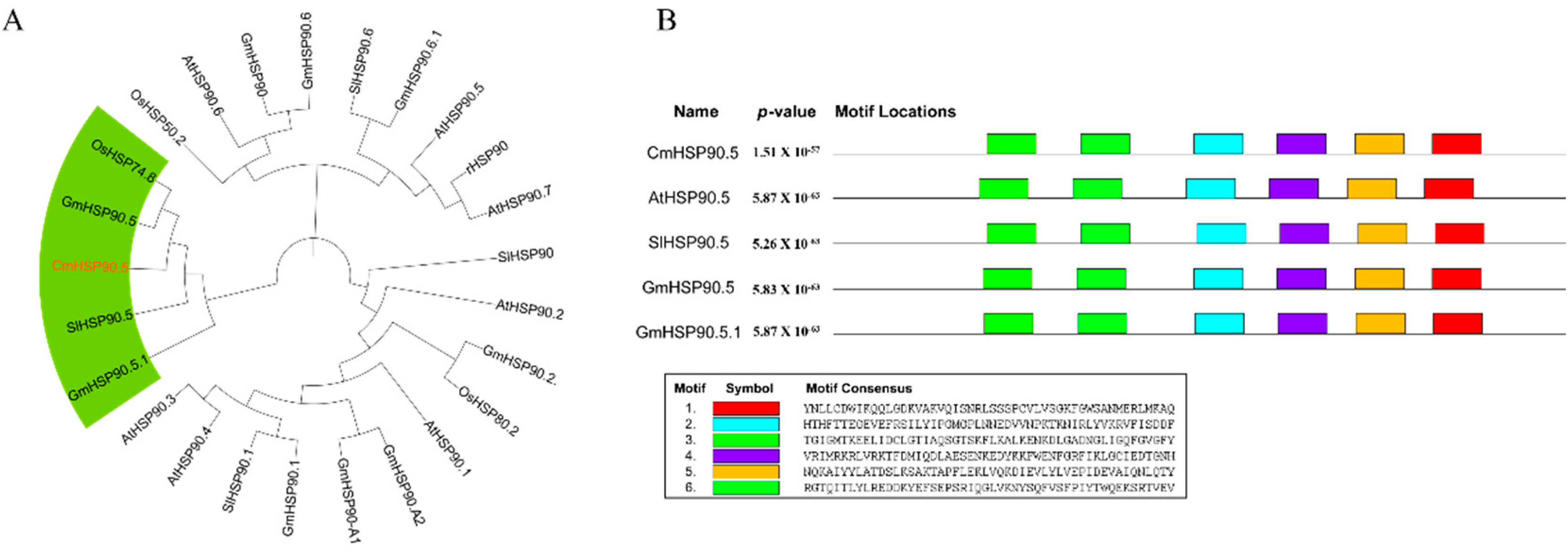
3.1. Identification and Characterization of CmHSP90.5
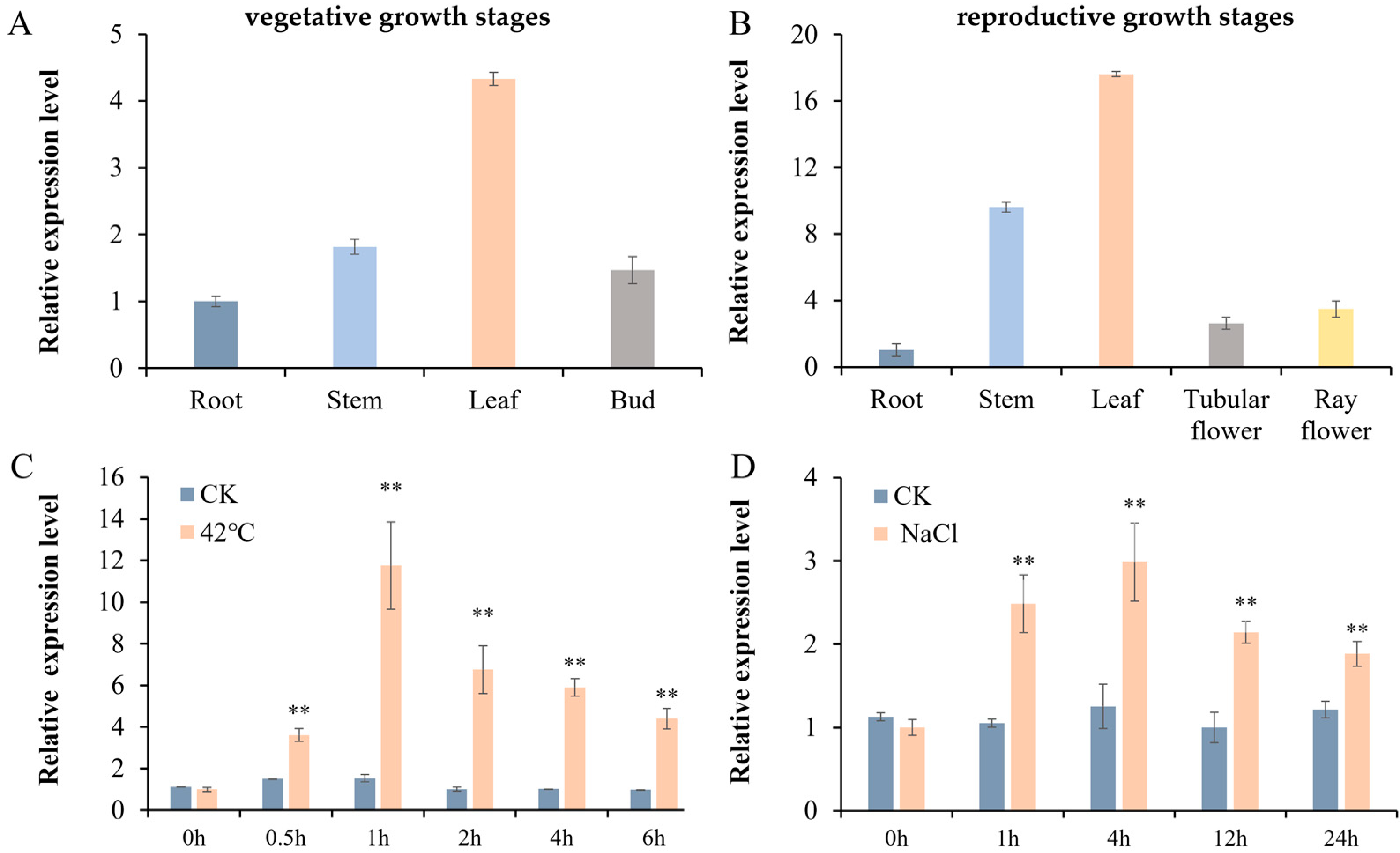
3.2. Expression Profiles of CmHSP90.5 in Different Tissues of Chrysanthemum and in Response to Heat and Salt Treatment
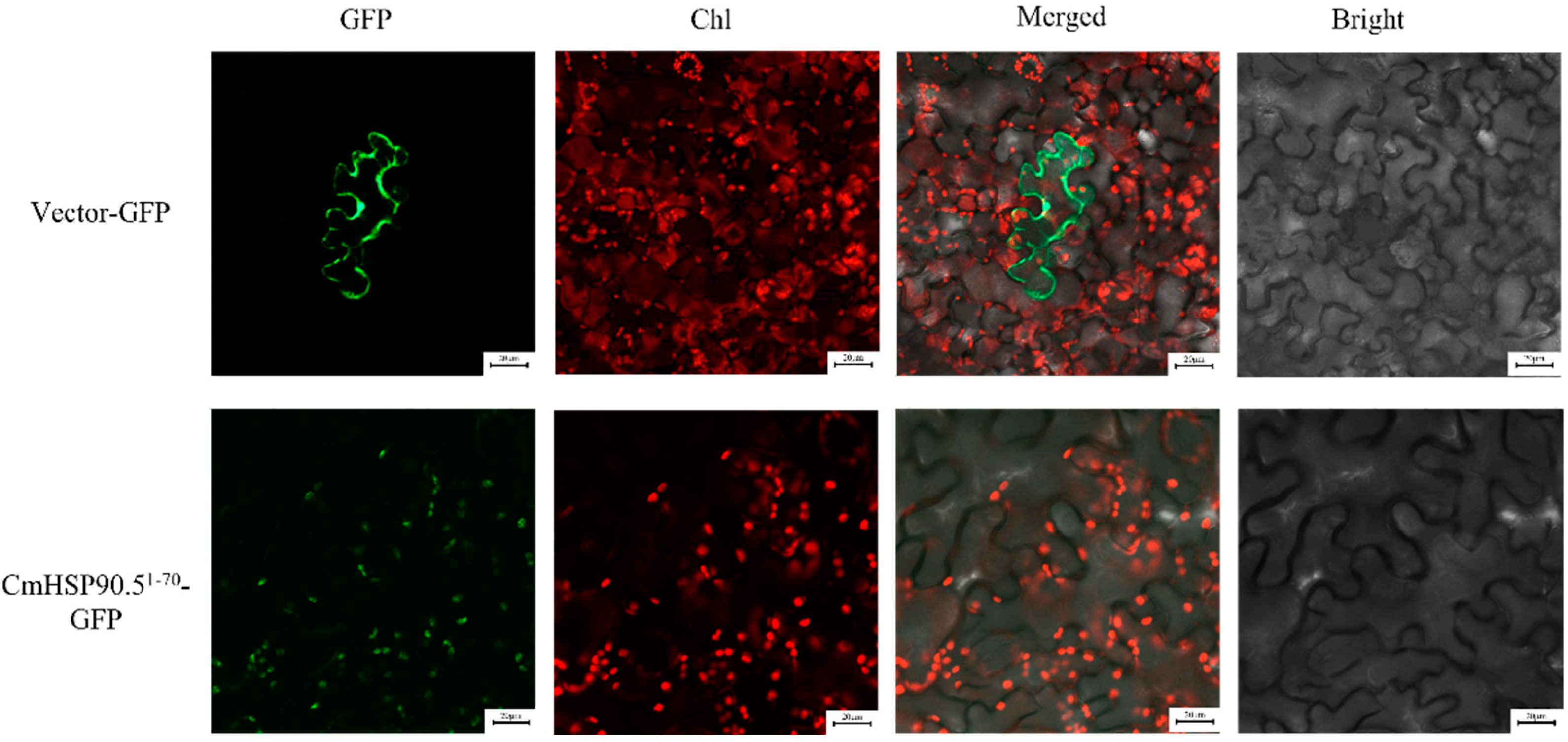
3.3. Subcellular Localization of CmHSP90.5
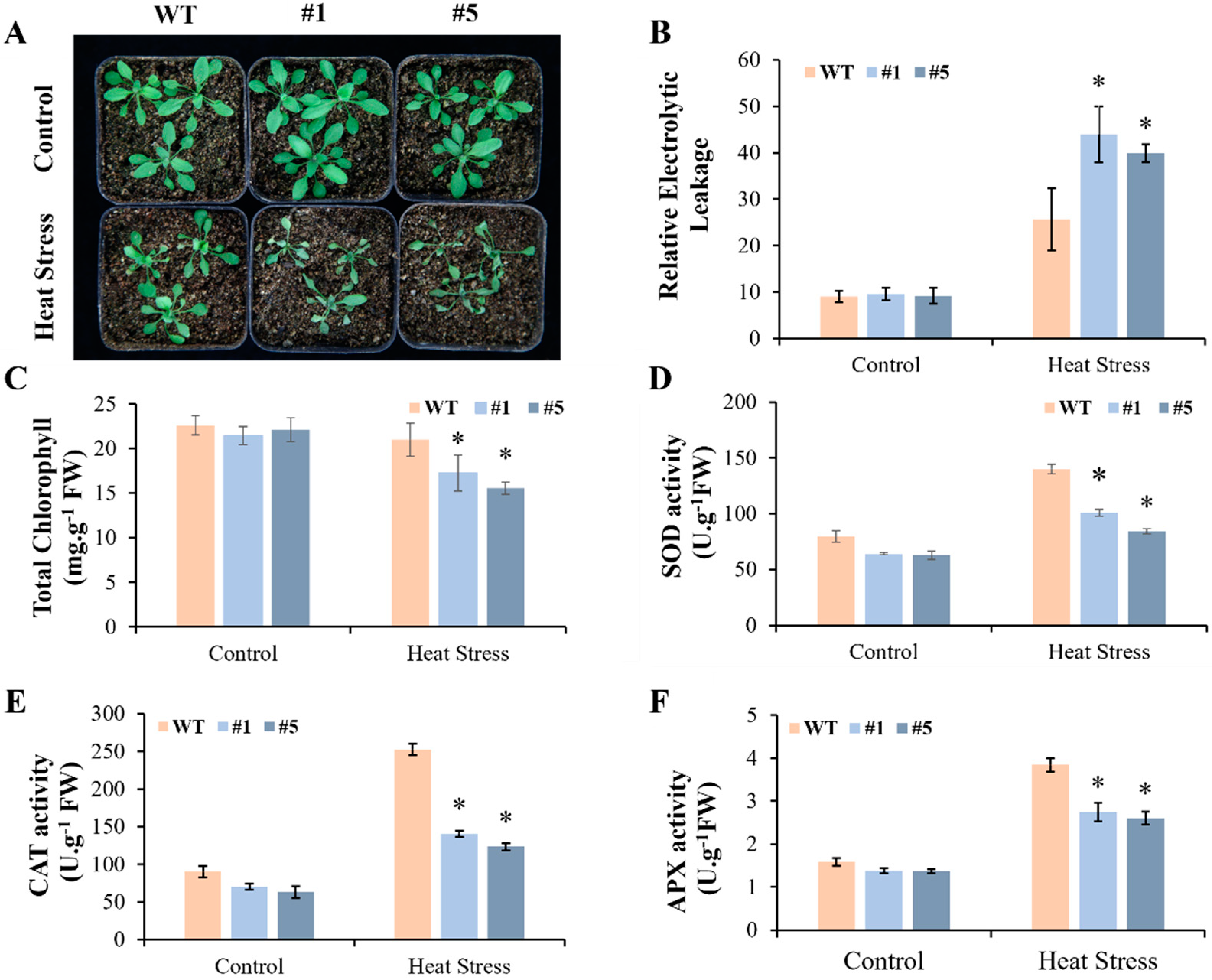
3.4. Overexpression of CmHSP90.5 Reduced Heat Tolerance in Transgenic Arabidopsis Plants
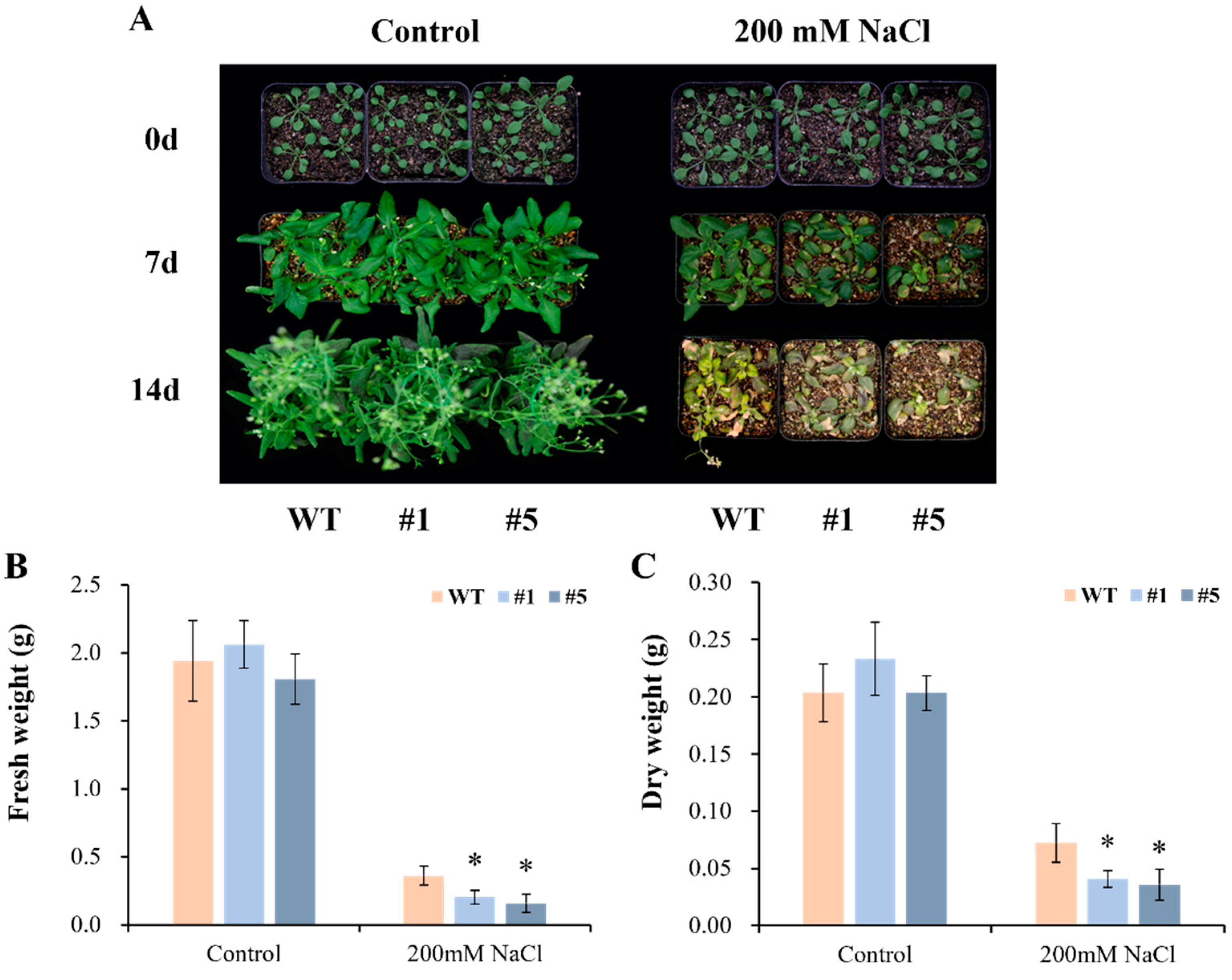
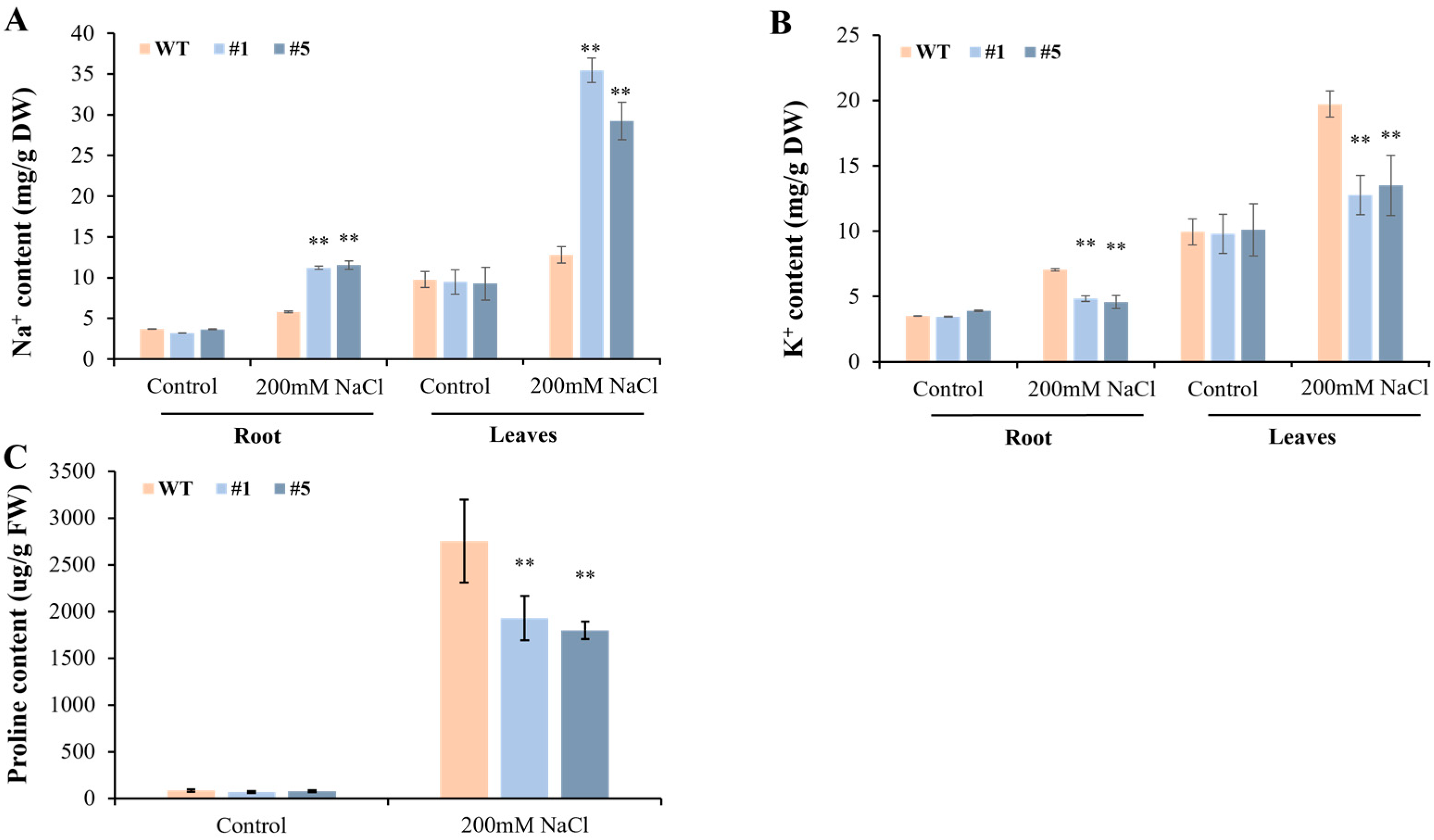
3.5. Overexpression of CmHSP90.5 Reduced Salt Tolerance in Transgenic Arabidopsis Plants
4. Discussion
4.1. CmHSP90.5 Is a Heat Shock Protein Family Member and Is Chloroplast-Localized
4.2. CmHSP90.5 Confers Sensitivity to Heat Stress by Regulating Antioxidants and Heat Shock Proteins
4.3. CmHSP90.5 Confers Salt Stress Sensitivity by Regulating Ion Homeostasis and Osmosis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Parihar, P.; Singh, S.; Singh, R.; Singh, V.P.; Prasad, S.M. Effect of salinity stress on plants and its tolerance strategies: A review. Environ. Sci. Pollut. Res. Int. 2015, 22, 4056–4075. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, K.; Fujit, M. Extreme Temperature Responses, Oxidative Stress and Antioxidant Defense in Plants. In Abiotic Stress—Plant Responses and Applications in Agriculture; Intechopen: London, UK, 2013. [Google Scholar]
- Hasanuzzaman, M.; Hossain, M.A.; da Silva, J.A.T. Response and Tolerance to Abiotic Oxidative Stress: Antioxidant Defense Is a Key Factor. In Crop Stress and its Management: Perspectives and Strategies; Springer: Dordrecht, The Netherlands, 2012; pp. 261–315. [Google Scholar]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasanuzzaman, M.; Nahar, K.; Alam, M.M.; Roychowdhury, R.; Fujita, M. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 2013, 14, 9643–9684. [Google Scholar] [CrossRef]
- Wang, X.; Cai, J.; Liu, F.; Jin, M.; Yu, H.; Jiang, D.; Wollenweber, B.; Dai, T.; Cao, W. Pre-anthesis high temperature acclimation alleviates the negative effects of post-anthesis heat stress on stem stored carbohydrates remobilization and grain starch accumulation in wheat. J. Cereal. Sci. 2012, 55, 331–336. [Google Scholar] [CrossRef]
