Genome-Wide Analysis of the NBS-LRR Gene Family and SSR Molecular Markers Development in Solanaceae
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome and Transcriptome Data Acquisition of Solanaceae Species
2.2. Construction of Solanaceae Species Phylogenetic Trees and Identification of Gene Duplication Types
2.3. Identification of NBS-LRR Family Genes and Distribution on Chromosomes
2.4. Developmental Tree Construction and Domain Analysis of the NBS-LRR Genes
2.5. Prediction of NBS-LRR Family Protein Interactions
2.6. Analysis of the Expression Patterns
2.7. Development of SSR Molecular Markers
3. Results
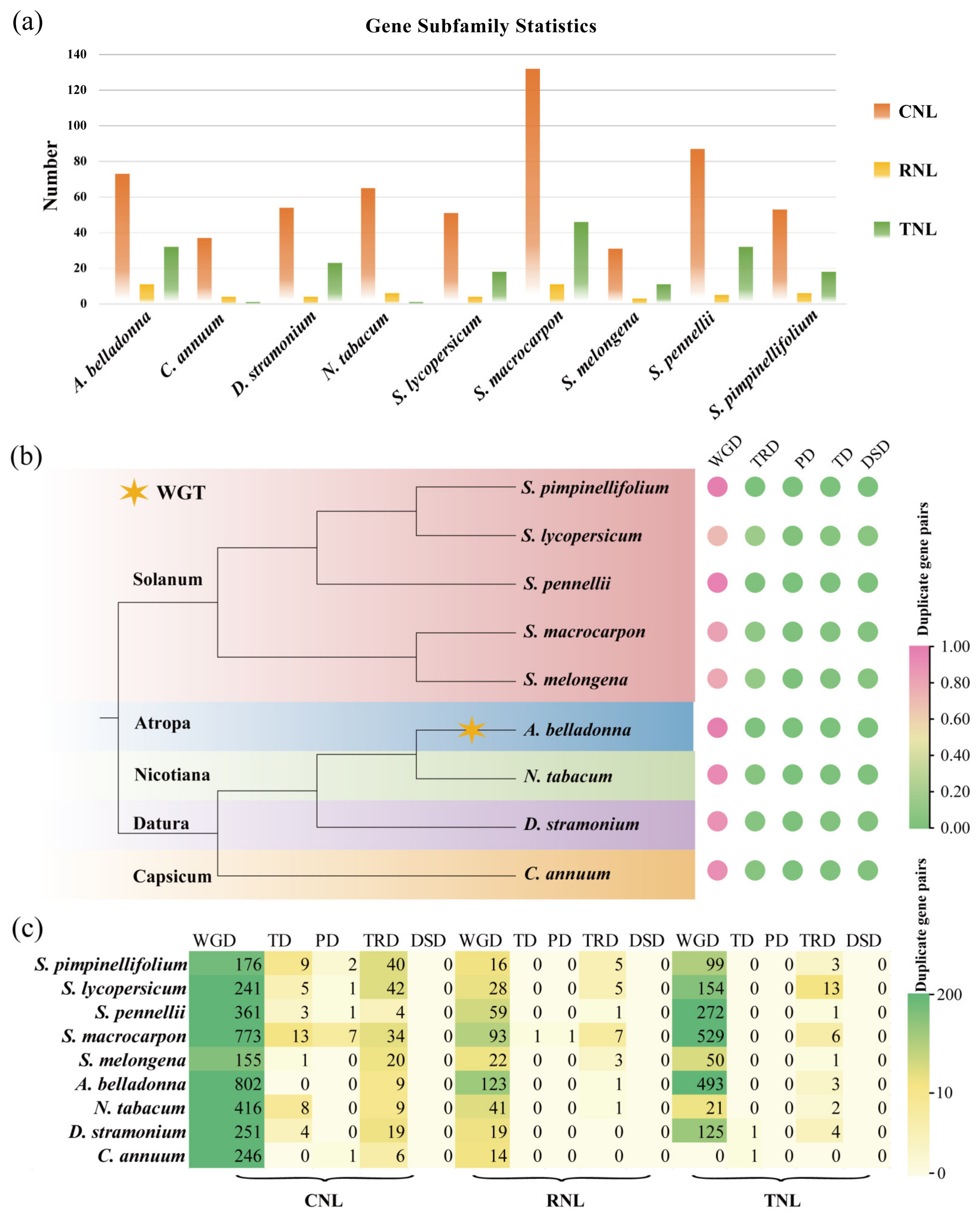
3.1. Identification, Classification, Gene Duplication Analysis of NBS-LRR Family Genes in Solanaceae
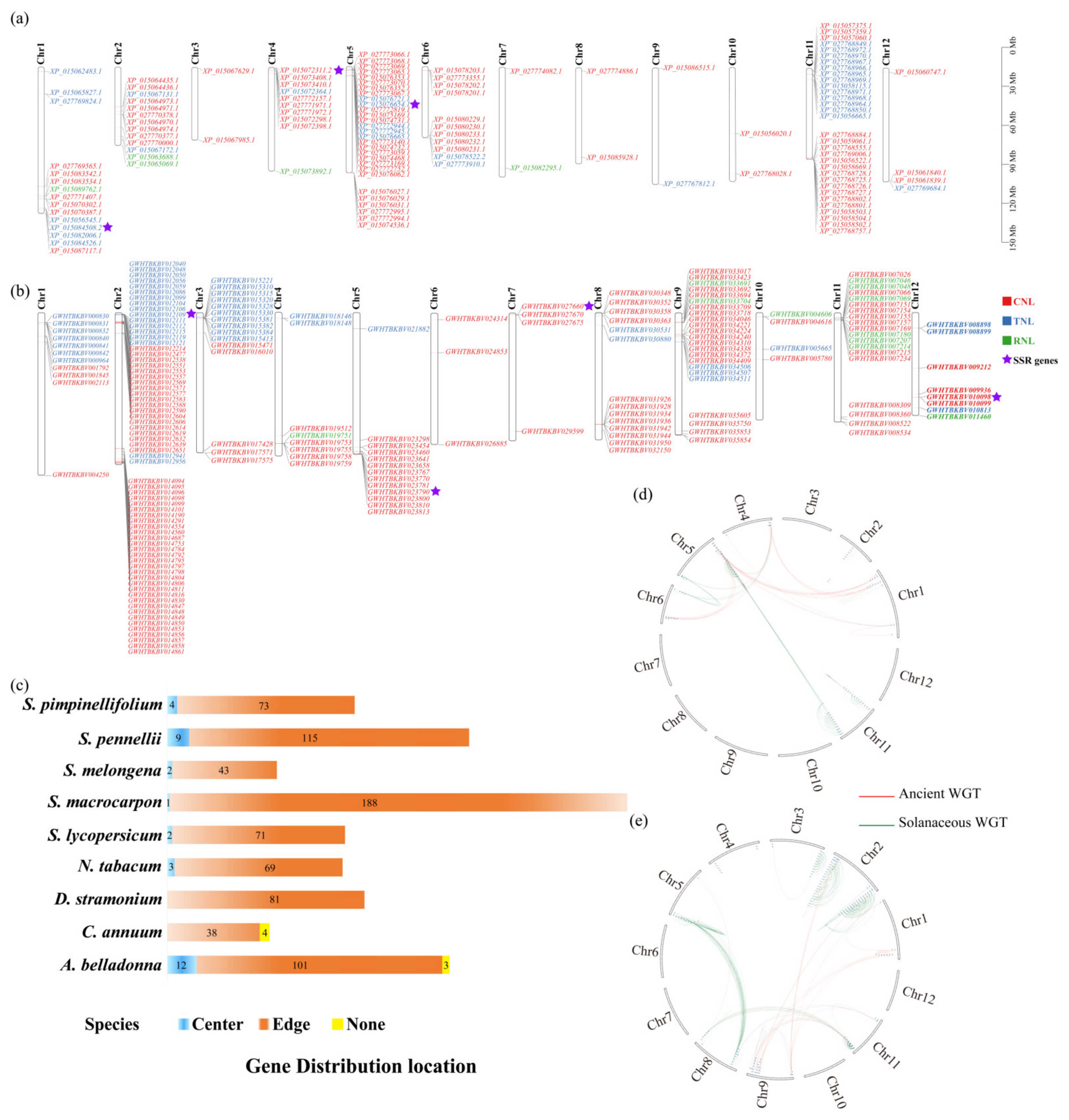
3.2. Chromosomal Distribution and Expansion Events of NBS-LRR Family Genes
3.3. Phylogenetic Analysis and Identification of Conserved Motifs Within the NBS-LRR Genes
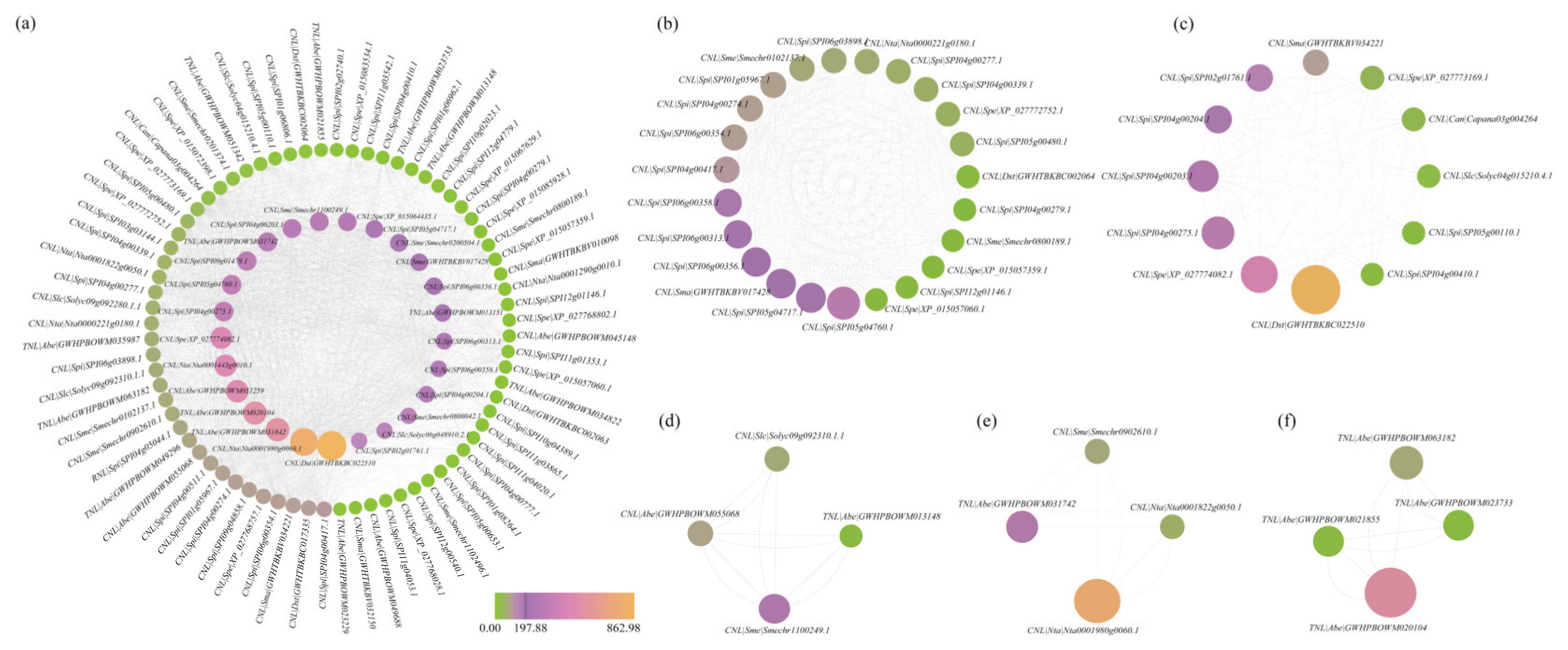
3.4. Comprehensive Analysis of PPI Networks Within the NBS-LRR Family Genes
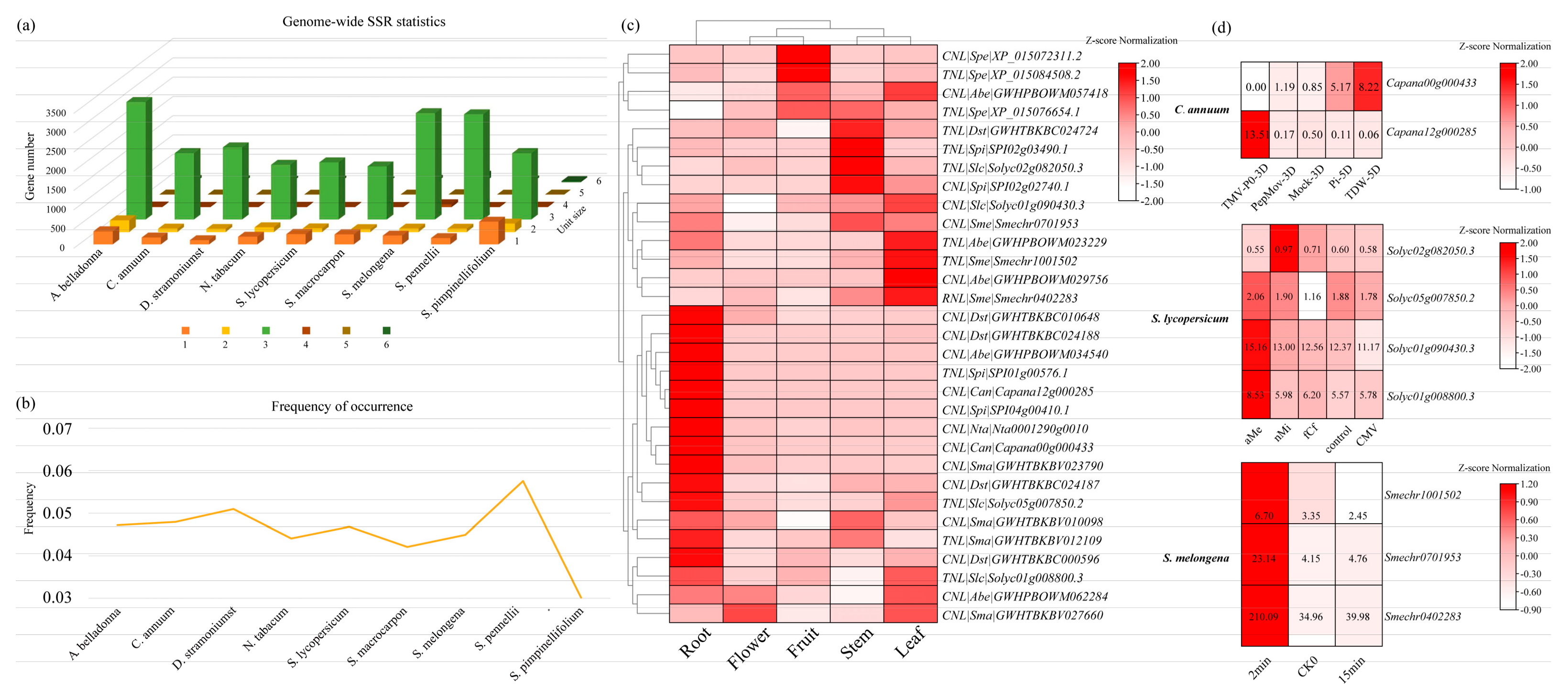
3.5. Development and Characterization of SSR Markers in All Genes from Solanaceae Species
3.6. Identification and Expression Analysis of NBS-LRR Family Genes Containing SSR Markers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gebhardt, C. The historical role of species from the Solanaceae plant family in genetic research. Theor. Appl. Genet. 2016, 129, 2281–2294. [Google Scholar] [CrossRef] [PubMed]
- Barchi, L.; Pietrella, M.; Venturini, L.; Minio, A.; Toppino, L.; Acquadro, A.; Andolfo, G.; Aprea, G.; Avanzato, C.; Bassolino, L.; et al. A chromosome-anchored eggplant genome sequence reveals key events in Solanaceae evolution. Sci. Rep. 2019, 9, 11769. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Niu, Y.; Li, Q.; Xia, L.; Wang, C.; Luo, S.; Li, N.; Xuan, S.; Wang, Y.; Shen, S.; et al. Mapping and identification of genes responsible for less-photosensitive fruit coloration in eggplant. Veg. Res. 2023, 3, 32. [Google Scholar] [CrossRef]
