Rotational Strip Bean and Celery Intercropping Alters the Microbial Community to Improve Crop Yield and Soil Nutrients
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Treatments
2.2. Bean and Celery Yields
2.3. Photosynthesis Property
2.4. Soil Sampling
2.5. Soil Nutrients
2.6. Soil DNA Extraction and Quantitative PCR
2.7. High-Throughput Amplicon Sequencing and Data Processing
2.8. Statistical Analysis
3. Results
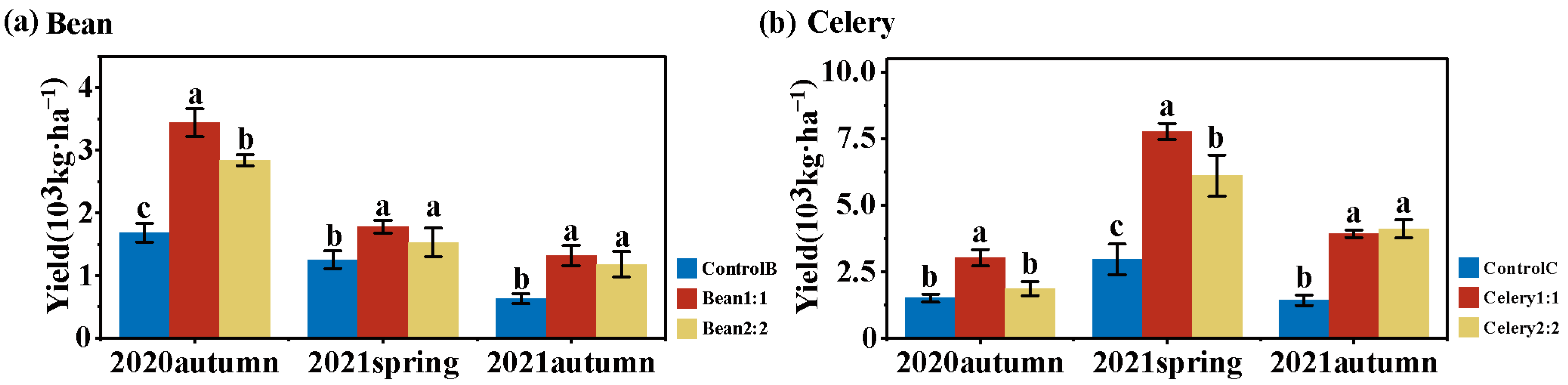
3.1. Yields of Bean and Celery under Rotational Strip Intercropping
3.2. Illuminance and Net Photosynthetic Rate of Rotational Strip Bean and Celery Intercropping
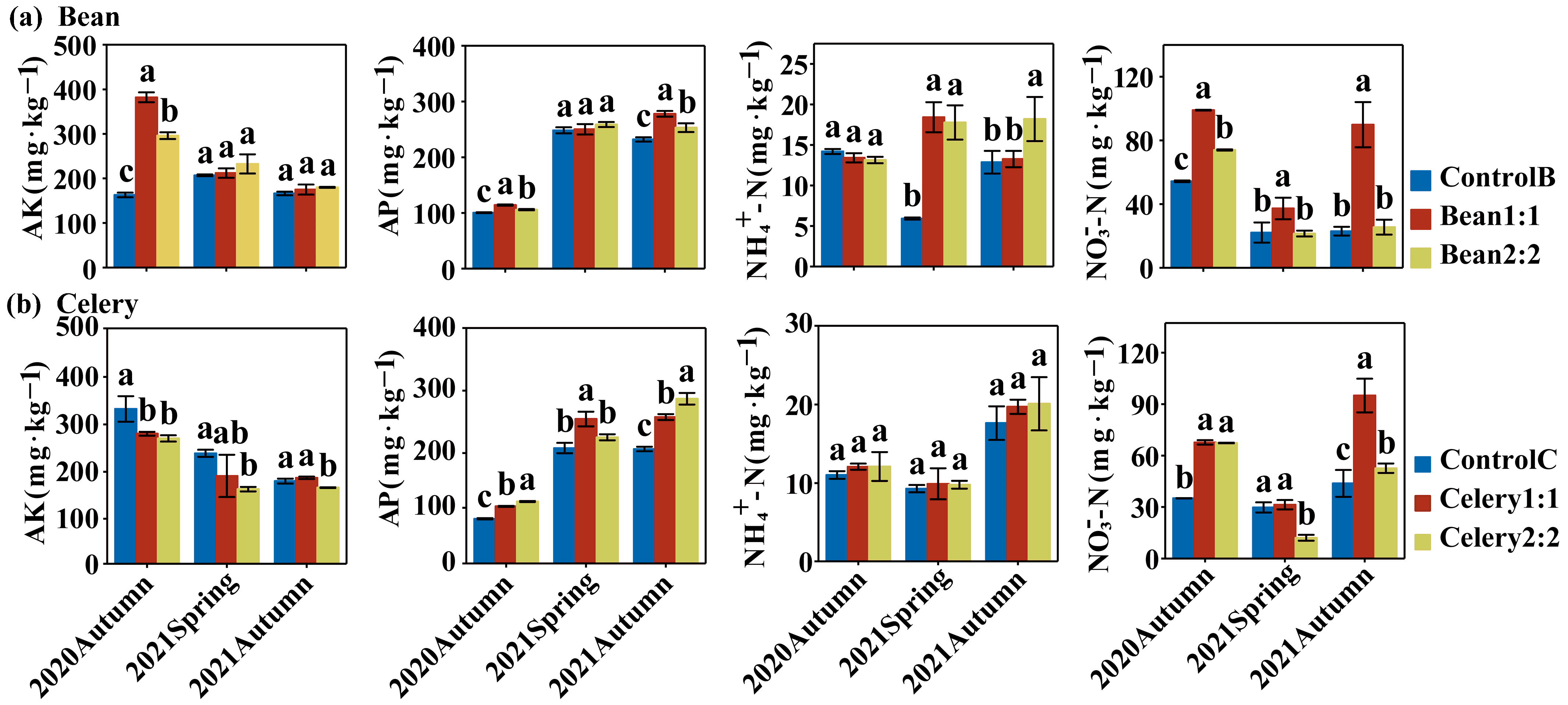
3.3. Soil Nutrients of Rotational Strip Bean and Celery Intercropping
3.4. Soil Microbial Community Abundances and Diversity
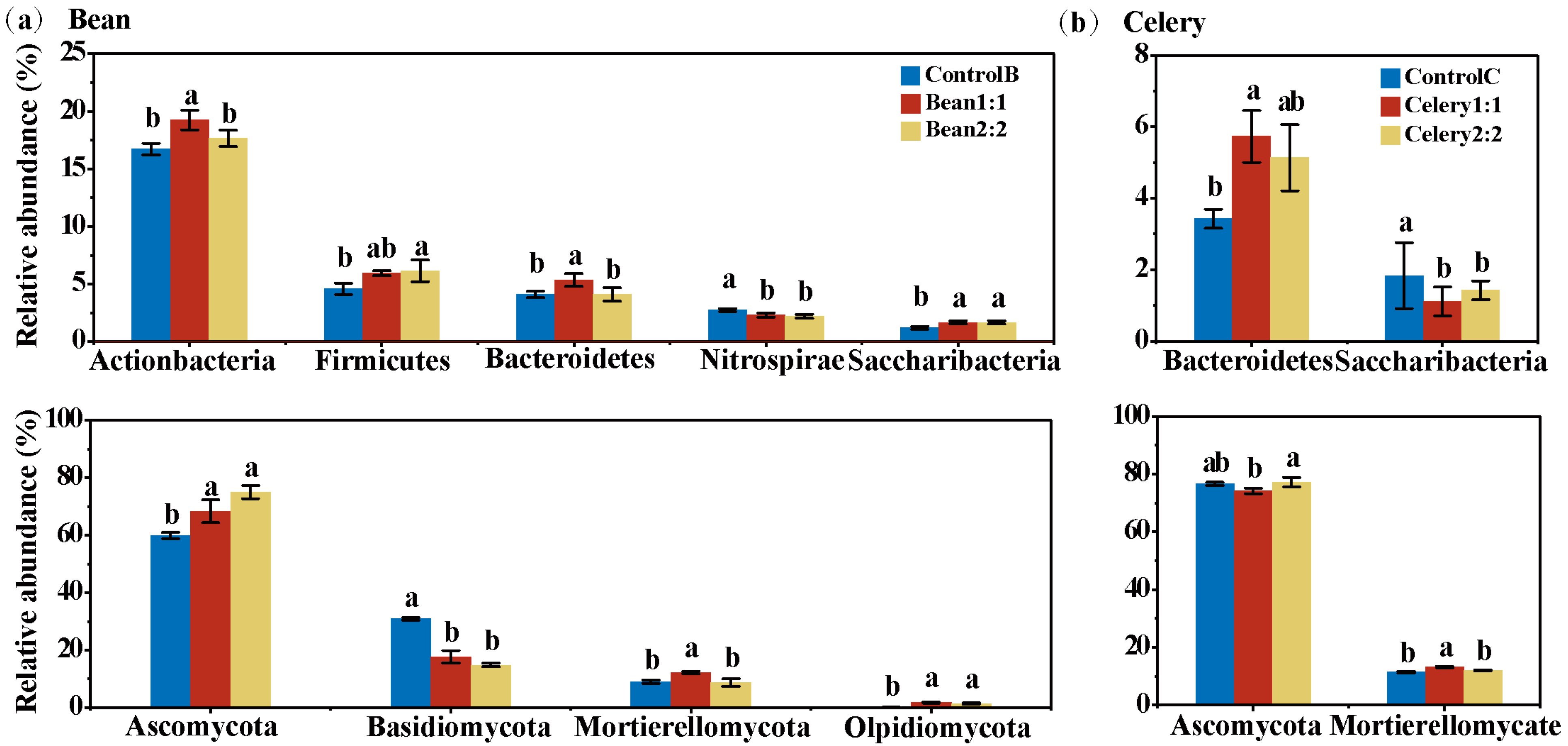
3.5. Bacterial Community Composition in Rotational Strip Intercropping
3.6. Fungi Community Composition of in Rotational Strip Intercropping
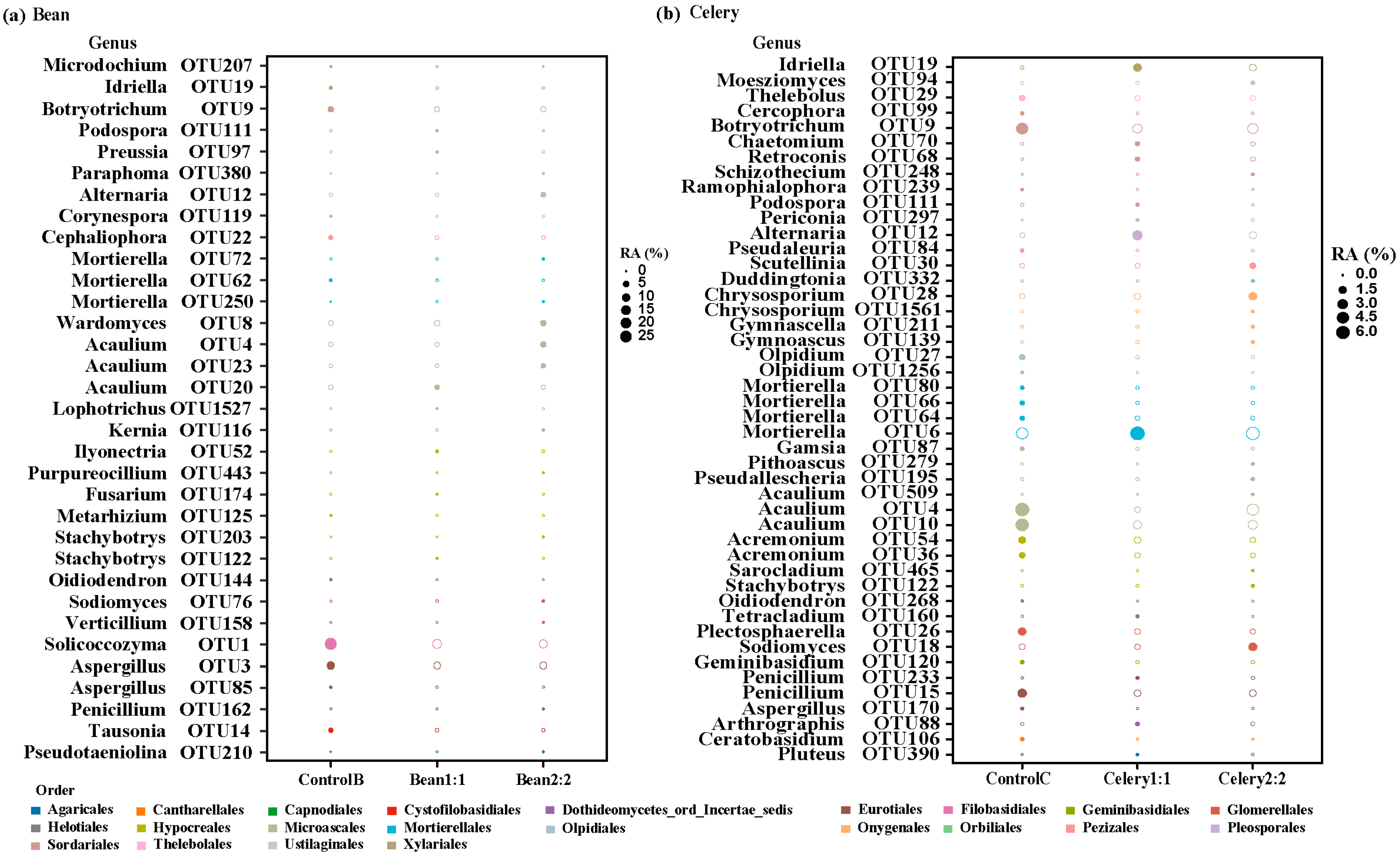
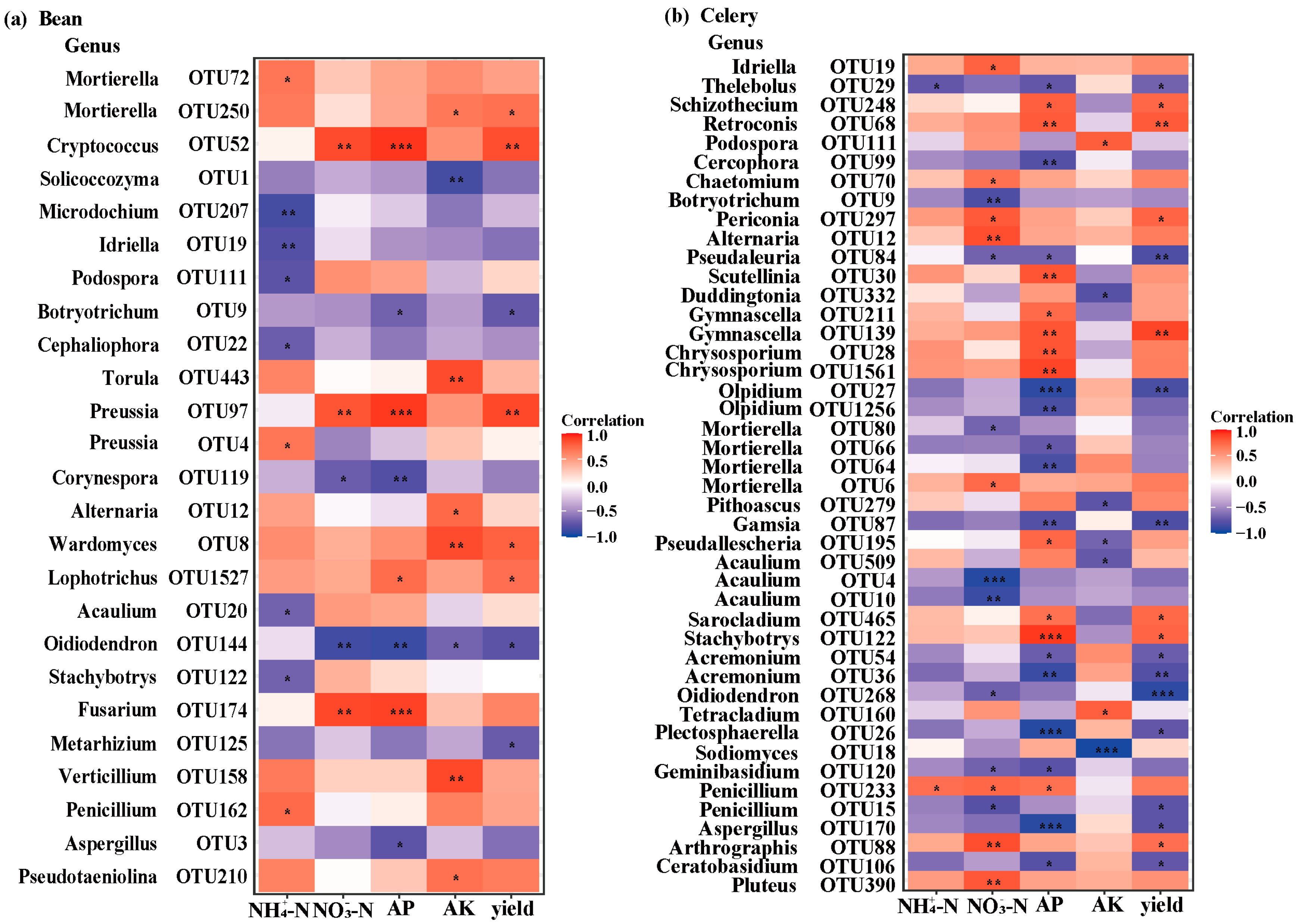
3.7. Correlation Analysis of Sensitive OTUs with Soil Nutrients and Yield
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tilman, D.; Reich, P.B.; Knops, J.M.H. Biodiversity and ecosystem stability in a decade-long grassland experiment. Nature 2006, 441, 629–632. [Google Scholar] [CrossRef] [PubMed]
- Loreau, M.; De Mazancourt, C.; Duffy, E. Biodiversity and ecosystem stability: A synthesis of underlying mechanisms. Ecol. Lett. 2013, 16, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Bowles, T.M.; Mooshammer, M.; Socolar, Y.; Calderón, F.; Cavigelli, M.A.; Culman, S.W.; Deen, W.; Drury, C.F.; Garcia, A.G.Y.; Gaudin, A.C.M.; et al. Long-Term Evidence Shows that Crop-Rotation Diversification Increases Agricultural Resilience to Adverse Growing Conditions in North America. One Earth 2020, 2, 284–293. [Google Scholar] [CrossRef]
- Garland, G.; Edlinger, A.; Banerjee, S.; Degrune, F.; Garcia-Palacios, P.; Pescador, D.S.; Herzog, C.; Romdhane, S.; Saghai, A.; Spor, A.; et al. Crop cover is more important than rotational diversity for soil multifunctionality and cereal yields in European cropping systems. Nat. Food 2021, 2, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Chadfield, V.G.A.; Hartley, S.E.; Redeker, K.R. Associational resistance through intercropping reduces yield losses to soil-borne pests and diseases. New Phytol. 2022, 235, 2393–2405. [Google Scholar] [CrossRef]