- Bui, D.C.; Lee, Y.; Lim, J.Y.; Fu, M.; Kim, J.C.; Choi, G.J.; Son, H.; Lee, Y.W. Heat shock protein 90 is required for sexual and asexual development, virulence, and heat shock response in Fusarium graminearum. Sci. Rep. 2016, 6, 28154. [Google Scholar] [CrossRef] [Green Version]
- Merck, K.B.; Groenen, P.J.; Voorter, C.E.; de Haard-Hoekman, W.A.; Horwitz, J.; Bloemendal, H.; de Jong, W.W. Structural and functional similarities of bovine alpha-crystallin and mouse small heat-shock protein. A family of chaperones. J. Biol. Chem. 1993, 268, 1046–1052. [Google Scholar] [CrossRef]
- Picard, D. Heat-shock protein 90, a chaperone for folding and regulation. Cell Mol. Life Sci. 2002, 59, 1640–1648. [Google Scholar] [CrossRef]
- Wayne, N.; Mishra, P.; Bolon, D.N. Hsp90 and client protein maturation. Methods Mol. Biol. 2011, 787, 33–44. [Google Scholar] [PubMed] [Green Version]
- Krishna, P.; Gloor, G. The Hsp90 family of proteins in Arabidopsis thaliana. Cell Stress Chaperones 2001, 6, 238–246. [Google Scholar] [CrossRef]
- Sangster, T.A.; Queitsch, C. The HSP90 chaperone complex, an emerging force in plant development and phenotypic plasticity. Curr. Opin. Plant Biol. 2005, 8, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.S.; Li, Z.Y.; Chen, Y.; Chen, M.; Li, L.C.; Ma, Y.Z. Heat shock protein 90 in plants: Molecular mechanisms and roles in stress responses. Int. J. Mol. Sci. 2012, 13, 15706–15723. [Google Scholar] [CrossRef] [PubMed]
- Wegele, H.; Müller, L.; Buchner, J. Hsp70 and Hsp90—A relay team for protein folding. Rev. Physiol. Biochem. Pharmacol. 2004, 151, 1–44. [Google Scholar] [PubMed]
- Young, J.C.; Moarefi, I.; Hartl, F.U. Hsp90: A specialized but essential protein-folding tool. J. Cell Biol. 2001, 154, 267–273. [Google Scholar] [CrossRef]