- Kumar, M.; Chandran, D.; Tomar, M.; Bhuyan, D.J.; Grasso, S.; Sá, A.G.; Carciofi, B.A.; Radha; Dhumal, S.; Singh, S.; et al. Valorization Potential of Tomato (Solanum lycopersicum L.) Seed: Nutraceutical Quality, Food Properties, Safety Aspects, and Application as a Health-Promoting Ingredient in Foods. Horticulturae 2022, 8, 265. [Google Scholar] [CrossRef]
- Akbar, S. Atropa belladonna L. (Solanaceae). In Handbook of 200 Medicinal Plants: A Comprehensive Review of Their Traditional Medical Uses and Scientific Justifications; Akbar, S., Ed.; Springer International Publishing: Cham, Switzerland, 2020; pp. 373–379. [Google Scholar]
- Sharma, M.; Dhaliwal, I.; Rana, K.; Delta, A.K.; Kaushik, P. Phytochemistry, Pharmacology, and Toxicology of Datura Species-A Review. Antioxidants 2021, 10, 1291. [Google Scholar] [CrossRef]
- Edwards, K.D.; Fernandez-Pozo, N.; Drake-Stowe, K.; Humphry, M.; Evans, A.D.; Bombarely, A.; Allen, F.; Hurst, R.; White, B.; Kernodle, S.P.; et al. A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genom. 2017, 18, 448. [Google Scholar] [CrossRef]
- Polashock, J.; Zelzion, E.; Fajardo, D.; Zalapa, J.; Georgi, L.; Bhattacharya, D.; Vorsa, N. The American cranberry: First insights into the whole genome of a species adapted to bog habitat. BMC Plant Biol. 2014, 14, 165. [Google Scholar] [CrossRef]
- Bai, C.; Wu, C.; Ma, L.; Fu, A.; Zheng, Y.; Han, J.; Li, C.; Yuan, S.; Zheng, S.; Gao, L.; et al. Transcriptomics and metabolomics analyses provide insights into postharvest ripening and senescence of tomato fruit under low temperature. Hortic. Plant J. 2023, 9, 109–121. [Google Scholar] [CrossRef]
- Li, N.; Wang, Y.; Zheng, R.; Song, X. Research progress on biological functions of lncRNAs in major vegetable crops. Veg. Res. 2022, 2, 14. [Google Scholar] [CrossRef]
- Kim, S.; Park, M.; Yeom, S.-I.; Kim, Y.-M.; Lee, J.M.; Lee, H.-A.; Seo, E.; Choi, J.; Cheong, K.; Kim, K.-T.; et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 2014, 46, 270–278. [Google Scholar] [CrossRef]
