ddRADseq Applications for Petunia × hybrida Clonal Line Breeding: Genotyping and Variant Identification for Target-Specific Assays
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. ddRADseq Library Preparation and Sequencing
2.3. ddRADseq Data Processing
2.4. Genetic Distance, Structure and Statistic Analyses
2.5. SNP Subset Selection for Core Set and Location in CDSs
3. Results
3.1. Genetic Distance and Structure Analysis
3.2. Genetic Statistics and AMOVA
3.3. GD Analysis Comparison Between Total and Core Set SNP Profiles
3.4. GD Analysis Comparison Between Total and CDS-Located SNP Profiles
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aswath, C.; Bose, T.K.; Bhatia, R.; Saha, T.N.; Dutta, K. Commercial Flowers; Daya Publishing House, Ed.; Daya Publishing: Delhi, India, 2021; Volume 4, ISBN 9789354614163. [Google Scholar]
- Chen, Q.X.C.; Warner, R.M. Identification of QTL for Plant Architecture and Flowering Performance Traits in a Multi-Environment Evaluation of a Petunia axillaris × P. exserta Recombinant Inbred Line Population. Horticulturae 2022, 8, 1006. [Google Scholar] [CrossRef]
- USDA. Floriculture Crops 2020 Summary; USDA: Washington, DC, USA, 2021. [Google Scholar]
- Gebhardt, C. The Historical Role of Species from the Solanaceae Plant Family in Genetic Research. Theor. Appl. Genet. 2016, 129, 2281. [Google Scholar] [CrossRef]
- Sink, K.C. Petunia; Springer: Berlin/Heidelberg, Germany, 1984; Volume 9. [Google Scholar] [CrossRef]
- Geitmann, A. Petunia: Evolutionary, Developmental and Physiological Genetics. Ann. Bot. 2011, 107, vi–vii. [Google Scholar] [CrossRef]
- Guo, Y.; Warner, R.M. Dissecting Genetic Diversity and Genomic Background of Petunia cultivars with Contrasting Growth Habits. Hortic. Res. 2020, 7, 155. [Google Scholar] [CrossRef]
- Hermann, K.; Klahre, U.; Venail, J.; Brandenburg, A.; Kuhlemeier, C. The Genetics of Reproductive Organ Morphology in Two Petunia Species with Contrasting Pollination Syndromes. Planta 2015, 241, 1241–1254. [Google Scholar] [CrossRef]
- Rodrigues, D.M.; Caballero-Villalobos, L.; Turchetto, C.; Assis Jacques, R.; Kuhlemeier, C.; Freitas, L.B. Do We Truly Understand Pollination Syndromes in Petunia as Much as We Suppose? AoB Plants 2018, 10, ply057. [Google Scholar] [CrossRef] [PubMed]
- Regalado, J.J.; Carmona-Martín, E.; Querol, V.; Veléz, C.G.; Encina, C.L.; Pitta-Alvarez, S.I. Production of Compact Petunias through Polyploidization. Plant Cell Tissue Organ Cult. 2017, 129, 61–71. [Google Scholar] [CrossRef]
- Dermen, H. Polyploidy in Petunia. Am. J. Bot. 1931, 18, 250–261. [Google Scholar] [CrossRef]
- Robbins, T.P.; Harbord, R.M.; Sonneveld, T.; Clarke, K. The Molecular Genetics of Self-Incompatibility in Petunia hybrida. Ann. Bot. 2000, 85, 105–112. [Google Scholar] [CrossRef][Green Version]
- Williams, J.S.; Wu, L.; Li, S.; Sun, P.; Kao, T.-H.; Robbins, T.P.; Sims, T.L. Insight into S-RNase-Based Self-Incompatibility in Petunia: Recent Findings and Future Directions. Front. Plant Sci. 2015, 6, 41. [Google Scholar] [CrossRef]
- Yue, Y.; Du, J.; Li, Y.; Thomas, H.R.; Frank, M.H.; Wang, L.; Hu, H. Insight into the Petunia Dof Transcription Factor Family Reveals a New Regulator of Male-Sterility. Ind. Crops Prod. 2021, 161, 113196. [Google Scholar] [CrossRef]
- Nivison, H.T.; Hanson, M.R. Identification of a Mitochondrial Protein Associated with Cytoplasmic Male Sterility in Petunia. Plant Cell 1989, 1, 1121–1130. [Google Scholar] [CrossRef][Green Version]
- Farinati, S.; Draga, S.; Betto, A.; Palumbo, F.; Vannozzi, A.; Lucchin, M.; Barcaccia, G. Current Insights and Advances into Plant Male Sterility: New Precision Breeding Technology Based on Genome Editing Applications. Front. Plant Sci. 2023, 14, 1223861. [Google Scholar] [CrossRef]
- Gerats, T.; Vandenbussche, M. A Model System for Comparative Research: Petunia. Trends Plant Sci. 2005, 10, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Melzer, R.; Janssen, B.J.; Dornelas, M.C.; Vandenbussche, M.; Chambrier, P.; Rodrigues Bento, S.; Morel, P. Petunia, Your Next Supermodel? Front. Plant Sci. 2016, 7, 72. [Google Scholar] [CrossRef]
- Meyer, P.; Heidmann, I.; Forkmann, G.; Saedler, H. A New Petunia Flower Colour Generated by Transformation of a Mutant with a Maize Gene. Nature 1987, 330, 677–678. [Google Scholar] [CrossRef] [PubMed]
- Mahmood-ur-Rahman, M.; Ali, I.; Husnain, T.; Riazuddin, S. RNA Interference: The Story of Gene Silencing in Plants and Humans. Elsevier Biotechnol. Adv. 2008, 26, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Napoli, C.; Lemieux, C.; Jorgensen, R. Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in Trans. Plant Cell 1990, 2, 279–289. [Google Scholar] [CrossRef]
- Cao, Z.; Guo, Y.; Yang, Q.; He, Y.; Fetouh, M.I.; Warner, R.M.; Deng, Z. Genome-Wide Identification of Quantitative Trait Loci for Important Plant and Flower Traits in Petunia Using a High-Density Linkage Map and an Interspecific Recombinant Inbred Population Derived from Petunia integrifolia and P. axillaris. Hortic. Res. 2019, 6, 27. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, W.; Ruan, Y.; Dai, B.; Yang, T.; Gou, T.; Liu, C.; Ning, G.; Liu, G.; Yu, Y.; et al. Construction of a High-Density Genetic Map and Mapping of Double Flower Genes in Petunia. Sci. Hortic. 2024, 329, 112988. [Google Scholar] [CrossRef]
- Bossolini, E.; Klahre, U.; Brandenburg, A.; Reinhardt, D.; Kuhlemeier, C.; Belzile, F. High Resolution Linkage Maps of the Model Organism Petunia Reveal Substantial Synteny Decay with the Related Genome of Tomato. Genome 2011, 54, 327–340. [Google Scholar] [CrossRef]
- Bombarely, A.; Moser, M.; Amrad, A.; Bao, M.; Bapaume, L.; Barry, C.S.; Bliek, M.; Boersma, M.R.; Borghi, L.; Bruggmann, R.; et al. Insight into the Evolution of the Solanaceae from the Parental Genomes of Petunia hybrida. Nat. Plants 2016, 2, 16074. [Google Scholar] [CrossRef]
- Saei, A.; Hunter, D.; Hilario, E.; David, C.; Ireland, H.; Esfandiari, A.; King, I.; Grierson, E.; Wang, L.; Boase, M.; et al. Chromosome-Level Genome Assembly and Annotation of Petunia hybrida. Sci. Data 2024, 12, 1262. [Google Scholar] [CrossRef] [PubMed]
- Kashikar, S.G.; Khalatkar, A.S. Breeding for Flower Colour in Petunia Hybrida Hort. In Proceedings of the Symposium on Vegetable and Flower seed Production 111; International Society for Horticultural Science (ISHS): Forli, Italy, 1981; pp. 35–40. [Google Scholar]
- Man-Zhu, D.; Se-Ping, B. Advances in Genetics and Breeding of Petunia hybrida Vilm. Chin. Bull. Bot. 2004, 21, 385. [Google Scholar]
- Masclaux-Daubresse, C.; Daniel-Vedele, F.; Dechorgnat, J.; Chardon, F.; Gaufichon, L.; Suzuki, A. Nitrogen Uptake, Assimilation and Remobilization in Plants: Challenges for Sustainable and Productive Agriculture. Ann. Bot. 2010, 105, 1141–1157. [Google Scholar] [CrossRef] [PubMed]
- Lütken, H.; Clarke, J.L.; Müller, R. Genetic Engineering and Sustainable Production of Ornamentals: Current Status and Future Directions. Plant Cell Rep. 2012, 31, 1141–1157. [Google Scholar] [CrossRef]
- CPVO Legislation in Force. Available online: https://cpvo.europa.eu/en/about-us/law-and-practice/legislation-in-force (accessed on 27 October 2024).