- Wahbi, S.; Prin, Y.; Thioulouse, J.; Sanguin, H.; Baudoin, E.; Maghraoui, T.; Oufdou, K.; Le Roux, C.; Galiana, A.; Hafidi, M.; et al. Impact of Wheat/Faba Bean Mixed Cropping or Rotation Systems on Soil Microbial Functionalities. Front. Plant Sci. 2016, 7, 1364. [Google Scholar] [CrossRef]
- Luo, C.; Lv, J.; Guo, Z.; Dong, Y. Intercropping of Faba Bean with Wheat Under Different Nitrogen Levels Reduces Faba Bean Rust and Consequent Yield Loss. Plant Dis. 2022, 106, 2370–2379. [Google Scholar] [CrossRef]
- Ma, H.; Yu, X.; Yu, Q.; Wu, H.; Zhang, H.; Pang, J.; Gao, Y. Maize/alfalfa intercropping enhances yield and phosphorus acquisition. Field Crops Res. 2023, 303, 109136. [Google Scholar] [CrossRef]
- Mesfin, S.; Gebresamuel, G.; Haile, M.; Zenebe, A. Potentials of legumes rotation on yield and nitrogen uptake of subsequent wheat crop in northern Ethiopia. Heliyon 2023, 9, e16126. [Google Scholar] [CrossRef]
- Yu, T.; Nie, J.; Zang, H.; Zeng, Z.; Yang, Y. Peanut-based Rotation Stabilized Diazotrophic Communities and Increased Subsequent Wheat Yield. Microb. Ecol. 2023, 86, 2447–2460. [Google Scholar] [CrossRef]
- Lu, J.; Liu, Y.; Zou, X.; Zhang, X.; Yu, X.; Wang, Y.; Si, T. Rotational strip peanut/cotton intercropping improves agricultural production through modulating plant growth, root exudates, and soil microbial communities. Agric. Ecosyst. Environ. 2024, 359, 108767. [Google Scholar] [CrossRef]
- Zou, X.X.; Shi, P.X.; Zhang, C.J.; Si, T.; Wang, Y.F.; Zhang, X.J.; Yu, X.N.; Wang, H.X.; Wang, M.L. Rotational strip intercropping of maize and peanuts has multiple benefits for agricultural production in the northern agropastoral ecotone region of China. Eur. J. Agron. 2021, 129, 126304. [Google Scholar] [CrossRef]
- Chi, B.; Zhang, Y.; Zhang, D.; Zhang, X.; Dai, J.; Dong, H. Wide-strip intercropping of cotton and peanut combined with strip rotation increases crop productivity and economic returns. Field Crops Res. 2019, 243, 107617. [Google Scholar] [CrossRef]
- Han, Y.; Dong, Q.; Zhang, K.; Sha, D.; Jiang, C.; Yang, X.; Liu, X.; Zhang, H.; Wang, X.; Guo, F.; et al. Maize-peanut rotational strip intercropping improves peanut growth and soil properties by optimizing microbial community diversity. Peerj 2022, 10, e13777. [Google Scholar] [CrossRef] [PubMed]