- Liu, F.; Yu, H.; Deng, Y.; Zheng, J.; Liu, M.; Ou, L.; Yang, B.; Dai, X.; Ma, Y.; Feng, S.; et al. PepperHub, an Informatics Hub for the Chili Pepper Research Community. Mol. Plant 2017, 10, 1129–1132. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, D.; Hu, Y.; Li, H.; Ramstein, G.; Zhou, S.; Zhang, X.; Bao, Z.; Zhang, Y.; Song, B.; et al. Phylogenomic discovery of deleterious mutations facilitates hybrid potato breeding. Cell 2023, 186, 2313–2328.e15. [Google Scholar] [CrossRef] [PubMed]
- Gürbüz, N.; Uluişik, S.; Frary, A.; Frary, A.; Doğanlar, S. Health benefits and bioactive compounds of eggplant. Food Chem. 2018, 268, 602–610. [Google Scholar] [CrossRef] [PubMed]
- Ezechukwu, C.S.; Mbegbu, E.C.; Nwani, C.D.; Onoja, S.O.; Orji, E.A.; Ugwu, G.C.; Nnamonu, E.I.; Ugwu, G.N. Spermicidal and antioxidant potency of Solanum macrocarpon L. (African eggplant) leaf ethanol extract in albino rats. Comp. Clin. Pathol. 2024, 33, 367–377. [Google Scholar] [CrossRef]
- Bolger, A.; Scossa, F.; Bolger, M.E.; Lanz, C.; Maumus, F.; Tohge, T.; Quesneville, H.; Alseekh, S.; Sørensen, I.; Lichtenstein, G.; et al. The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat. Genet. 2014, 46, 1034–1038. [Google Scholar] [CrossRef]
- Han, H.; Li, X.; Li, T.; Chen, Q.; Zhao, J.; Zhai, H.; Deng, L.; Meng, X.; Li, C. Chromosome-level genome assembly of Solanum pimpinellifolium. Sci. Data 2024, 11, 577. [Google Scholar] [CrossRef]
- Doganlar, S.; Frary, A.; Daunay, M.C.; Lester, R.N.; Tanksley, S.D. A comparative genetic linkage map of eggplant (Solanum melongena) and its implications for genome evolution in the solanaceae. Genetics 2002, 161, 1697–1711. [Google Scholar] [CrossRef]
- Ellis, J.; Dodds, P.; Pryor, T. Structure, function and evolution of plant disease resistance genes. Curr. Opin. Plant Biol. 2000, 3, 278–284. [Google Scholar] [CrossRef]
- Du, J.; Wang, B.; Huang, M.; Chen, X.; Nie, L.; Wang, T.; Chen, H.; Song, B. Advancements in unraveling and enhancing bacterial wilt resistance in Solanaceous crops. Veg. Res. 2023, 3, 29. [Google Scholar] [CrossRef]
- Gassmann, W.; Hinsch, M.E.; Staskawicz, B.J. The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 1999, 20, 265–277. [Google Scholar] [CrossRef]