- UPOV Lex. Available online: https://upovlex.upov.int/en/convention (accessed on 27 October 2024).
- CPVO Protocol for Tests on Distinctness, Uniformity and Stability. Petunia Juss. × Petchoa J.M.H. Shaw. Available online: https://cpvo.europa.eu/sites/default/files/documents/petunia_2.pdf (accessed on 9 May 2025).
- UPOV Guidelines for the Conduct of Tests for Distinctness, Uniformity and Stability-Petunia (Petunia Juss.). Available online: https://www.google.com/url?sa=t&source=web&rct=j&opi=89978449&url=https://cpvo.europa.eu/sites/default/files/documents/petunia_2.pdf&ved=2ahUKEwj1uNWP7qaSAxVyhv0HHQz9OpIQFnoECBwQAQ&usg=AOvVaw2AfowugxhJGzXpqgTvDkKK (accessed on 15 January 2026).
- Yu, J.K.; Chung, Y.S. Plant Variety Protection: Current Practices and Insights. Genes 2021, 12, 1127. [Google Scholar] [CrossRef]
- Gilliland, T.J.; Annicchiarico, P.; Julier, B.; Ghesquière, M. A Proposal for Enhanced EU Herbage VCU and DUS Testing Procedures. Grass Forage Sci. 2020, 75, 227–241. [Google Scholar] [CrossRef]
- CPVO Guidance on the Use of Biochemical and Molecular Markers in the Examination of Distinctiveness, Uniformity and Stability (DUS). Available online: https://www.upov.int/edocs/tgpdocs/en/tgp_15.pdf (accessed on 4 May 2025).
- Patella, A.; Scariolo, F.; Palumbo, F.; Barcaccia, G. Genetic Structure of Cultivated Varieties of Radicchio (Cichorium intybus L.): A Comparison between F1 Hybrids and Synthetics. Plants 2019, 8, 213. [Google Scholar] [CrossRef]
- Seki, K.; Komatsu, K.; Hiraga, M.; Tanaka, K.; Uno, Y.; Matsumura, H. Development of PCR-Based Marker for Resistance to Fusarium Wilt Race 2 in Lettuce (Lactuca sativa L.). Euphytica 2021, 217, 126. [Google Scholar] [CrossRef]
- Zhigunov, A.V.; Ulianich, P.S.; Lebedeva, M.V.; Chang, P.L.; Nuzhdin, S.V.; Potokina, E.K. Development of F1 Hybrid Population and the High-Density Linkage Map for European Aspen (Populus tremula L.) Using RADseq Technology. BMC Plant Biol. 2017, 17, 180. [Google Scholar] [CrossRef]
- Peterson, B.K.; Weber, J.N.; Kay, E.H.; Fisher, H.S.; Hoekstra, H.E. Double Digest RADseq: An Inexpensive Method for De Novo SNP Discovery and Genotyping in Model and Non-Model Species. PLoS ONE 2012, 7, e37135. [Google Scholar] [CrossRef]
- Chiurugwi, T.; Kemp, S.; Powell, W.; Hickey, L.T. Speed Breeding Orphan Crops. Theor. Appl. Genet. 2019, 132, 607–616. [Google Scholar] [CrossRef]
- Johnson, M.; Deshpande, S.; Vetriventhan, M.; Upadhyaya, H.D.; Wallace, J.G. Genome-Wide Population Structure Analyses of Three Minor Millets: Kodo Millet, Little Millet, and Proso Millet. Plant Genome 2019, 12, 190021. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.; Hackett, C.; Hedley, P.; Liu, H.; Milne, L.; Bayer, M.; Marshall, D.; Jorgensen, L.; Gordon, S.; Brennan, R. The Use of Genotyping by Sequencing in Blackcurrant (Ribes nigrum): Developing High-Resolution Linkage Maps in Species without Reference Genome Sequences. Mol. Breed. 2014, 33, 835–849. [Google Scholar] [CrossRef]
- Praveen, P.; Gopal, R.; Ramakrishnan, U. The Population Structure of Invasive Lantana Camara Is Shaped by Its Mating System. eLife 2024, 14, RP104988. [Google Scholar] [CrossRef]
- Zhang, X.J.; Liu, X.F.; Liu, D.T.; Cao, Y.R.; Li, Z.H.; Ma, Y.P.; Ma, H. Genetic Diversity and Structure of Rhododendron meddianum, a Plant Species with Extremely Small Populations. Plant Divers. 2021, 43, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, T.M.; Nazareno, A.G. One Step Away From Extinction: A Population Genomic Analysis of A Narrow Endemic, Tropical Plant Species. Front. Plant Sci. 2021, 12, 730258. [Google Scholar] [CrossRef]
- Ramirez-Ramirez, A.R.; Bidot-Martínez, I.; Mirzaei, K.; Rasoamanalina Rivo, O.L.; Menéndez-Grenot, M.; Clapé-Borges, P.; Espinosa-Lopez, G.; Bertin, P. Comparing the Performances of SSR and SNP Markers for Population Analysis in Theobroma cacao L., as Alternative Approach to Validate a New DdRADseq Protocol for Cacao Genotyping. PLoS ONE 2024, 19, e0304753. [Google Scholar] [CrossRef]
- Molina, C.; Aguirre, N.C.; Vera, P.A.; Filippi, C.V.; Puebla, A.F.; Poltri, S.N.M.; Paniego, N.B.; Acevedo, A. DdRADseq-Mediated Detection of Genetic Variants in Sugarcane. Plant Mol. Biol. 2023, 111, 205–219. [Google Scholar] [CrossRef]
- McLean-Rodríguez, F.D.; Costich, D.E.; Camacho-Villa, T.C.; Pè, M.E.; Dell’Acqua, M. Genetic Diversity and Selection Signatures in Maize Landraces Compared across 50 Years of in Situ and Ex Situ Conservation. Heredity 2021, 126, 913–928. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Guo, J.; Yu, B.; Chen, W.; Zhang, H.; Zhou, X.; Chen, Y.; Huang, L.; Liu, N.; Ren, X.; et al. Construction of DdRADseq-Based High-Density Genetic Map and Identification of Quantitative Trait Loci for Trans-Resveratrol Content in Peanut Seeds. Front. Plant Sci. 2021, 12, 644402. [Google Scholar] [CrossRef]
- Martina, M.; Acquadro, A.; Gulino, D.; Brusco, F.; Rabaglio, M.; Portis, E.; Lanteri, S. First Genetic Maps Development and QTL Mining in Ranunculus asiaticus L. through DdRADseq. Front. Plant Sci. 2022, 13, 1009206. [Google Scholar] [CrossRef]
- Ksouri, N.; Sánchez, G.; Forcada, C.F.I.; Contreras-Moreira, B.; Gogorcena, Y. DdRAD-Seq-Derived SNPs Reveal Novel Association Signatures for Fruit-Related Traits in Peach. bioRxiv 2023. [Google Scholar] [CrossRef]
- Daemi-Saeidabad, M.; Shojaeiyan, A.; Vivian-Smith, A.; Stenøien, H.K.; Falahati-Anbaran, M. The Taxonomic Significance of DdRADseq Based Microsatellite Markers in the Closely Related Species of Heracleum (Apiaceae). PLoS ONE 2020, 15, e0232471. [Google Scholar] [CrossRef]
- Reinula, I.; Träger, S.; Hernández-Agramonte, I.M.; Helm, A.; Aavik, T. Landscape Genetic Analysis Suggests Stronger Effects of Past than Current Landscape Structure on Genetic Patterns of Primula veris. Divers. Distrib. 2021, 27, 1648–1662. [Google Scholar] [CrossRef]
