miRNA in Molecular Diagnostics
Abstract
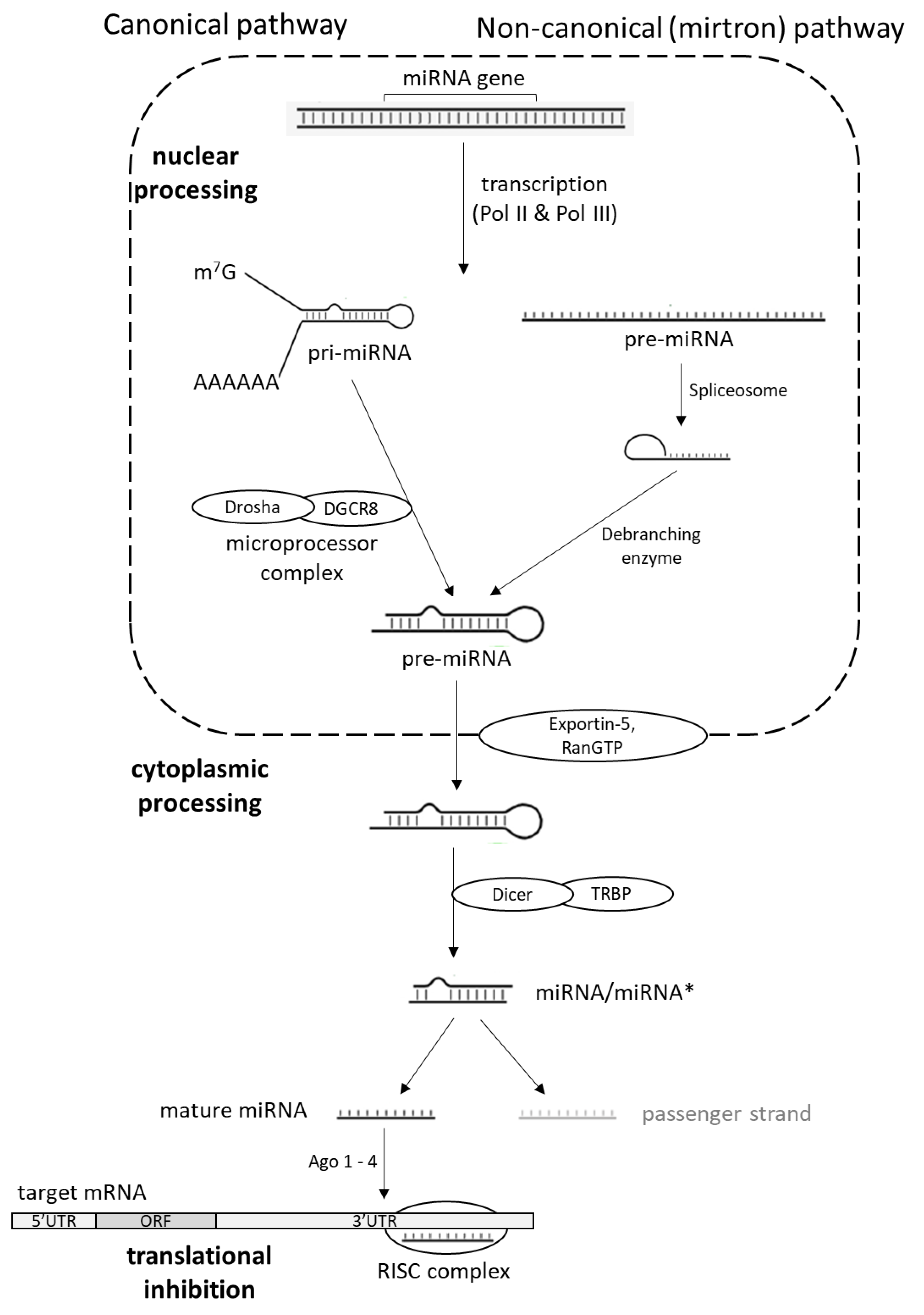
:1. MicroRNA Biogenesis
2. miRNA in Malignant Tumors
2.1. miRNAs in Leukemia and Lymphoma
2.2. miRNA in Brain Tumors
2.3. miRNA in Lung Cancer
2.4. miRNA in Breast Carcinoma
2.5. miRNA in Bladder and Renal Carcinoma
2.6. miRNA in Colon, Hepatocellular and Gastric Carcinoma
2.7. miRNA in Cervical Carcinoma, Testicular Tumors, and Prostate Cancer
2.8. miRNA in Skin Tumors
2.9. miRNA in Other Tumors
3. miRNA in Viral Diseases
3.1. DNA Viruses
3.1.1. Herpesviruses
3.1.2. Polyomaviruses
3.1.3. Papillomavirus
3.1.4. Adenoviruses
3.1.5. Hepadnaviridae
3.2. RNA Viruses
3.2.1. Flaviviridae
3.2.2. Retroviruses
3.2.3. Influenza Virus
3.2.4. Coronaviruses
4. Methods for miRNA Detection
4.1. Microarrays
4.2. Quantitative Real Time Polymerase Chain Reaction
4.3. In Situ Hybridization
4.4. Northern Blotting
4.5. Next Generation Sequencing
5. miRNA Usage in Current Molecular Diagnostics
6. Future Perspective
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, V.; Han, J.; Siomi, M. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Slack, F.; Basson, M.; Pasquinelli, A.; Bettinger, J.; Rougvie, A.; Horvitz, H.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Lau, N.C.; Lim, L.P.; Weinstein, E.G.; Bartel, D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001, 294, 858–862. [Google Scholar] [CrossRef] [PubMed]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef]
- Gulyaeva, L.F.; Kushlinskiy, N.E. Regulatory mechanisms of microRNA expression. J. Transl. Med. 2016, 14, 143. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Meyer, J.; Borkhardt, A.; Tuschl, T. New microRNAs from mouse and human. RNA 2003, 9, 175–179. [Google Scholar] [CrossRef]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Lee, Y.; Yeom, K.H.; Kim, Y.K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef] [PubMed]
- Okada, C.; Yamashita, E.; Lee, S.J.; Shibata, S.; Katahira, J.; Nakagawa, A.; Yoneda, Y.; Tsukihara, T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science 2009, 326, 1275–1279. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Croce, C.M. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012, 4, 143–159. [Google Scholar] [CrossRef]
- Hutvágner, G.; McLachlan, J.; Pasquinelli, A.E.; Bálint, E.; Tuschl, T.; Zamore, P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838. [Google Scholar] [CrossRef]