- Kourelis, J.; van der Hoorn, R.A.L. Defended to the Nines: 25 Years of Resistance Gene Cloning Identifies Nine Mechanisms for R Protein Function. Plant Cell 2018, 30, 285–299. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.X.; Yano, M.; Yamanouchi, U.; Iwamoto, M.; Monna, L.; Hayasaka, H.; Katayose, Y.; Sasaki, T. The Pib gene for rice blast resistance belongs to the nucleotide binding and leucine-rich repeat class of plant disease resistance genes. Plant J. 1999, 19, 55–64. [Google Scholar] [CrossRef]
- Yu, T.; Ma, X.; Liu, Z.; Feng, X.; Wang, Z.; Ren, J.; Cao, R.; Zhang, Y.; Nie, F.; Song, X. TVIR: A comprehensive vegetable information resource database for comparative and functional genomic studies. Hortic. Res. 2022, 9, uhac213. [Google Scholar] [CrossRef] [PubMed]
- Ng, A.; Xavier, R.J. Leucine-rich repeat (LRR) proteins: Integrators of pattern recognition and signaling in immunity. Autophagy 2011, 7, 1082–1084. [Google Scholar] [CrossRef] [PubMed]
- Maruta, N.; Sorbello, M.; Lim, B.Y.J.; McGuinness, H.Y.; Shi, Y.; Ve, T.; Kobe, B. TIR domain-associated nucleotides with functions in plant immunity and beyond. Curr. Opin. Plant Biol. 2023, 73, 102364. [Google Scholar] [CrossRef]
- Dangl, J.L.; Jones, J.D.G. Plant pathogens and integrated defence responses to infection. Nature 2001, 411, 826–833. [Google Scholar] [CrossRef]
- Rairdan, G.J.; Collier, S.M.; Sacco, M.A.; Baldwin, T.T.; Boettrich, T.; Moffett, P. The coiled-coil and nucleotide binding domains of the Potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell 2008, 20, 739–751. [Google Scholar] [CrossRef]
- Li, Q.; Li, J.; Sun, J.-L.; Ma, X.-F.; Wang, T.-T.; Berkey, R.; Yang, H.; Niu, Y.-Z.; Fan, J.; Li, Y.; et al. Multiple Evolutionary Events Involved in Maintaining Homologs of Resistance to Powdery Mildew 8 in Brassica napus. Front. Plant Sci. 2016, 7, 1065. [Google Scholar] [CrossRef]
- Zhou, H.; Yan, F.; Hao, F.; Ye, H.; Yue, M.; Woeste, K.; Zhao, P.; Zhang, S. Pan-genome and transcriptome analyses provide insights into genomic variation and differential gene expression profiles related to disease resistance and fatty acid biosynthesis in eastern black walnut (Juglans nigra). Hortic. Res. 2023, 10, uhad015. [Google Scholar] [CrossRef]
- Collier, S.M.; Hamel, L.P.; Moffett, P. Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol. Plant Microbe Interact. 2011, 24, 918–931. [Google Scholar] [CrossRef]