- Zuehlke, A.; Johnson, J.L. Hsp90 and co-chaperones twist the functions of diverse client proteins. Biopolymers 2010, 93, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Jacob, P.; Hirt, H.; Bendahmane, A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 2017, 15, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Jarosz, D.F.; Lindquist, S. Hsp90 and environmental stress transform the adaptive value of natural genetic variation. Science 2010, 330, 1820–1824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, H.; Zhao, R.; Fan, P.; Wang, X.; Chen, X.; Li, Y. Overexpression of AtHsp90.2, AtHsp90.5 and AtHsp90.7 in Arabidopsis thaliana enhances plant sensitivity to salt and drought stresses. Planta 2009, 229, 955–964. [Google Scholar] [CrossRef]
- Wei, Y.; Liu, W.; Hu, W.; Yan, Y.; Shi, H. The chaperone MeHSP90 recruits MeWRKY20 and MeCatalase1 to regulate drought stress resistance in cassava. New Phytol. 2020, 226, 476–491. [Google Scholar] [CrossRef]
- Sable, A.; Rai, K.M.; Choudhary, A.; Yadav, V.K.; Agarwal, S.K.; Sawant, S.V. Inhibition of Heat Shock proteins HSP90 and HSP70 induce oxidative stress, suppressing cotton fiber development. Sci. Rep. 2018, 8, 3620. [Google Scholar] [CrossRef]
- Song, H.; Fan, P.; Li, Y. Overexpression of Organellar and Cytosolic AtHSP90 in Arabidopsis thaliana Impairs Plant Tolerance to Oxidative Stress. Plant Mol. Biol. Rep. 2009, 27, 342–349. [Google Scholar] [CrossRef]
- McLellan, C.A.; Turbyville, T.J.; Wijeratne, E.M.; Kerschen, A.; Vierling, E.; Queitsch, C.; Whitesell, L.; Gunatilaka, A.A. A rhizosphere fungus enhances Arabidopsis thermotolerance through production of an HSP90 inhibitor. Plant Physiol. 2007, 145, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa-Yokoi, A.; Tainaka, H.; Yoshida, E.; Tamoi, M.; Yabuta, Y.; Shigeoka, S. The 26S proteasome function and Hsp90 activity involved in the regulation of HsfA2 expression in response to oxidative stress. Plant Cell Physiol. 2010, 51, 486–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meiri, D.; Breiman, A. Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J. 2009, 59, 387–399. [Google Scholar] [CrossRef]