- Lee, Y.; Hur, I.; Park, S.Y.; Kim, Y.K.; Suh, M.R.; Kim, V.N. The role of PACT in the RNA silencing pathway. EMBO J. 2006, 25, 522–532. [Google Scholar] [CrossRef]
- Meister, G.; Landthaler, M.; Patkaniowska, A.; Dorsett, Y.; Teng, G.; Tuschl, T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 2004, 15, 185–197. [Google Scholar] [CrossRef]
- Yoda, M.; Kawamata, T.; Paroo, Z.; Ye, X.; Iwasaki, S.; Liu, Q.; Tomari, Y. ATP-dependent human RISC assembly pathways. Nat. Struct. Mol. Biol. 2010, 17, 17–23. [Google Scholar] [CrossRef]
- Choe, J.; Cho, H.; Lee, H.C.; Kim, Y.K. microRNA/Argonaute 2 regulates nonsense-mediated messenger RNA decay. EMBO Rep. 2010, 11, 380–386. [Google Scholar] [CrossRef] [Green Version]
- Nishi, K.; Nishi, A.; Nagasawa, T.; Ui-Tei, K. Human TNRC6A is an Argonaute navigator protein for microRNA-mediated gene silencing in the nucleus. RNA 2013, 19, 17–35. [Google Scholar] [CrossRef] [PubMed]
- Ruby, J.G.; Jan, C.H.; Bartel, D.P. Intronic microRNA precursors that bypass Drosha processing. Nature 2007, 448, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Okamura, K.; Hagen, J.W.; Duan, H.; Tyler, D.M.; Lai, E.C. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 2007, 130, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Berezikov, E.; Chung, W.J.; Willis, J.; Cuppen, E.; Lai, E.C. Mammalian mirtron genes. Mol. Cell 2007, 28, 328–336. [Google Scholar] [CrossRef]
- Babiarz, J.E.; Ruby, J.G.; Wang, Y.; Bartel, D.P.; Blelloch, R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008, 22, 2773–2785. [Google Scholar] [CrossRef]
- Musilova, K.; Mraz, M. MicroRNAs in B-cell lymphomas: How a complex biology gets more complex. Leukemia 2015, 29, 1004–1017. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Calin, G.A.; Cimmino, A.; Fabbri, M.; Ferracin, M.; Wojcik, S.E.; Shimizu, M.; Taccioli, C.; Zanesi, N.; Garzon, R.; Aqeilan, R.I.; et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc. Natl. Acad. Sci. USA 2008, 105, 5166–5171. [Google Scholar] [CrossRef]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef] [Green Version]