- Lv, Q.; Chi, B.; He, N.; Zhang, D.; Dai, J.; Zhang, Y.; Dong, H. Cotton-Based Rotation, Intercropping, and Alternate Intercropping Increase Yields by Improving Root-Shoot Relations. Agronomy 2023, 13, 413. [Google Scholar] [CrossRef]
- Zou, X.; Liu, Y.; Huang, M.; Li, F.; Si, T.; Wang, Y.; Yu, X.; Zhang, X.; Wang, H.; Shi, P. Rotational strip intercropping of maize and peanut enhances productivity by improving crop photosynthetic production and optimizing soil nutrients and bacterial communities. Field Crops Res. 2023, 291, 108770. [Google Scholar] [CrossRef]
- Sudini, H.; Liles, M.R.; Arias, C.R.; Bowen, K.L.; Huettel, R.N. Exploring Soil Bacterial Communities in Different Peanut-Cropping Sequences Using Multiple Molecular Approaches. Phytopathology 2011, 101, 819–827. [Google Scholar] [CrossRef]
- Guo, F.; Wang, M.; Si, T.; Wang, Y.; Zhao, H.; Zhang, X.; Yu, X.; Wan, S.; Zou, X. Maize-peanut intercropping led to an optimization of soil from the perspective of soil microorganism. Arch. Agron. Soil Sci. 2021, 67, 1986–1999. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, J.; Wu, F. Soil microbial communities in cucumber monoculture and rotation systems and their feedback effects on cucumber seedling growth. Plant Soil 2017, 415, 507–520. [Google Scholar] [CrossRef]
- Liu, X.; Ren, X.; Tang, S.; Zhang, Z.; Huang, Y.; Sun, Y.; Gao, Z.; Ma, Z. Effects of Broccoli Rotation on Soil Microbial Community Structure and Physicochemical Properties in Continuous Melon Cropping. Agronomy 2023, 13, 2066. [Google Scholar] [CrossRef]
- Li, N.; Gao, D.; Zhou, X.; Chen, S.; Li, C.; Wu, F. Intercropping with Potato-Onion Enhanced the Soil Microbial Diversity of Tomato. Microorganisms 2020, 8, 834. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Shi, Y.; Wu, F.; Pan, K.; Zhou, X. Intercropping of wheat changed cucumber rhizosphere bacterial community composition and inhibited cucumber Fusarium wilt disease. Sci. Agric. 2020, 77, e20190005. [Google Scholar] [CrossRef]
- Zak, D.R.; Holmes, W.E.; White, D.C.; Peacock, A.D.; Tilman, D. Plant diversity, soil microbial communities, and ecosystem function: Are there any links? Ecology 2003, 84, 2042–2050. [Google Scholar] [CrossRef]
- Eisenhauer, N.; Dobies, T.; Cesarz, S.; Hobbie, S.E.; Meyer, R.J.; Worm, K.; Reich, P.B. Plant diversity effects on soil food webs are stronger than those of elevated CO2 and N deposition in a long-term grassland experiment. Proc. Natl. Acad. Sci. USA 2013, 110, 6889–6894. [Google Scholar] [CrossRef] [PubMed]