- Ma, X.; Wang, H.; Hu, Y.; Hu, Q.; Qian, R.; Zheng, J.; Zhang, X. DdRAD-Seq Data Reveals the Significant Population Structure and Potential Causes of Population Divergence in Bougainvillea. Sci. Rep. 2025, 15, 34200. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Huang, S.; Huang, G.; Wu, X.; Jin, H.; Liu, Y.; Xiao, Y.; Zhou, R. Genetic Relatedness and Association Mapping of Horticulturally Valuable Traits for the Ceiba Plants Using DdRAD Sequencing. Hortic. Plant J. 2023, 9, 826–836. [Google Scholar] [CrossRef]
- Betto, A.; Palumbo, F.; Riommi, D.; Vannozzi, A.; Barcaccia, G. Harnessing Genomics for Breeding Lantana camara L.: Genotyping and Ploidy Testing of Clonal Lines Through DdRADseq Applications. Int. J. Mol. Sci. 2025, 26, 4898. [Google Scholar] [CrossRef]
- Roy, S.C.; Moitra, K.; De Sarker, D. Assessment of Genetic Diversity among Four Orchids Based on DdRAD Sequencing Data for Conservation Purposes. Physiol. Mol. Biol. Plants 2017, 23, 169–183. [Google Scholar] [CrossRef]
- Moreau, E.P.; Honig, J.A.; Molnar, T.J. High-Density Linkage Mapping and Identification of Quantitative Trait Loci Associated with Powdery Mildew Resistance in Flowering Dogwood (Cornus florida). Horticulturae 2022, 8, 405. [Google Scholar] [CrossRef]
- Kim, H.; Yoon, J.B.; Lee, J. Development of Fluidigm SNP Type Genotyping Assays for Marker-Assisted Breeding of Chili Pepper (Capsicum annuum L.). Hortic. Sci. Technol. 2017, 35, 465–479. [Google Scholar] [CrossRef]
- Scaglione, D.; Pinosio, S.; Marroni, F.; Di Centa, E.; Fornasiero, A.; Magris, G.; Scalabrin, S.; Cattonaro, F.; Taylor, G.; Morgante, M. Single Primer Enrichment Technology as a Tool for Massive Genotyping: A Benchmark on Black Poplar and Maize. Ann. Bot. 2019, 124, 543–551. [Google Scholar] [CrossRef]
- Sun, X.; Liu, D.; Zhang, X.; Li, W.; Liu, H.; Hong, W.; Jiang, C.; Guan, N.; Ma, C.; Zeng, H.; et al. SLAF-Seq: An Efficient Method of Large-Scale De Novo SNP Discovery and Genotyping Using High-Throughput Sequencing. PLoS ONE 2013, 8, 58700. [Google Scholar] [CrossRef]
- Yang, Q.; Zhang, J.; Shi, X.; Chen, L.; Qin, J.; Zhang, M.; Yang, C.; Song, Q.; Yan, L.; Yang, Q.; et al. Development of SNP Marker Panels for Genotyping by Target Sequencing (GBTS) and Its Application in Soybean. Mol. Breed. 2023, 43, 26–27. [Google Scholar] [CrossRef]
- Dou, T.; Wang, C.; Ma, Y.; Chen, Z.; Zhang, J.; Guo, G. CoreSNP: An Efficient Pipeline for Core Marker Profile Selection from Genome-Wide SNP Datasets in Crops. BMC Plant Biol. 2023, 23, 580. [Google Scholar] [CrossRef]
- Sung, C.J.; Kulkarni, R.; Hillhouse, A.; Simpson, C.E.; Cason, J.; Burow, M.D. Reduced-Cost Genotyping by Resequencing in Peanut Breeding Programs Using Tecan Allegro Targeted Resequencing V2. Genes 2024, 15, 1364. [Google Scholar] [CrossRef]
- Broccanello, C.; Chiodi, C.; Funk, A.; Mcgrath, J.M.; Panella, L.; Stevanato, P. Comparison of Three PCR-Based Assays for SNP Genotyping in Plants. Plant Methods 2018, 14, 28. [Google Scholar] [CrossRef]
- Park, J.S.; Kang, M.Y.; Shim, E.J.; Oh, J.H.; Seo, K.I.; Kim, K.S.; Sim, S.C.; Chung, S.M.; Park, Y.; Lee, G.P.; et al. Genome-Wide Core Sets of SNP Markers and Fluidigm Assays for Rapid and Effective Genotypic Identification of Korean Cultivars of Lettuce (Lactuca sativa L.). Hortic. Res. 2022, 9, uhac119. [Google Scholar] [CrossRef]
- Chen, X.; Wang, Q.; Wang, F.; Wu, X.; Pan, Y.; Xue, L.; Duan, Y.; Wang, S.; Guan, Y.; Zhao, K.; et al. SNP Fingerprint Database and Makers Screening for Current Phalaenopsis Cultivars. Ornam. Plant Res. 2025, 5, e011. [Google Scholar] [CrossRef]
- Patzer, L.; Thomsen, T.; Wamhoff, D.; Schulz, D.F.; Linde, M.; Debener, T. Development of a Robust SNP Marker Set for Genotyping Diverse Gene Bank Collections of Polyploid Roses. BMC Plant Biol. 2024, 24, 1076. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Shen, A.; Tan, Y.; Liu, L.; Li, S.; Tan, Z. Development of KASP Markers, SNP Fingerprinting and Population Genetic Analysis of Cymbidium ensifolium (L.) Sw. Germplasm Resources in China. Front. Plant Sci. 2024, 15, 1460603. [Google Scholar] [CrossRef]
- Abed, A.; Légaré, G.; Pomerleau, S.; St-Cyr, J.; Boyle, B.; Belzile, F.J. Genotyping-by-Sequencing on the Ion Torrent Platform in Barley. Methods Mol. Biol. 2019, 1900, 233–252. [Google Scholar] [CrossRef]
- Li, H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M. Twelve Years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Catchen, J.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An Analysis Tool Set for Population Genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef] [PubMed]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R Package for Genetic Analysis of Populations with Clonal, Partially Clonal, and/or Sexual Reproduction. PeerJ 2014, 2014, e281. [Google Scholar] [CrossRef]
- Nei, M. Analysis of Gene Diversity in Subdivided Populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.A.M.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-Generation PLINK: Rising to the Challenge of Larger and Richer Datasets. Gigascience 2015, 4, s13742-015-0047-8. [Google Scholar] [CrossRef]
- Manichaikul, A.; Mychaleckyj, J.C.; Rich, S.S.; Daly, K.; Sale, M.; Chen, W.M. Robust Relationship Inference in Genome-Wide Association Studies. Bioinformatics 2010, 26, 2867–2873. [Google Scholar] [CrossRef]
- Sinnwell, J.P.; Therneau, T.M.; Schaid, D.J. The Kinship2 R Package for Pedigree Data. Hum. Hered. 2014, 78, 91–93. [Google Scholar] [CrossRef]
- Van Der Ploeg, A. Drawing Non-Layered Tidy Trees in Linear Time. Softw. Pract. Exp. 2014, 44, 1467–1484. [Google Scholar] [CrossRef]
- Gruber, B.; Unmack, P.J.; Berry, O.F.; Georges, A. Dartr: An r Package to Facilitate Analysis of SNP Data Generated from Reduced Representation Genome Sequencing. Mol. Ecol. Resour. 2018, 18, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Goudet, J. Hierfstat, a Package for R to Compute and Test Hierarchical F-Statistics. Mol. Ecol. Notes 2005, 5, 184–186. [Google Scholar] [CrossRef]
- Gabelli, G. Minimal Discriminating Marker Set Finder. Available online: https://github.com/Gabelberg/minimal_discriminating_marker_set/blob/main/README.md (accessed on 15 January 2026).