- Qi, D.; Innes, R.W. Recent Advances in Plant NLR Structure, Function, Localization, and Signaling. Front. Immunol. 2013, 4, 348. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.C.; Grover, A.; Kahl, G. Mining microsatellites in eukaryotic genomes. Trends Biotechnol. 2007, 25, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Li, N.; Li, X.; Lu, Y.; Feng, D.; Zhang, X.; Gu, A.; Ge, Y.; Tabusam, J.; Liu, M.; et al. Genome-wide development and utilization of Simple Sequence Repeats in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Veg. Res. 2022, 2, 9. [Google Scholar] [CrossRef]
- Sharma, J.; Manjunatha, N.; Pokhare, S.S.; Patil, P.G.; Agarrwal, R.; Chakranarayan, M.G.; Aralimar, A.; Devagire, P.; Marathe, R.A. Genetic Diversity and Streptomycin Sensitivity in Xanthomonas axonopodis pv. punicae Causing Oily Spot Disease in Pomegranates. Horticulturae 2022, 8, 441. [Google Scholar] [CrossRef]
- Zhu, Y.; Ma, T.; Lin, Y.; Peng, Y.; Huang, Y.; Jiang, J. SSR molecular marker developments and genetic diversity analysis of Zanthoxylum nitidum (Roxb.) DC. Sci. Rep. 2023, 13, 20767. [Google Scholar] [CrossRef]
- Ahmad, A.; Wang, J.-D.; Pan, Y.-B.; Sharif, R.; Gao, S.-J. Development and Use of Simple Sequence Repeats (SSRs) Markers for Sugarcane Breeding and Genetic Studies. Agronomy 2018, 8, 260. [Google Scholar] [CrossRef]
- Bhattarai, G.; Shi, A.; Kandel, D.R.; Solís-Gracia, N.; da Silva, J.A.; Avila, C.A. Genome-wide simple sequence repeats (SSR) markers discovered from whole-genome sequence comparisons of multiple spinach accessions. Sci. Rep. 2021, 11, 9999. [Google Scholar] [CrossRef]
- Lin, X.; Jia, Y.; Heal, R.; Prokchorchik, M.; Sindalovskaya, M.; Olave-Achury, A.; Makechemu, M.; Fairhead, S.; Noureen, A.; Heo, J.; et al. Solanum americanum genome-assisted discovery of immune receptors that detect potato late blight pathogen effectors. Nat. Genet. 2023, 55, 1579–1588. [Google Scholar] [CrossRef]
- Bradeen, J.M. On the Value of Wild Solanum Species for Improved Crop Disease Resistance: Resistances to Nematodes and Viruses. In The Wild Solanums Genomes; Carputo, D., Aversano, R., Ercolano, M.R., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 95–118. [Google Scholar]