- Steinauer, K.; Chatzinotas, A.; Eisenhauer, N. Root exudate cocktails: The link between plant diversity and soil microorganisms? Ecol. Evol. 2016, 6, 7387–7396. [Google Scholar] [CrossRef] [PubMed]
- Falkowski, P.G.; Fenchel, T.; Delong, E.F. The microbial engines that drive Earth’s biogeochemical cycles. Science 2008, 320, 1034–1039. [Google Scholar] [CrossRef] [PubMed]
- Martiny JB, H.; Eisen, J.A.; Penn, K.; Allison, S.D.; Horner-Devine, M.C. Drivers of bacterial β-diversity depend on spatial scale. Proc. Natl. Acad. Sci. USA 2011, 108, 7850–7854. [Google Scholar] [CrossRef] [PubMed]
- Wagg, C.; Bender, S.F.; Widmer, F.; van der Heijden, M.G.A. Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc. Natl. Acad. Sci. USA 2014, 111, 5266–5270. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Xu, Y.; Zhang, J.; Hao, X.; Lu, Y. Core Microbiota in Agricultural Soils and Their Potential Associations with Nutrient Cycling. Msystems 2019, 4, 10–1128. [Google Scholar] [CrossRef]
- Gao, H.; Tian, G.; Rahman, M.K.U.; Wu, F. Cover Crop Species Composition Alters the Soil Bacterial Community in a Continuous Pepper Cropping System. Front. Microbiol. 2022, 12, 789034. [Google Scholar] [CrossRef]
- Hu, H.Y.; Li, H.; Hao, M.M.; Ren, Y.N.; Zhang, M.K.; Liu, R.Y.; Zhang, Y.; Li, G.; Chen, J.S.; Ning, T.Y.; et al. Nitrogen fixation and crop productivity enhancements co-driven by intercrop root exudates and key rhizosphere bacteria. J. Appl. Ecol. 2021, 58, 2243–2255. [Google Scholar] [CrossRef]
- Wongkiew, S.; Polprasert, C.; Noophan, P.; Koottatep, T.; Kanokkantapong, V.; Surendra, K.C.; Khanal, S.K. Effects of vermicompost leachate on nitrogen, phosphorus, and microbiome in a food waste bioponic system. J. Environ. Manag. 2023, 339, 117860. [Google Scholar] [CrossRef] [PubMed]
- Muyzer, G.; Dewaak, E.C.; Uitterlinden, A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 2010, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.G.; Zhang, J.Y.; Rahman, M.K.U.; Gao, D.M.; Wei, Z.; Wu, F.Z. p-Coumaric can alter the composition of cucumber rhizosphere microbial communities and induce negative plant-microbial interactions. Biol. Fertil. Soils 2018, 54, 363–372. [Google Scholar] [CrossRef]
- Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Koljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Hartman, K.; van der Heijden, M.G.A.; Wittwer, R.A.; Banerjee, S.; Walser, J.C.; Schlaeppi, K. Cropping practices manipulate abundance patterns of root and soil microbiome members paving the way to smart farming. Microbiome 2018, 6, 1–14. [Google Scholar]
- Tilman, D. Benefits of intensive agricultural intercropping. Nat. Plants 2020, 6, 604–605. [Google Scholar] [CrossRef]
- Yang, X.; Xiong, J.; Du, T.; Ju, X.; Gan, Y.; Li, S.; Xia, L.; Shen, Y.; Pacenka, S.; Steenhuis, T.S.; et al. Diversifying crop rotation increases food production, reduces net greenhouse gas emissions and improves soil health. Nat. Commun. 2024, 15, 198. [Google Scholar] [CrossRef] [PubMed]
- Furey, G.N.; Tilman, D. Plant biodiversity and the regeneration of soil fertility. Proc. Natl. Acad. Sci. USA 2021, 118, e2111321118. [Google Scholar] [CrossRef]
- Abidemi, A.A.; Ewulo, B.S.; Aiyelari, O.P.; Hu, J. Effects of NPK Fertilizer and Vine Care on Soil Chemical Properties and Cucumber (Cucumis sativus L.) Growth and Yield Parameters. Int. J. Plant Soil Sci. 2021, 33, 136–151. [Google Scholar]
- Cappelli, S.L.; Domeignoz-Horta, L.A.; Loaiza, V.; Laine, A.L. Plant biodiversity promotes sustainable agriculture directly and via belowground effects. Trends Plant Sci. 2022, 27, 674–687. [Google Scholar] [CrossRef]
- Zhang, X.; Mauzerall, D.L.; Davidson, E.A.; Kanter, D.R.; Cai, R. The Economic and Environmental Consequences of Implementing Nitrogen-Efficient Technologies and Management Practices in Agriculture. J. Environ. Qual. 2015, 44, 312–324. [Google Scholar] [CrossRef] [PubMed]
- Kermah, M.; Franke, A.C.; Adjei-Nsiah, S.; Ahiabor, B.D.K.; Abaidoo, R.C.; Giller, K.E. Maize-grain legume intercropping for enhanced resource use efficiency and crop productivity in the Guinea savanna of northern Ghana. Field Crops Res. 2017, 213, 38–50. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Chen, S.; Huang, Z.; Huang, T.; Tang, X.; He, L.; Li, Z.; Xiong, J.; Zhong, R.; Jiang, J.; et al. Effects of Intercropping and Nitrogen Application on Soil Fertility and Microbial Communities in Peanut Rhizosphere Soil. Agronomy 2024, 14, 635. [Google Scholar] [CrossRef]
- Jiao, N.; Wang, J.; Ma, C.; Zhang, C.; Guo, D.; Zhang, F.; Jensen, E.S. The importance of aboveground and belowground interspecific interactions in determining crop growth and advantages of peanut/maize intercropping. Crop J. 2021, 9, 1460–1469. [Google Scholar] [CrossRef]