- Krzywinski, M.; Schein, J.; Birol, İ.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Wang, B.; Yang, X.; Jia, Y.; Xu, Y.; Jia, P.; Dang, N.; Wang, S.; Xu, T.; Zhao, X.; Gao, S.; et al. High-Quality Arabidopsis Thaliana Genome Assembly with Nanopore and HiFi Long Reads. Genom. Proteom. Bioinform. 2022, 20, 4–13. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A Graphical Gene-Set Enrichment Tool for Animals and Plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Krishnan, G.S.; Singh, A.K.; Waters, D.L.E.; Henry, R.J. Molecular Markers for Harnessing Heterosis. Mol. Markers Plants 2012, 119–136. [Google Scholar] [CrossRef]
- Caballero, A.; Villanueva, B.; Druet, T. On the Estimation of Inbreeding Depression Using Different Measures of Inbreeding from Molecular Markers. Evol. Appl. 2020, 14, 416–428. [Google Scholar] [CrossRef]
- Nguyen, T.K.; Ha, S.T.T.; Lim, J.H. Analysis of Chrysanthemum Genetic Diversity by Genotyping-by-Sequencing. Hortic. Environ. Biotechnol. 2020, 61, 903–913. [Google Scholar] [CrossRef]
- Karikari, B.; Li, S.; Bhat, J.A.; Cao, Y.; Kong, J.; Yang, J.; Gai, J.; Zhao, T. Genome-Wide Detection of Major and Epistatic Effect QTLs for Seed Protein and Oil Content in Soybean Under Multiple Environments Using High-Density Bin Map. Int. J. Mol. Sci. 2019, 20, 979. [Google Scholar] [CrossRef]
- Yu, Q.; Ling, Y.; Xiong, Y.; Zhao, W.; Xiong, Y.; Dong, Z.; Yang, J.; Zhao, J.; Zhang, X.; Ma, X. RAD-Seq as an Effective Strategy for Heterogenous Variety Identification in Plants—A Case Study in Italian Ryegrass (Lolium multiflorum). BMC Plant Biol. 2022, 22, 231. [Google Scholar] [CrossRef]
- Peltier, D.; Farcy, E.; Dulieu, H.; Bervillé, A. Origin, Distribution and Mapping of RAPD Markers from Wild Petunia Species in Petunia hybrida Hort Lines. Theor. Appl. Genet. 1994, 88, 637–645. [Google Scholar] [CrossRef]
- Galliot, C.; Hoballah, M.E.; Kuhlemeier, C.; Stuurman, J. Genetics of Flower Size and Nectar Volume in Petunia Pollination Syndromes. Planta 2006, 225, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Strommer, J.; Peters, J.; Zethof, J.; De Keukeleire, P.; Gerats, T. AFLP Maps of Petunia hybrida: Building Maps When Markers Cluster. Theor. Appl. Genet. 2002, 105, 1000–1009. [Google Scholar] [CrossRef] [PubMed]
- Vallejo, V.A.; Tychonievich, J.; Lin, W.K.; Wangchu, L.; Barry, C.S.; Warner, R.M. Identification of QTL for Crop Timing and Quality Traits in an Interspecific Petunia Population. Mol. Breed. 2015, 35, 2. [Google Scholar] [CrossRef]
- Liu, C.; He, Y.; Gou, T.; Li, X.; Ning, G.; Bao, M. Identification of Molecular Markers Associated with the Double Flower Trait in Petunia hybrida. Sci. Hortic. 2016, 206, 43–50. [Google Scholar] [CrossRef]
- Klahre, U.; Gurba, A.; Hermann, K.; Saxenhofer, M.; Bossolini, E.; Guerin, P.M.; Kuhlemeier, C. Pollinator Choice in Petunia Depends on Two Major Genetic Loci for Floral Scent Production. Curr. Biol. 2011, 21, 730–739. [Google Scholar] [CrossRef]
- Pezzi, P.H.; Guzmán-Rodriguez, S.; Giudicelli, G.C.; Turchetto, C.; Bombarely, A.; Freitas, L.B. A Convoluted Tale of Hybridization between Two Petunia Species from a Transitional Zone in South America. Perspect. Plant Ecol. Evol. Syst. 2022, 56, 125688. [Google Scholar] [CrossRef]
- Caballero-Villalobos, L.; Silva-Arias, G.A.; Turchetto, C.; Giudicelli, G.C.; Petzold, E.; Bombarely, A.; Freitas, L.B. Neutral and Adaptive Genomic Variation in Hybrid Zones of Two Ecologically Diverged Petunia Species (Solanaceae). Bot. J. Linn. Soc. 2021, 196, 100–122. [Google Scholar] [CrossRef]
- Guo, Y.; Wiegert-Rininger, K.E.; Vallejo, V.A.; Barry, C.S.; Warner, R.M. Transcriptome-Enabled Marker Discovery and Mapping of Plastochron-Related Genes in Petunia spp. BMC Genom. 2015, 16, 726. [Google Scholar] [CrossRef]
- Guo, Y.; Lin, W.K.; Chen, Q.; Vallejo, V.A.; Warner, R.M. Genetic Determinants of Crop Timing and Quality Traits in Two Interspecific Petunia Recombinant Inbred Line Populations. Sci. Rep. 2017, 7, 3200. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A Robust, Simple Genotyping-by-Sequencing (GBS) Approach for High Diversity Species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef] [PubMed]
- Berardi, A.E.; Esfeld, K.; Jäggi, L.; Mandel, T.; Cannarozzi, G.M.; Kuhlemeier, C. Complex Evolution of Novel Red Floral Color in Petunia. Plant Cell 2021, 33, 2273–2295. [Google Scholar] [CrossRef] [PubMed]
- Arnaud-Haond, S.; Duarte, C.M.; Alberto, F.; Serrão, E.A. Standardizing Methods to Address Clonality in Population Studies. Mol. Ecol. 2007, 16, 5115–5139. [Google Scholar] [CrossRef] [PubMed]
- Clifton, M.D.; D’Ombrain, T.H.; Watts-Parker, B.D.; Wills, A.R.; Hoebee, S.E. Genomic Evidence for a Hybrid Origin of the Critically Endangered Shrub Callistemon kenmorrisonii (Myrtaceae) and Persistence Involving Extreme Clonality. Conserv. Genet. 2025, 26, 1011–1028. [Google Scholar] [CrossRef]
- Lasso, E. The Importance of Setting the Right Genetic Distance Threshold for Identification of Clones Using Amplified Fragment Length Polymorphism: A Case Study with Five Species in the Tropical Plant Genus Piper. Mol. Ecol. Resour. 2008, 8, 74–82. [Google Scholar] [CrossRef]
- Flesher, K.N.; Jurgensen, M.F.; Gailing, O. Comparison of Phenotypic and Genetic Clone Delineation in Quaking Aspen, Populus tremuloides. Trees-Struct. Funct. 2016, 30, 1657–1667. [Google Scholar] [CrossRef]
- Coste, C.F.D.; Ronget, V.; Ramirez-Loza, J.-P.; Cubaynes, S.; Pavard, S.; Bienvenu, F. The Kinship Matrix: Inferring the Kinship Structure of a Population from Its Demography. Ecol. Lett. 2021, 24, 2750–2762. [Google Scholar] [CrossRef]
- Thompson, E.A. Identity by Descent: Variation in Meiosis, Across Genomes, and in Populations. Genetics 2013, 194, 301–326. [Google Scholar] [CrossRef]
- McMaster, E.S.; Yap, J.Y.S.; Chen, S.H.; Sherieff, A.; Bate, M.; Brown, I.; Jones, M.; Rossetto, M. On the Edge: Conservation Genomics of the Critically Endangered Dwarf Mountain Pine Pherosphaera fitzgeraldii. Basic Appl. Ecol. 2024, 80, 61–71. [Google Scholar] [CrossRef]
- Rueda, M.A.R.; Briceño-Pinzón, I.D.; Miguel, L.A.; Martínez, J.L.Q.; Padua, L.N.; de Lima Ribeiro, R.H.; Martins, V.S.; de Souza, L.C.; da Silva Junior, A.L.; de Souza Marçal, T. Multivariate Analysis of Genetic Variability in Advanced Potato Clones Under Tropical Conditions. Agric. Res. 2025, 1–10. [Google Scholar] [CrossRef]
- Woldeyohannes, A.B.; Iohannes, S.D.; Miculan, M.; Caproni, L.; Ahmed, J.S.; de Sousa, K.; Desta, E.A.; Fadda, C.; Pè, M.E.; Dell’acqua, M. Data-Driven, Participatory Characterization of Farmer Varieties Discloses Teff Breeding Potential under Current and Future Climates. Elife 2022, 11, e80009. [Google Scholar] [CrossRef] [PubMed]
- McMaster, E.S.; Lu-Irving, P.; van der Merwe, M.M.; Ho, S.Y.W.; Rossetto, M. Evaluating Kinship Estimation Methods for Reduced-Representation SNP Data in Non-Model Species. Mol. Ecol. Resour. 2025, 25, e70038. [Google Scholar] [CrossRef]
- Scariolo, F.; Palumbo, F.; Farinati, S.; Barcaccia, G. Pipeline to Design Inbred Lines and F1 Hybrids of Leaf Chicory (Radicchio) Using Male Sterility and Genotyping-by-Sequencing. Plants 2023, 12, 1242. [Google Scholar] [CrossRef]
- UPOV Explanatory Notes on Essentially Derived Varieties Under the 1991 Act of the UPOV Convention. Available online: https://www.upov.int/edocs/expndocs/en/upov_exn_edv.pdf (accessed on 4 May 2025).