- Aviezer-Hagai, K.; Skovorodnikova, J.; Galigniana, M.; Farchi-Pisanty, O.; Maayan, E.; Bocovza, S.; Efrat, Y.; von Koskull-Doring, P.; Ohad, N.; Breiman, A. Arabidopsis immunophilins ROF1 (AtFKBP62) and ROF2 (AtFKBP65) exhibit tissue specificity, are heat-stress induced, and bind HSP90. Plant Mol. Biol. 2007, 63, 237–255. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhang, Y.; Kieffer, M.; Yu, H.; Kepinski, S.; Estelle, M. HSP90 regulates temperature-dependent seedling growth in Arabidopsis by stabilizing the auxin co-receptor F-box protein TIR1. Nat. Commun. 2016, 7, 10269. [Google Scholar] [CrossRef]
- Watanabe, E.; Mano, S.; Hara-Nishimura, I.; Nishimura, M.; Yamada, K. HSP90 stabilizes auxin receptor TIR1 and ensures plasticity of auxin responses. Plant Signal. Behav. 2017, 12, e1311439. [Google Scholar] [CrossRef]
- Xu, J.; Xue, C.; Xue, D.; Zhao, J.; Gai, J.; Guo, N.; Xing, H. Overexpression of GmHsp90s, a heat shock protein 90 (Hsp90) gene family cloning from soybean, decrease damage of abiotic stresses in Arabidopsis thaliana. PLoS ONE 2013, 8, e69810. [Google Scholar] [CrossRef]
- Xiang, J.; Chen, X.; Hu, W.; Xiang, Y.; Yan, M.; Wang, J. Overexpressing heat-shock protein OsHSP50.2 improves drought tolerance in rice. Plant Cell Rep. 2018, 37, 1585–1595. [Google Scholar] [CrossRef]
- Zhou, L.J.; Geng, Z.; Wang, Y.; Wang, Y.; Liu, S.; Chen, C.; Song, A.; Jiang, J.; Chen, S.; Chen, F. A novel transcription factor CmMYB012 inhibits flavone and anthocyanin biosynthesis in response to high temperatures in chrysanthemum. Hortic. Res. 2021, 8, 248. [Google Scholar] [CrossRef]
- Li, F.; Zhang, H.; Zhao, H.; Gao, T.; Song, A.; Jiang, J.; Chen, F.; Chen, S. Chrysanthemum CmHSFA4 gene positively regulates salt stress tolerance in transgenic chrysanthemum. Plant Biotechnol. J. 2018, 16, 1311–1321. [Google Scholar] [CrossRef] [Green Version]
- An, J.; Song, A.; Guan, Z.; Jiang, J.; Chen, F.; Lou, W.; Fang, W.; Liu, Z.; Chen, S. The over-expression of Chrysanthemum crassum CcSOS1 improves the salinity tolerance of chrysanthemum. Mol. Biol. Rep. 2014, 41, 4155–4162. [Google Scholar] [CrossRef]
- Wang, K.; Wu, Y.H.; Tian, X.Q.; Bai, Z.Y.; Liang, Q.Y.; Liu, Q.L.; Pan, Y.Z.; Zhang, L.; Jiang, B.B. Overexpression of DgWRKY4 Enhances Salt Tolerance in Chrysanthemum Seedlings. Front. Plant Sci. 2017, 8, 1592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, W.; He, M.; Liu, J.; Ma, X.; Zhang, Y.; Dai, S.; Zhou, Y. Overexpression of Chrysanthemum lavandulifolium ClCBF1 in Chrysanthemum morifolium ‘White Snow’ improves the level of salinity and drought tolerance. Plant Physiol. Biochem. 2018, 124, 50–58. [Google Scholar] [CrossRef]