- van der Putten, W.H.; Bardgett, R.D.; Bever, J.D.; Bezemer, T.M.; Casper, B.B.; Fukami, T.; Kardol, P.; Klironomos, J.N.; Kulmatiski, A.; Schweitzer, J.A.; et al. Plant-soil feedbacks: The past, the present and future challenges. J. Ecol. 2013, 101, 265–276. [Google Scholar] [CrossRef]
- Nannipieri, P.; Hannula, S.E.; Pietramellara, G.; Schloter, M.; Sizmur, T.; Pathan, S.I. Legacy effects of rhizodeposits on soil microbiomes: A perspective. Soil Biol. Biochem. 2023, 184, 109107. [Google Scholar] [CrossRef]
- Yan, X.; Niu, J.; Li, Y.; Sun, S.; Li, X.; Jin, K. Species-specific soil legacy effects on ecosystem multifunctionality via regulating plant overyielding. Land Degrad. Dev. 2023, 34, 1235–1245. [Google Scholar] [CrossRef]
- Li, L.; Tilman, D.; Lambers, H.; Zhang, F.S. Plant diversity and overyielding: Insights from belowground facilitation of intercropping in agriculture. New Phytol. 2014, 203, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yang, Z.; Liu, J.; Li, X.; Wang, X.; Dai, C.; Zhang, T.; Carrion, V.J.; Wei, Z.; Cao, F.; et al. Crop rotation and native microbiome inoculation restore soil capacity to suppress a root disease. Nat. Commun. 2023, 14, 8126. [Google Scholar] [PubMed]
- Rice, A.V.; Currah, R.S. Two new species of Pseudogymnoascus with Geomyces anamorphs and their phylogenetic relationship with Gymnostellatospora. Mycologia 2006, 98, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Si, H.; He, J.; Fan, L.; Li, L. The shifts of maize soil microbial community and networks are related to soil properties under different organic fertilizers. Rhizosphere 2021, 19, 100388. [Google Scholar] [CrossRef]
- Qin, J.; Bian, C.; Duan, S.; Wang, W.; Li, G.; Jin, L. Effects of different rotation cropping systems on potato yield, rhizosphere microbial community and soil biochemical properties. Front. Plant Sci. 2022, 13, 999730. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, L. Effects of additives on the co-composting of forest residues with cattle manure. Bioresour. Technol. 2023, 368, 128384. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Shen, S.; Wan, C.; Wang, S.; Yang, F.; Zhang, K.; Gao, W. Organic fertilizer substitution over six years improves the productivity of garlic, bacterial diversity, and microbial communities network complexity. Appl. Soil Ecol. 2023, 182, 104718. [Google Scholar]