- Zhang, F.; Qiu, F.; Zeng, J.; Xu, Z.; Tang, Y.; Zhao, T.; Gou, Y.; Su, F.; Wang, S.; Sun, X.; et al. Revealing evolution of tropane alkaloid biosynthesis by analyzing two genomes in the Solanaceae family. Nat. Commun. 2023, 14, 1446. [Google Scholar] [CrossRef]
- Ichihara, H.; Yamada, M.; Kohara, M.; Hirakawa, H.; Ghelfi, A.; Tamura, T.; Nakaya, A.; Nakamura, Y.; Shirasawa, S.; Yamashita, S.; et al. Plant GARDEN: A portal website for cross-searching between different types of genomic and genetic resources in a wide variety of plant species. BMC Plant Biol. 2023, 23, 391. [Google Scholar] [CrossRef]
- Wang, X.; Gao, L.; Jiao, C.; Stravoravdis, S.; Hosmani, P.S.; Saha, S.; Zhang, J.; Mainiero, S.; Strickler, S.R.; Catala, C.; et al. Genome of Solanum pimpinellifolium provides insights into structural variants during tomato breeding. Nat. Commun. 2020, 11, 5817. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Shen, S.; Li, C.; Zhang, C.; Chen, X.; Fu, Y.; Yu, T.; Zhou, R.; Liu, D.; Yang, Q.Y.; et al. SoIR: A comprehensive Solanaceae information resource for comparative and functional genomic study. Nucleic Acids Res. 2024, gkae1040. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Wang, J.; Wang, W.; Hu, T.; Hu, H.; Bao, C. A high-quality chromosome-level genome assembly reveals genetics for important traits in eggplant. Hortic. Res. 2020, 7, 153. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.H.; Vogel, A.; Denton, A.K.; Istace, B.; Wormit, A.; van de Geest, H.; Bolger, M.E.; Alseekh, S.; Maß, J.; Pfaff, C.; et al. De Novo Assembly of a New Solanum pennellii Accession Using Nanopore Sequencing. Plant Cell 2017, 29, 2336–2348. [Google Scholar] [CrossRef]
- Feng, S.; Liu, Z.; Chen, H.; Li, N.; Yu, T.; Zhou, R.; Nie, F.; Guo, D.; Ma, X.; Song, X. PHGD: An integrative and user-friendly database for plant hormone-related genes. Imeta 2024, 3, e164. [Google Scholar] [CrossRef]