- Li, P.; Song, A.; Gao, C.; Wang, L.; Wang, Y.; Sun, J.; Jiang, J.; Chen, F.; Chen, S. Chrysanthemum WRKY gene CmWRKY17 negatively regulates salt stress tolerance in transgenic chrysanthemum and Arabidopsis plants. Plant Cell Rep. 2015, 34, 1365–1378. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Zhang, L.; Ren, Y.; Lu, B.; Yang, C.; Feng, Z.; Liu, Z.; Chen, J.; Ma, W.; Wang, Y.; Yu, X.; et al. FLOURY ENDOSPERM7 encodes a regulator of starch synthesis and amyloplast development essential for peripheral endosperm development in rice. J. Exp. Bot. 2016, 67, 633–647. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Song, H.; Zhou, Z.; Shi, N.; Ying, Q.; Wang, H. Functional characterization of AtHsp90.3 in Saccharomyces cerevisiae and Arabidopsis thaliana under heat stress. Biotechnol. Lett. 2010, 32, 979–987. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Zhang, W.; Gao, T.; Li, P.; Tian, C.; Song, A.; Jiang, J.; Guan, Z.; Fang, W.; Chen, F.; Chen, S. Chrysanthemum CmWRKY53 negatively regulates the resistance of chrysanthemum to the aphid Macrosiphoniella sanborni. Hortic. Res. 2020, 7, 109. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. Cell Mol. Biol. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, T.; Sakuma, Y.; Todaka, D.; Maruyama, K.; Qin, F.; Mizoi, J.; Kidokoro, S.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis heat-shock transcription factor HsfA3 in the transcriptional cascade downstream of the DREB2A stress-regulatory system. Biochem. Bioph. Res. Commun. 2008, 368, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Wei, H.; Ding, Y.; Li, W.; Liu, Z.; Chen, L.; Tang, S.; Ding, C.; Jiang, Y.; Li, G. Melatonin regulates antioxidant strategy in response to continuous salt stress in rice seedlings. Plant Physiol. Biochem. 2021, 165, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Fankhauser, C.; Casal, J.J. Phenotypic characterization of a photomorphogenic mutant. Plant J. 2004, 39, 747–760. [Google Scholar] [CrossRef] [PubMed]