- Noli, E.; Teriaca, M.S.; Conti, S. Criteria for the Definition of Similarity Thresholds for Identifying Essentially Derived Varieties. Plant Breed. 2013, 132, 525–531. [Google Scholar] [CrossRef]
- Jamali, S.H.; Cockram, J.; Hickey, L.T. Insights into Deployment of DNA Markers in Plant Variety Protection and Registration. Theor. Appl. Genet. 2019, 132, 1911–1929. [Google Scholar] [CrossRef] [PubMed]
- ISF View on Intellectual Property. Available online: https://worldseed.org/wp-content/uploads/2021/10/ISF-View-on-Intellectual-Property-2012-amended-2021.pdf (accessed on 16 January 2026).
- Van Eeuwijk, F.A.; Law, J.R. Statistical Aspects of Essential Derivation, with Illustrations Based on Lettuce and Barley. Euphytica 2004, 137, 129–137. [Google Scholar] [CrossRef]
- ISF Guidelines for Handling Disputes on Essential Derivation of Maize Lines. Available online: https://worldseed.org/wp-content/uploads/2015/10/ISF_Guidelines_Disputes_EDV_Maize_2014.pdf (accessed on 10 January 2026).
- Rousselle, Y.; Jones, E.; Charcosset, A.; Moreau, P.; Robbins, K.; Stich, B.; Knaak, C.; Flament, P.; Karaman, Z.; Martinant, J.P.; et al. Study on Essential Derivation in Maize: III. Selection and Evaluation of a Panel of Single Nucleotide Polymorphism Loci for Use in European and North American Germplasm. Crop Sci. 2015, 55, 1170–1180. [Google Scholar] [CrossRef]
- Yang, X.; Fang, Z.; Cao, Y.; Tian, L.; Peng, H.; Zhang, Y.; Xu, J.; Zhou, J.; Huo, H.; Qi, D.; et al. Development and Application of a Multiple Nucleotide Polymorphism (MNP)-Based Molecular Identification System for Pear Cultivars. BMC Plant Biol. 2026, 26, 15. [Google Scholar] [CrossRef]
- COGEM Update on Unauthorised Genetically Modified Garden Petunia Varieties. Available online: https://cogem.net/en/publication/update-on-unauthorised-genetically-modified-garden-petunia-varieties/ (accessed on 19 May 2025).
- Entani, T.; Takayama, S.; Iwano, M.; Shiba, H.; Che, F.S.; Isogai, A. Relationship between Polyploidy and Pollen Self-Incompatibility Phenotype in Petunia hybrida Vilm. Biosci. Biotechnol. Biochem. 1999, 63, 1882–1888. [Google Scholar] [CrossRef]
- Chen, Y.; Jiang, P.; Wilde, H.D. A Self-Pollinating Mutant of Petunia hybrida. Sci. Hortic. 2014, 177, 10–13. [Google Scholar] [CrossRef]
- Thurow, L.B.; Gasic, K.; Bassols Raseira, M.D.C.; Bonow, S.; Marques Castro, C. Genome-Wide SNP Discovery through Genotyping by Sequencing, Population Structure, and Linkage Disequilibrium in Brazilian Peach Breeding Germplasm. Tree Genet. Genomes 2020, 16, 10. [Google Scholar] [CrossRef]
- Ksouri, N.; Sánchez, G.; Font i Forcada, C.; Contreras-Moreira, B.; Gogorcena, Y. A Reproducible DdRAD-Seq Protocol Reveals Novel Genomic Association Signatures for Fruit-Related Traits in Peach. Plant Methods 2025, 21, 101. [Google Scholar] [CrossRef]