- Yu, T.; Bai, Y.; Liu, Z.; Wang, Z.; Yang, Q.; Wu, T.; Feng, S.; Zhang, Y.; Shen, S.; Li, Q.; et al. Large-scale analyses of heat shock transcription factors and database construction based on whole-genome genes in horticultural and representative plants. Hortic. Res. 2022, 9, uhac035. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene duplication and evolution in recurring polyploidization–diploidization cycles in plants. Genome Biol. 2019, 20, 38. [Google Scholar] [CrossRef]
- Song, X.; Zhang, Y.; Li, C.; Li, N.; Shen, S.; Yu, T.; Liu, Z.; Zhou, R.; Cao, R.; Ma, X.; et al. Two major duplication events shaped the transcription factor repertoires in Solanaceae species. Sci. Hortic. 2024, 323, 112484. [Google Scholar] [CrossRef]
- Li, P.; Quan, X.; Jia, G.; Xiao, J.; Cloutier, S.; You, F.M. RGAugury: A pipeline for genome-wide prediction of resistance gene analogs (RGAs) in plants. BMC Genom. 2016, 17, 852. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Jiao, B.; Yang, Y.; Shan, L.; Li, T.; Li, X.; Xi, Z.; Wang, X.; Liu, J. WGDI: A user-friendly toolkit for evolutionary analyses of whole-genome duplications and ancestral karyotypes. Mol. Plant 2022, 15, 1841–1851. [Google Scholar] [CrossRef] [PubMed]
- Rozewicki, J.; Li, S.; Amada, K.M.; Standley, D.M.; Katoh, K. MAFFT-DASH: Integrated protein sequence and structural alignment. Nucleic Acids Res. 2019, 47, W5–W10. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef] [PubMed]
- Jaillon, O.; Aury, J.-M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar] [PubMed]
- Song, X.; Liu, H.; Shen, S.; Huang, Z.; Yu, T.; Liu, Z.; Yang, Q.; Wu, T.; Feng, S.; Zhang, Y.; et al. Chromosome-level pepino genome provides insights into genome evolution and anthocyanin biosynthesis in Solanaceae. Plant J. 2022, 110, 1128–1143. [Google Scholar] [CrossRef] [PubMed]
- Sumathi, M.; Yasodha, R. Microsatellite resources of Eucalyptus: Current status and future perspectives. Bot. Stud. 2014, 55, 73. [Google Scholar] [CrossRef]
- Victoria, F.C.; da Maia, L.C.; de Oliveira, A.C. In silico comparative analysis of SSR markers in plants. BMC Plant Biol. 2011, 11, 15. [Google Scholar] [CrossRef]
- Rengel, D.; Clemente, H.S.; Servant, F.; Ladouce, N.; Paux, E.; Wincker, P.; Couloux, A.; Sivadon, P.; Grima-Pettenati, J. A new genomic resource dedicated to wood formation in Eucalyptus. BMC Plant Biol. 2009, 9, 36. [Google Scholar] [CrossRef]