- Yang, Y.; Xu, N.; Zhang, Z.; Lei, C.; Chen, B.; Qin, G.; Qiu, D.; Lu, T.; Qian, H. Deciphering Microbial Community and Nitrogen Fixation in the Legume Rhizosphere. J. Agric. Food Chem. 2024, 72, 5659–5670. [Google Scholar] [CrossRef]
- Tang, C.; Sun, B.; Zeeshan, M.; Li, J.; Zhang, X. Funneliformis mosseae-induced changes of rhizosphere microbial community structure enhance Capsicum annuum L. plant growth and fruit yield. Soil Sci. Soc. Am. J. 2023, 87, 843–855. [Google Scholar] [CrossRef]
- Novello, G.; Gamalero, E.; Bona, E.; Boatti, L.; Mignone, F.; Massa, N.; Cesaro, P.; Lingua, G.; Berta, G. The Rhizosphere Bacterial Microbiota of Vitis vinifera cv. Pinot Noir in an Integrated Pest Management Vineyard. Front. Microbiol. 2017, 8, 1528. [Google Scholar] [CrossRef] [PubMed]
- Masuda, Y.; Yamanaka, H.; Xu, Z.X.; Shiratori, Y.; Aono, T.; Amachi, S.; Senoo, K.; Itoh, H. Diazotrophic Anaeromyxobacter Isolates from Soils. Appl. Environ. Microbiol. 2020, 86, e00956-20. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Liu, Y.; Yao, D.; Wang, N.; Ye, X.; Cui, Z.; Wang, H. Phylogenetic diversity of stochasticity-dominated predatory myxobacterial community drives multi-nutrient cycling in typical farmland soils. Sci. Total Environ. 2023, 871, 161680. [Google Scholar] [CrossRef] [PubMed]
- Samac, D.A.; Kinkel, L.L. Suppression of the root-lesion nematode (Pratylenchus penetrans) in alfalfa (Medicago sativa) by Streptomyces spp. Plant Soil 2001, 235, 35–44. [Google Scholar] [CrossRef]
- Law, J.W.F.; Ser, H.L.; Khan, T.M.; Chuah, L.H.; Pusparajah, P.; Chan, K.G.; Goh, B.H.; Lee, L.H. The Potential of Streptomyces as Biocontrol Agents against the Rice Blast Fungus, Magnaporthe oryzae (Pyricularia oryzae). Front. Microbiol. 2017, 8, 215552. [Google Scholar] [CrossRef]
- Van Der Heijden, M.G.A.; Bardgett, R.D.; Van Straalen, N.M. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef]
- Bender, S.F.; Van Der Heijden, M.G.A. Soil biota enhance agricultural sustainability by improving crop yield, nutrient uptake and reducing nitrogen leaching losses. J. Appl. Ecol. 2015, 52, 228–239. [Google Scholar] [CrossRef]
- Chen, J.; Qiao, M.; Yang, Y.; Gao, Z.; Yang, Z.; Lin, W. Exogenous Streptomyces spp. benefit naked oat growth under dry farming conditions by modifying rhizosphere bacterial communities. Appl. Soil Ecol. 2023, 189, 104946. [Google Scholar] [CrossRef]