- Wang, Y.; Lv, H.; Xiang, X.; Yang, A.; Feng, Q.; Dai, P.; Li, Y.; Jiang, X.; Liu, G.; Zhang, X. Construction of a SNP Fingerprinting Database and Population Genetic Analysis of Cigar Tobacco Germplasm Resources in China. Front. Plant Sci. 2021, 12, 618133. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Thiel, T.; Sretenovic-Rajicic, T.; Baum, M.; Valkoun, J.; Guo, P.; Grando, S.; Ceccarelli, S.; Graner, A. Identification and Validation of a Core Set of Informative Genic SSR and SNP Markers for Assaying Functional Diversity in Barley. Mol. Breed. 2008, 22, 1–13. [Google Scholar] [CrossRef]
- Kuang, M.; Wei, S.J.; Wang, Y.Q.; Zhou, D.Y.; Ma, L.; Fang, D.; Yang, W.H.; Ma, Z.Y. Development of a Core Set of SNP Markers for the Identification of Upland Cotton Cultivars in China. J. Integr. Agric. 2016, 15, 954–962. [Google Scholar] [CrossRef]
- Yang, G.; Chen, S.; Chen, L.; Sun, K.; Huang, C.; Zhou, D.; Huang, Y.; Wang, J.; Liu, Y.; Wang, H.; et al. Development of a Core SNP Arrays Based on the KASP Method for Molecular Breeding of Rice. Rice 2019, 12, 21. [Google Scholar] [CrossRef]
- Meng, B.; Wang, S.; Li, W.X.; Guo, Z.; Tang, J. QTL Mapping of Fusarium Ear Rot Resistance Using Genotyping by Target Sequencing (GBTS) in Maize. J. Appl. Genet. 2025, 66, 787–796. [Google Scholar] [CrossRef]
- Yu, H.J.; Jeong, Y.M.; Lee, Y.J.; Yim, B.; Cho, A.; Mun, J.H. Marker Integration and Development of Fluidigm/KASP Assays for High-Throughput Genotyping of Radish. Hortic. Environ. Biotechnol. 2020, 61, 767–777. [Google Scholar] [CrossRef]
- Nagano, S.; Hirao, T.; Takashima, Y.; Matsushita, M.; Mishima, K.; Takahashi, M.; Iki, T.; Ishiguri, F.; Hiraoka, Y. SNP Genotyping with Target Amplicon Sequencing Using a Multiplexed Primer Panel and Its Application to Genomic Prediction in Japanese Cedar, Cryptomeria japonica (L.f.) D.Don. Forests 2020, 11, 898. [Google Scholar] [CrossRef]
- Guo, Z.; Wang, H.; Tao, J.; Ren, Y.; Xu, C.; Wu, K.; Zou, C.; Zhang, J.; Xu, Y. Development of Multiple SNP Marker Panels Affordable to Breeders through Genotyping by Target Sequencing (GBTS) in Maize. Mol. Breed. 2019, 39, 37. [Google Scholar] [CrossRef]
- Ayalew, H.; Tsang, P.W.; Chu, C.; Wang, J.; Liu, S.; Chen, C.; Ma, X.F. Comparison of TaqMan, KASP and RhAmp SNP Genotyping Platforms in Hexaploid Wheat. PLoS ONE 2019, 14, e0217222. [Google Scholar] [CrossRef]
- Pootakham, W.; Sonthirod, C.; Naktang, C.; Jomchai, N.; Sangsrakru, D.; Tangphatsornruang, S. Effects of Methylation-Sensitive Enzymes on the Enrichment of Genic SNPs and the Degree of Genome Complexity Reduction in a Two-Enzyme Genotyping-by-Sequencing (GBS) Approach: A Case Study in Oil Palm (Elaeis guineensis). Mol. Breed. 2016, 36, 154. [Google Scholar] [CrossRef]
- Brazier, T.; Glémin, S. Diversity and Determinants of Recombination Landscapes in Flowering Plants. PLoS Genet. 2022, 18, e1010141. [Google Scholar] [CrossRef]
- Zou, M.; Shabala, S.; Zhao, C.; Zhou, M. Molecular Mechanisms and Regulation of Recombination Frequency and Distribution in Plants. Theor. Appl. Genet. 2024, 137, 86. [Google Scholar] [CrossRef]