- Yong, Z.; Tang, H.; Luo, Y. Variation in Antioxidant Enzyme Activities of Two Strawberry Cultivars with Short-term Low Temperature Stress. World J. Agric. Sci. 2008, 4, 458–462. [Google Scholar]
- Gao, X.; Ren, Z.; Zhao, Y.; Zhang, H. Overexpression of SOD2 increases salt tolerance of Arabidopsis. Plant Physiol. 2003, 133, 1873–1881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, C.; Haslbeck, M.; Babujee, L.; Jahn, O.; Reumann, S. Identification and characterization of a stress-inducible and a constitutive small heat-shock protein targeted to the matrix of plant peroxisomes. Plant Physiol. 2006, 141, 47–60. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Van Montagu, M.; Verbruggen, N. Small heat shock proteins and stress tolerance in plants. Biochim. Biophys Acta 2002, 1577, 1–9. [Google Scholar] [CrossRef]
- Han, G.; Yuan, F.; Guo, J.; Zhang, Y.; Sui, N.; Wang, B. AtSIZ1 improves salt tolerance by maintaining ionic homeostasis and osmotic balance in Arabidopsis. Plant Sci. 2019, 285, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Terasawa, K.; Minami, M.; Minami, Y. Constantly updated knowledge of Hsp90. J. Biochem. 2005, 137, 443–447. [Google Scholar] [CrossRef]
- Stebbins, C.E.; Russo, A.A.; Schneider, C.; Rosen, N.; Hartl, F.U.; Pavletich, N.P. Crystal structure of an Hsp90-geldanamycin complex: Targeting of a protein chaperone by an antitumor agent. Cell 1997, 89, 239–250. [Google Scholar] [CrossRef] [Green Version]
- Grenert, J.P.; Johnson, B.D.; Toft, D.O. The importance of ATP binding and hydrolysis by hsp90 in formation and function of protein heterocomplexes. J. Biol. Chem. 1999, 274, 17525–17533. [Google Scholar] [CrossRef] [Green Version]
- Oh, S.E.; Yeung, C.; Babaei-Rad, R.; Zhao, R. Cosuppression of the chloroplast localized molecular chaperone HSP90.5 impairs plant development and chloroplast biogenesis in Arabidopsis. BMC Res. Notes 2014, 7, 643. [Google Scholar] [CrossRef] [Green Version]