- Zhuang, Y.; Zhou, X.; Wang, S. Genetic diversity of NBS–LRR class disease-resistance gene analogs in cultivated and wild eggplants. Plant Syst. Evol. 2012, 298, 1399–1406. [Google Scholar] [CrossRef]
- Belkhadir, Y.; Subramaniam, R.; Dangl, J.L. Plant disease resistance protein signaling: NBS–LRR proteins and their partners. Curr. Opin. Plant Biol. 2004, 7, 391–399. [Google Scholar] [CrossRef]
- Qian, L.-H.; Zhou, G.-C.; Sun, X.-Q.; Lei, Z.; Zhang, Y.-M.; Xue, J.-Y.; Hang, Y.-Y. Distinct Patterns of Gene Gain and Loss: Diverse Evolutionary Modes of NBS-Encoding Genes in Three Solanaceae Crop Species. G3 Genes|Genomes|Genet. 2017, 7, 1577–1585. [Google Scholar] [CrossRef]
- Singh, A.K.; Singh, P.K.; Arya, M.; Singh, N.K.; Singh, U.S. Molecular Screening of Blast Resistance Genes in Rice using SSR Markers. Plant Pathol. J. 2015, 31, 12–24. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, Y.; Han, D.; Kang, Z.; Li, G.; Cao, A.; Chen, P. SSR and STS markers for wheat stripe rust resistance gene Yr26. Euphytica 2008, 159, 359–366. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, X.; Li, C.; Liu, Z.; Zhou, R.; Shen, S.; Yu, T.; Jia, L.; Li, N. Genome-Wide Analysis of the NBS-LRR Gene Family and SSR Molecular Markers Development in Solanaceae. Horticulturae 2024, 10, 1293. https://doi.org/10.3390/horticulturae10121293
Song X, Li C, Liu Z, Zhou R, Shen S, Yu T, Jia L, Li N. Genome-Wide Analysis of the NBS-LRR Gene Family and SSR Molecular Markers Development in Solanaceae. Horticulturae. 2024; 10(12):1293. https://doi.org/10.3390/horticulturae10121293
Chicago/Turabian StyleSong, Xiaoming, Chunjin Li, Zhuo Liu, Rong Zhou, Shaoqin Shen, Tong Yu, Li Jia, and Nan Li. 2024. "Genome-Wide Analysis of the NBS-LRR Gene Family and SSR Molecular Markers Development in Solanaceae" Horticulturae 10, no. 12: 1293. https://doi.org/10.3390/horticulturae10121293
APA StyleSong, X., Li, C., Liu, Z., Zhou, R., Shen, S., Yu, T., Jia, L., & Li, N. (2024). Genome-Wide Analysis of the NBS-LRR Gene Family and SSR Molecular Markers Development in Solanaceae. Horticulturae, 10(12), 1293. https://doi.org/10.3390/horticulturae10121293