- Tiley, G.P.; Burleigh, G. The Relationship of Recombination Rate, Genome Structure, and Patterns of Molecular Evolution across Angiosperms. BMC Evol. Biol. 2015, 15, 194. [Google Scholar] [CrossRef]
- Park, M.; Jo, S.H.; Kwon, J.K.; Park, J.; Ahn, J.H.; Kim, S.; Lee, Y.H.; Yang, T.J.; Hur, C.G.; Kang, B.C.; et al. Comparative Analysis of Pepper and Tomato Reveals Euchromatin Expansion of Pepper Genome Caused by Differential Accumulation of Ty3/Gypsy-like Elements. BMC Genom. 2011, 12, 85. [Google Scholar] [CrossRef]
- Fuentes, R.R.; De Ridder, D.; Van Dijk, A.D.J.; Peters, S.A. Domestication Shapes Recombination Patterns in Tomato. Mol. Biol. Evol. 2022, 39, msab287. [Google Scholar] [CrossRef]
- Guyot, R.; Cheng, X.; Su, Y.; Cheng, Z.; Schlagenhauf, E.; Keller, B.; Ling, H.Q. Complex Organization and Evolution of the Tomato Pericentromeric Region at the FER Gene Locus. Plant Physiol. 2005, 138, 1205–1215. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mammadov, J.; Aggarwal, R.; Buyyarapu, R.; Kumpatla, S. SNP Markers and Their Impact on Plant Breeding. Int. J. Plant Genom. 2012, 2012, 728398. [Google Scholar] [CrossRef] [PubMed]
- Patrick, R.M.; Huang, X.Q.; Dudareva, N.; Li, Y. Dynamic Histone Acetylation in Floral Volatile Synthesis and Emission in Petunia Flowers. J. Exp. Bot. 2021, 72, 3704–3722. [Google Scholar] [CrossRef]
- Fu, J.; Huang, S.; Qian, J.; Qing, H.; Wan, Z.; Cheng, H.; Zhang, C. Genome-Wide Identification of Petunia HSF Genes and Potential Function of PhHSF19 in Benzenoid/Phenylpropanoid Biosynthesis. Int. J. Mol. Sci. 2022, 23, 2974. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Liu, X.; Jiang, L.; Qing, H.; Qian, J.; Li, Z.; Zhang, C.; Fu, J. Genome-Wide Identification of Petunia C2H2 Zinc Finger Family Genes and Their Potential Roles in Floral Volatile Benzenoids/Phenylpropanoids Metabolism. Planta 2026, 263, 43. [Google Scholar] [CrossRef]
- Bednarczyk, D.; Skaliter, O.; Kerzner, S.; Masci, T.; Shklarman, E.; Shor, E.; Vainstein, A. The Homeotic Gene PhDEF Regulates Production of Volatiles in Petunia Flowers by Activating EOBI and EOBII. Plant Cell 2025, 37, koaf027. [Google Scholar] [CrossRef]
- Sasse, J.; Schlegel, M.; Borghi, L.; Ullrich, F.; Lee, M.; Liu, G.W.; Giner, J.L.; Kayser, O.; Bigler, L.; Martinoia, E.; et al. Petunia hybrida PDR2 Is Involved in Herbivore Defense by Controlling Steroidal Contents in Trichomes. Plant Cell Environ. 2016, 39, 2725–2739. [Google Scholar] [CrossRef]
- Amano, I.; Kitajima, S.; Suzuki, H.; Koeduka, T.; Shitan, N. Transcriptome Analysis of Petunia axillaris Flowers Reveals Genes Involved in Morphological Differentiation and Metabolite Transport. PLoS ONE 2018, 13, e0198936. [Google Scholar] [CrossRef]
- Shor, E.; Vainstein, A. Petunia PHYTOCHROME INTERACTING FACTOR 4/5 Transcriptionally Activates Key Regulators of Floral Scent. Plant Mol. Biol. 2024, 114, 66. [Google Scholar] [CrossRef]
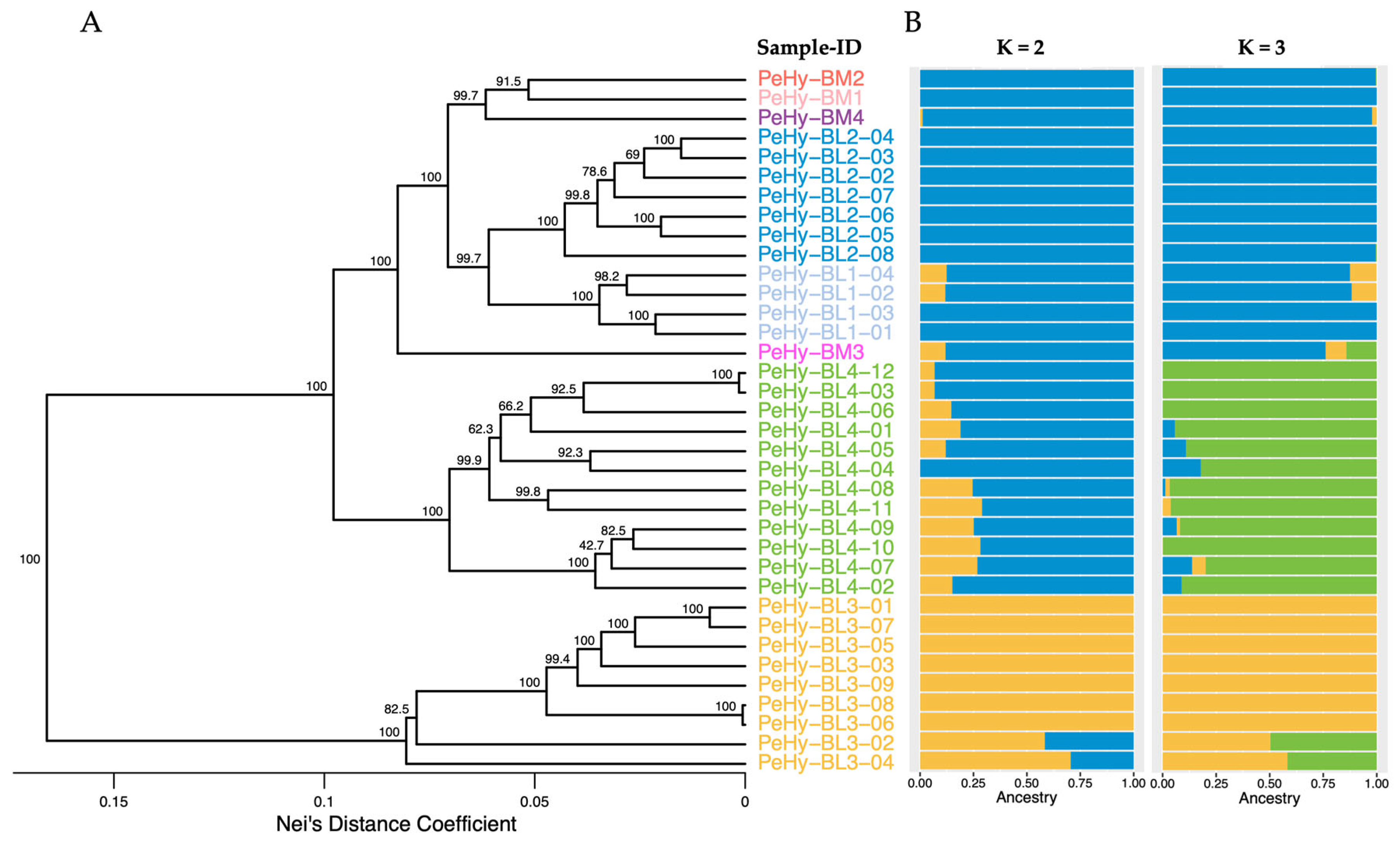
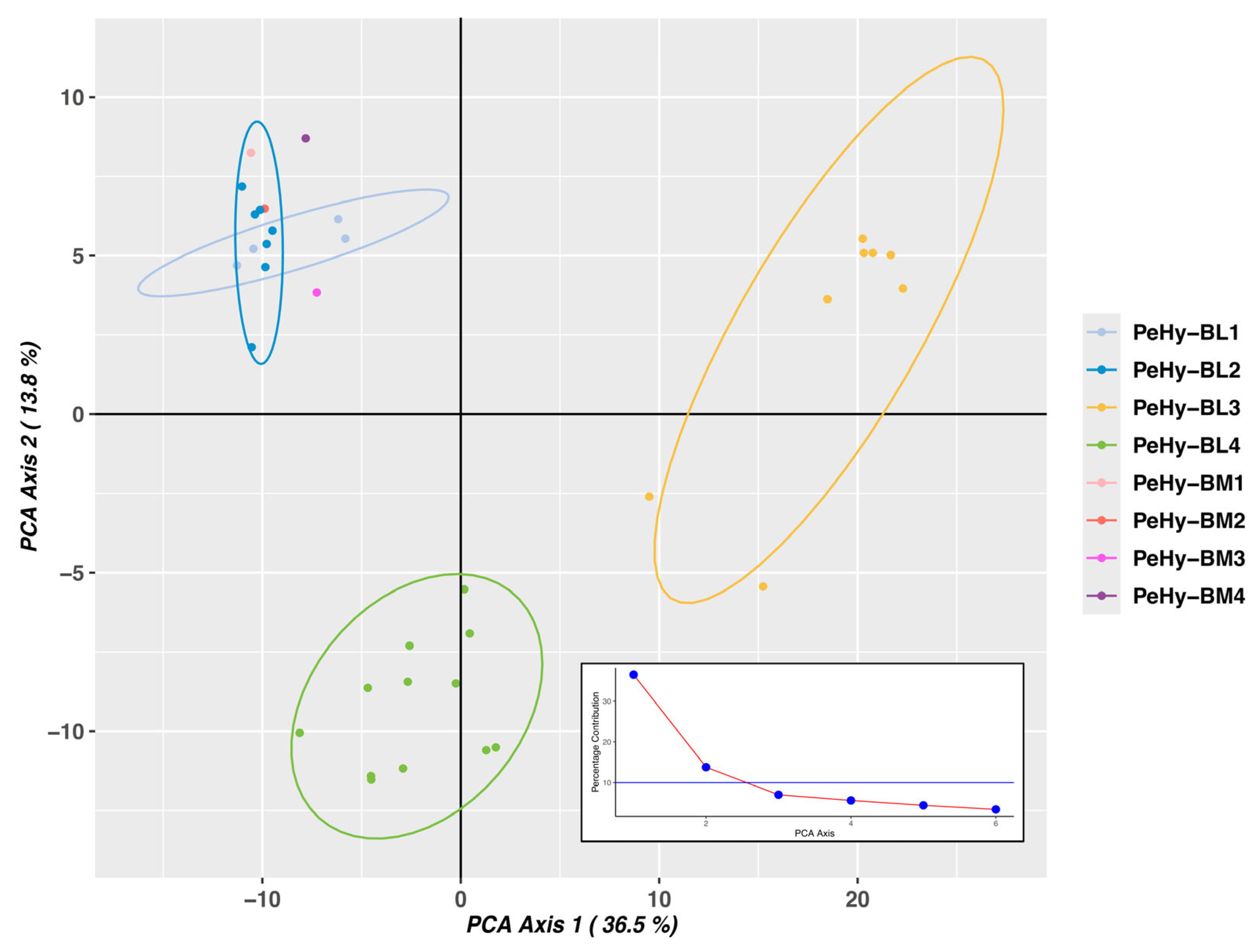
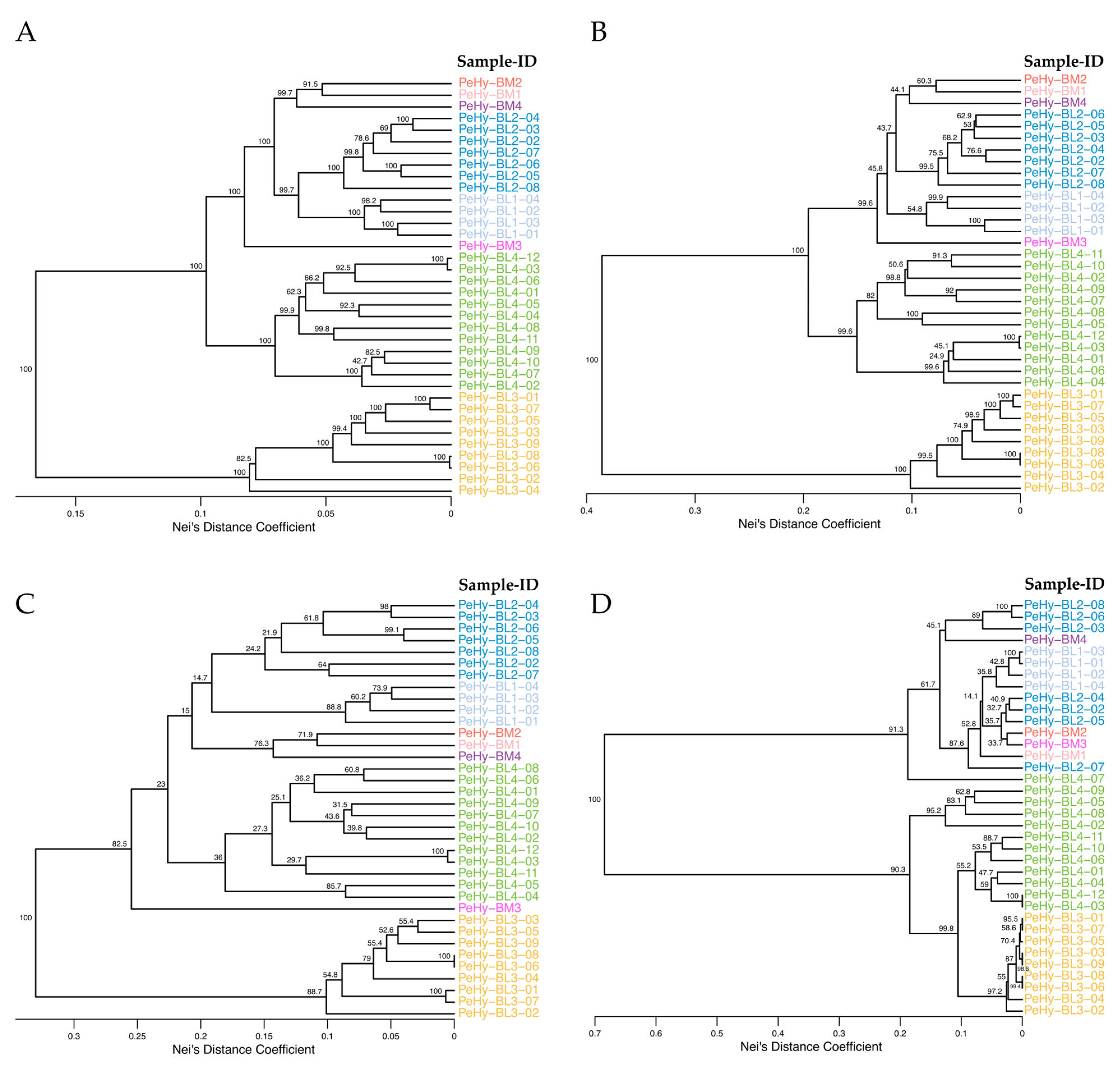
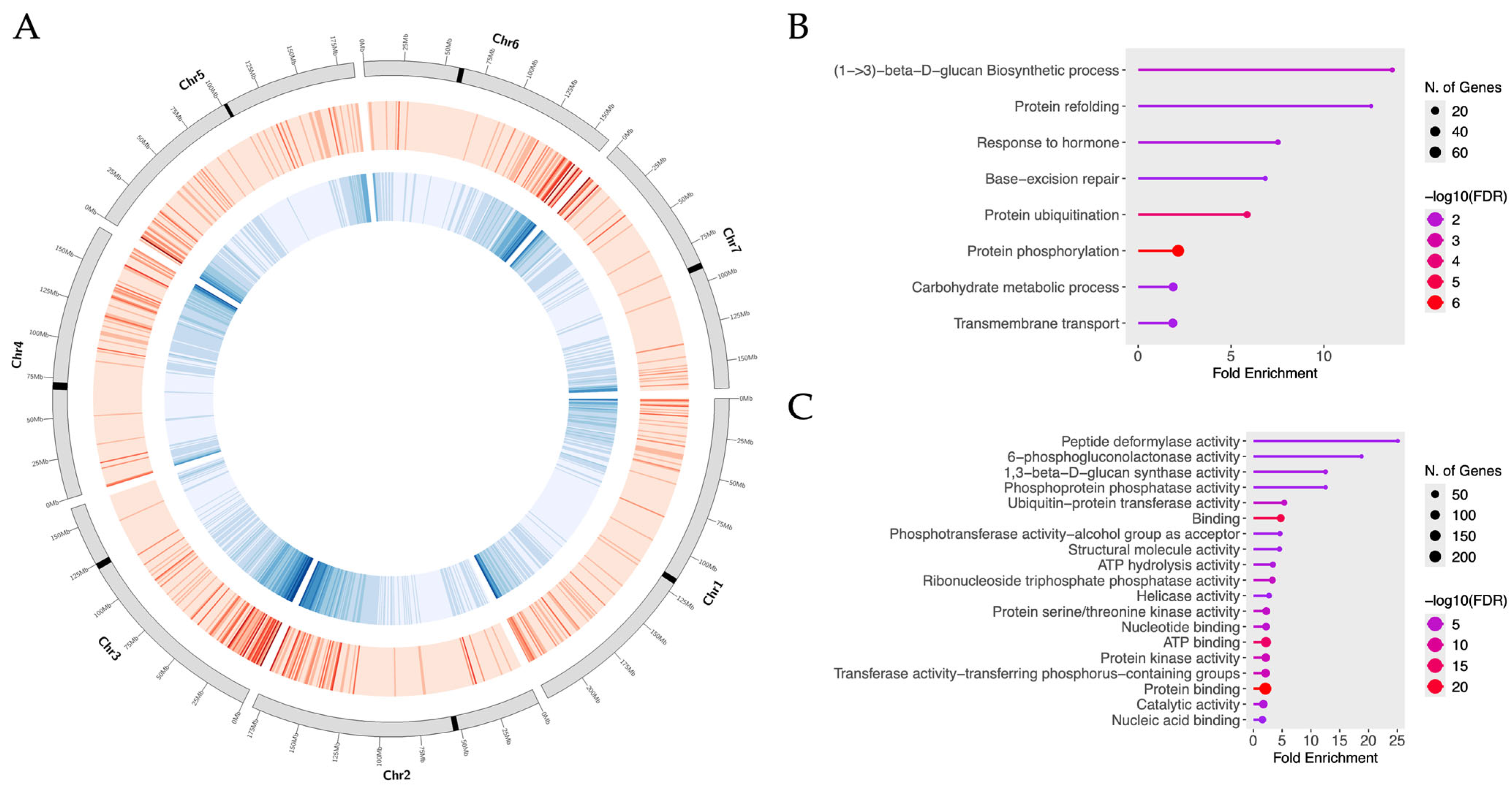