- Rathi, M.; Nandabalan, Y.K. Copper-tolerant rhizosphere bacteria-characterization and assessment of plant growth promoting factors. Environ. Sci. Pollut. Res. 2017, 24, 9723–9733. [Google Scholar] [CrossRef]
- Xiao, C.Q.; Chi, R.A.; He, H.; Zhang, W.X. Characterization of tricalcium phosphate solubilization by Stenotrophomonas maltophilia YC isolated from phosphate mines. J. Cent. South Univ. Technol. 2009, 16, 581–587. [Google Scholar] [CrossRef]
- Oteino, N.; Lally, R.D.; Kiwanuka, S.; Lloyd, A.; Ryan, D.; Germaine, K.J.; Dowling, D.N. Plant growth promotion induced by phosphate solubilizing endophytic Pseudomonas isolates. Front. Microbiol. 2015, 6, 745. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Wang, Z.; Wu, F.; Li, X.; Zhou, X. Litter Mixing Alters Microbial Decomposer Community to Accelerate Tomato Root Litter Decomposition. Microbiol. Spectr. 2022, 10, e00186-22. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Meng, S.; Wang, L.; Zhang, Z. Optimum Fermentation Condition of Soybean Curd Residue and Rice Bran by Preussia aemulans using Solid-State Fermentation Method. Int. J. Biol. 2015, 7, 66–74. [Google Scholar] [CrossRef][Green Version]
- Ma, A.; Zhuang, X.; Wu, J.; Cui, M.; Lv, D.; Liu, C.; Zhuang, G. Ascomycota Members Dominate Fungal Communities during Straw Residue Decomposition in Arable Soil. PLoS ONE 2013, 8, e66146. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Yuan, T.; Ibrahim, M.; Wu, F. Rotational Strip Bean and Celery Intercropping Alters the Microbial Community to Improve Crop Yield and Soil Nutrients. Horticulturae 2024, 10, 432. https://doi.org/10.3390/horticulturae10050432
Li S, Yuan T, Ibrahim M, Wu F. Rotational Strip Bean and Celery Intercropping Alters the Microbial Community to Improve Crop Yield and Soil Nutrients. Horticulturae. 2024; 10(5):432. https://doi.org/10.3390/horticulturae10050432
Chicago/Turabian StyleLi, Shuang, Tao Yuan, Musawar Ibrahim, and Fengzhi Wu. 2024. "Rotational Strip Bean and Celery Intercropping Alters the Microbial Community to Improve Crop Yield and Soil Nutrients" Horticulturae 10, no. 5: 432. https://doi.org/10.3390/horticulturae10050432
APA StyleLi, S., Yuan, T., Ibrahim, M., & Wu, F. (2024). Rotational Strip Bean and Celery Intercropping Alters the Microbial Community to Improve Crop Yield and Soil Nutrients. Horticulturae, 10(5), 432. https://doi.org/10.3390/horticulturae10050432