- Hahn, A.; Bublak, D.; Schleiff, E.; Scharf, K.D. Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 2011, 23, 741–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laloum, T.; Martin, G.; Duque, P. Alternative Splicing Control of Abiotic Stress Responses. Trends Plant Sci. 2018, 23, 140–150. [Google Scholar] [CrossRef] [Green Version]
- Moller, I.M.; Jensen, P.E.; Hansson, A. Oxidative modifications to cellular components in plants. Annu. Rev. Plant Biol. 2007, 58, 459–481. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Lu, Z.; Wang, L.; Jin, B. Plant Responses to Heat Stress: Physiology, Transcription, Noncoding RNAs, and Epigenetics. Int. J. Mol. Sci. 2020, 22, 117. [Google Scholar] [CrossRef]
- Matthews, C.; Arshad, M.; Hannoufa, A. Alfalfa response to heat stress is modulated by microRNA156. Physiol. Plant. 2019, 165, 830–842. [Google Scholar] [CrossRef]
- Hernandez, J.A.; Ferrer, M.A.; Jimenez, A.; Barcelo, A.R.; Sevilla, F. Antioxidant systems and O2−/H2O2 production in the apoplast of pea leaves. Its relation with salt-induced necrotic lesions in minor veins. Plant Physiol. 2001, 127, 817–831. [Google Scholar] [CrossRef]
- Katiyar-Agarwal, S.; Agarwal, M.; Grover, A. Heat-tolerant basmati rice engineered by over-expression of hsp101. Plant Mol. Biol. 2003, 51, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shkolnik, D.; Finkler, A.; Pasmanik-Chor, M.; Fromm, H. Calmodulin-Binding Transcription Activator 6: A Key Regulator of Na+ Homeostasis during Germination. Plant Physiol. 2019, 180, 1101–1118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, E.M.; Mascheroni, L.; Pompa, A.; Ragni, L.; Weimar, T.; Lilley, K.S.; Dupree, P.; Vitale, A. Plant endoplasmin supports the protein secretory pathway and has a role in proliferating tissues. Plant J. 2006, 48, 657–673. [Google Scholar] [CrossRef]
- Kant, S.; Kant, P.; Raveh, E.; Barak, S. Evidence that differential gene expression between the halophyte, Thellungiella halophila, and Arabidopsis thaliana is responsible for higher levels of the compatible osmolyte proline and tight control of Na+ uptake in T. halophila. Plant Cell Environ. 2006, 29, 1220–1234. [Google Scholar] [CrossRef]



| Primer Name | Sequence (5′-3′) |
|---|---|
| 35S | GACGCACAATCCCACTATCC |