| Pop-ID | N | Mn Bases | ≥Q20 Mn Bases | Mn Reads | MRL (bp) |
|---|---|---|---|---|---|
| PeHy-BL1 | 4 | 486.46 | 416.44 | 3.34 | 160 |
| PeHy-BL2 | 7 | 360.03 | 304.84 | 2.39 | 173 |
| PeHy-BL3 | 9 | 316.36 | 271.88 | 1.99 | 177 |
| PeHy-BL4 | 12 | 267.85 | 230.08 | 1.68 | 178 |
| PeHy-BM1 | 1 | 258.86 | 226.06 | 1.25 | 206 |
| PeHy-BM2 | 1 | 104.60 | 91.09 | 0.50 | 208 |
| PeHy-BM3 | 1 | 469.56 | 399.95 | 3.31 | 141 |
| PeHy-BM4 | 1 | 551.17 | 468.70 | 3.91 | 141 |
| Tot | 36 | 12,291.01 | 10,528.17 | 80.01 | – |
| Avg | 341.42 | 292.45 | 2.22 | 173 |
| GD | Cluster | Fst | ||||||
|---|---|---|---|---|---|---|---|---|
| PeHy-BL2 | ||||||||
| 0.07 | PeHy-BL1 | 0.23 | ||||||
| 0.11 | 0.10 | PeHy-BL4 | 0.23 | 0.22 | ||||
| 0.19 | 0.27 | 0.29 | PeHy-BL3 | 0.50 | 0.45 | 0.34 | ||
| PeHy-BL3 | PeHy-BL4 | PeHy-BL1 | PeHy-BL2 | PeHy-BL2 | PeHy-BL1 | PeHy-BL4 | PeHy-BL3 | |
| Pop ID | N | na | ne | Ho (%) | Hs (%) | Fis | PL (%) | PA (%) |
|---|---|---|---|---|---|---|---|---|
| PeHy-BL1 | 4 | 1.36 | 1.27 | 19.05 | 15.02 | −0.27 | 63.75 | 6.38 |
| PeHy-BL2 | 7 | 1.54 | 1.31 | 20.48 | 16.14 | −0.27 | 68.54 | 14.89 |
| PeHy-BL3 | 9 | 1.54 | 1.46 | 26.27 | 22.08 | −0.19 | 73.14 | 33.96 |
| PeHy-BL4 | 12 | 1.66 | 1.55 | 33.57 | 26.17 | −0.28 | 81.88 | 12.75 |
| Avg | 8 | 1.53 | 1.53 | 24.84 | 19.85 | −0.25 | 71.83 | 17.00 |
| StDev | 3 | 0.11 | 0.12 | 5.72 | 4.53 | 0.04 | 6.69 | 10.28 |
| Df | Sum Sq | Mean Sq | Est. Var. | Est. Var. (%) | |
|---|---|---|---|---|---|
| Between Subpopulations | 7 | 16,100 | 2300 | 490 | 58.19 |
| Within Subpopulations | 28 | 9855 | 352 | 352 | 41.81 |
| Total | 35 | 25,955 | 742 | 842 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Betto, A.; Scariolo, F.; Gabelli, G.; Riommi, D.; Farinati, S.; Vannozzi, A.; Palumbo, F.; Barcaccia, G. ddRADseq Applications for Petunia × hybrida Clonal Line Breeding: Genotyping and Variant Identification for Target-Specific Assays. Horticulturae 2026, 12, 160. https://doi.org/10.3390/horticulturae12020160
Betto A, Scariolo F, Gabelli G, Riommi D, Farinati S, Vannozzi A, Palumbo F, Barcaccia G. ddRADseq Applications for Petunia × hybrida Clonal Line Breeding: Genotyping and Variant Identification for Target-Specific Assays. Horticulturae. 2026; 12(2):160. https://doi.org/10.3390/horticulturae12020160
Chicago/Turabian StyleBetto, Angelo, Francesco Scariolo, Giovanni Gabelli, Damiano Riommi, Silvia Farinati, Alessandro Vannozzi, Fabio Palumbo, and Gianni Barcaccia. 2026. "ddRADseq Applications for Petunia × hybrida Clonal Line Breeding: Genotyping and Variant Identification for Target-Specific Assays" Horticulturae 12, no. 2: 160. https://doi.org/10.3390/horticulturae12020160
APA StyleBetto, A., Scariolo, F., Gabelli, G., Riommi, D., Farinati, S., Vannozzi, A., Palumbo, F., & Barcaccia, G. (2026). ddRADseq Applications for Petunia × hybrida Clonal Line Breeding: Genotyping and Variant Identification for Target-Specific Assays. Horticulturae, 12(2), 160. https://doi.org/10.3390/horticulturae12020160