| Actin2-F | GGTAACATTGTGCTCAGTGGTGG |
| Actin2-R | AACGACCTTAATCTTCATGCTGC |
| qAtHSP15.7-F | TGCCGGAGAATGTGAAAGTTG |
| qAtHSP15.7-R | CGATTTCGATGAAGTGTCCTTAGG |
| qAtHSP17.6c-F | AAGAATGACAAGTGGCACCGTG |
| qAtHSP17.6c-R | GGCTTTGATTTCCTCCATCTTAGC |
| qAtHSP101-F | TGCATTTAGCTGGTGCTTTGAT |
| qAtHSP101-R | CCACCGGCACTAGAGATTGC |
| qCmHSP90.5-F | TTATGAAATGATGGCAGTCGCTCTT |
| qCmHSP90.5-R | TCGGTCCTCACTTCTGATGGCTC |
| qAtSOS1-F | TTCATCATCCTCACAATGGCTCTAA |
| qAtSOS1-R | CCCTCATCAAGCATCTCCCAGTA |
| qAtHKT1;1-F | TCAGTGCATATGGAAACGTTGG |
| qAtHKT1;1-R | CAGCCACCATCGCTGATG |
| qAtP5CS1-F | TAGCACCCGAAGAGCCCCAT |
| qAtP5CS1-R | TTTCAGTTCCAACGCCAGTAGA |
| CmEF1α-F | TGTAACAAGATGGATGCCACAA |
| CmEF1α-R | TCGCCCTCAAACCCAGAAAT |
| CmHSP90.5-F | ATGGCTCCAGTTCTTAGCAGA |
| CmHSP90.5-R | TCAAGTACTCCATGGGTCGTCTT |
| pORE_R4-CmHSP90.5-F | CGGGATCCAATGGCTCCAGTTCTTAG |
| pORE_R4-CmHSP90.5-R | CGGAATTCTAGTACTCCATGGGTCGTC |
| pORE_R4-CmHSP90.51−70-F | CGGGATCCAATGGCTCCAGTTCTTAG |
| pORE_R4-CmHSP90.51−70-R | CGGAATTCTCTCACACCTAACAACAA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Wu, J.; Wang, Y.; Jiang, Y.; Li, F.; Chen, Y.; Jiang, J.; Wang, L.; Guan, Z.; Chen, F.; et al. Chrysanthemum CmHSP90.5 as a Tool to Regulate Heat and Salt Stress Tolerance. Horticulturae 2022, 8, 532. https://doi.org/10.3390/horticulturae8060532
Wang X, Wu J, Wang Y, Jiang Y, Li F, Chen Y, Jiang J, Wang L, Guan Z, Chen F, et al. Chrysanthemum CmHSP90.5 as a Tool to Regulate Heat and Salt Stress Tolerance. Horticulturae. 2022; 8(6):532. https://doi.org/10.3390/horticulturae8060532
Chicago/Turabian StyleWang, Xinhui, Jianpeng Wu, Yue Wang, Yuhan Jiang, Fei Li, Yu Chen, Jiafu Jiang, Likai Wang, Zhiyong Guan, Fadi Chen, and et al. 2022. "Chrysanthemum CmHSP90.5 as a Tool to Regulate Heat and Salt Stress Tolerance" Horticulturae 8, no. 6: 532. https://doi.org/10.3390/horticulturae8060532
APA StyleWang, X., Wu, J., Wang, Y., Jiang, Y., Li, F., Chen, Y., Jiang, J., Wang, L., Guan, Z., Chen, F., & Chen, S. (2022). Chrysanthemum CmHSP90.5 as a Tool to Regulate Heat and Salt Stress Tolerance. Horticulturae, 8(6), 532. https://doi.org/10.3390/horticulturae8060532