- Lovat, F.; Fassan, M.; Sacchi, D.; Ranganathan, P.; Palamarchuk, A.; Bill, M.; Karunasiri, M.; Gasparini, P.; Nigita, G.; Distefano, R.; et al. Knockout of both miR-15/16 loci induces acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 2018, 115, 13069–13074. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Thomson, J.M.; Hemann, M.T.; Hernando-Monge, E.; Mu, D.; Goodson, S.; Powers, S.; Cordon-Cardo, C.; Lowe, S.W.; Hannon, G.J.; et al. A microRNA polycistron as a potential human oncogene. Nature 2005, 435, 828–833. [Google Scholar] [CrossRef]
- Xiao, C.; Srinivasan, L.; Calado, D.P.; Patterson, H.C.; Zhang, B.; Wang, J.; Henderson, J.M.; Kutok, J.L.; Rajewsky, K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat. Immunol. 2008, 9, 405–414. [Google Scholar] [CrossRef]
- Psathas, J.N.; Doonan, P.J.; Raman, P.; Freedman, B.D.; Minn, A.J.; Thomas-Tikhonenko, A. The Myc-miR-17-92 axis amplifies B-cell receptor signaling via inhibition of ITIM proteins: A novel lymphomagenic feed-forward loop. Blood 2013, 122, 4220–4229. [Google Scholar] [CrossRef] [PubMed]
- Scherr, M.; Elder, A.; Battmer, K.; Barzan, D.; Bomken, S.; Ricke-Hoch, M.; Schröder, A.; Venturini, L.; Blair, H.J.; Vormoor, J.; et al. Differential expression of miR-17~92 identifies BCL2 as a therapeutic target in BCR-ABL-positive B-lineage acute lymphoblastic leukemia. Leukemia 2014, 28, 554–565. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.Y.; Oda, H.; Lai, M.; Skalsky, R.L.; Bethel, K.; Shepherd, J.; Kang, S.G.; Liu, W.H.; Sabouri-Ghomi, M.; Cullen, B.R.; et al. MicroRNA-17~92 plays a causative role in lymphomagenesis by coordinating multiple oncogenic pathways. EMBO J. 2013, 32, 2377–2391. [Google Scholar] [CrossRef] [PubMed]
- Cerna, K.; Oppelt, J.; Chochola, V.; Musilova, K.; Seda, V.; Pavlasova, G.; Radova, L.; Arigoni, M.; Calogero, R.A.; Benes, V.; et al. MicroRNA miR-34a downregulates FOXP1 during DNA damage response to limit BCR signalling in chronic lymphocytic leukaemia B cells. Leukemia 2019, 33, 403–414. [Google Scholar] [CrossRef]
- Mraz, M.; Chen, L.; Rassenti, L.Z.; Ghia, E.M.; Li, H.; Jepsen, K.; Smith, E.N.; Messer, K.; Frazer, K.A.; Kipps, T.J. miR-150 influences B-cell receptor signaling in chronic lymphocytic leukemia by regulating expression of GAB1 and FOXP1. Blood 2014, 124, 84–95. [Google Scholar] [CrossRef]
- Tano, N.; Kim, H.W.; Ashraf, M. microRNA-150 regulates mobilization and migration of bone marrow-derived mononuclear cells by targeting Cxcr4. PLoS ONE 2011, 6, e23114. [Google Scholar] [CrossRef]
- Vigorito, E.; Perks, K.L.; Abreu-Goodger, C.; Bunting, S.; Xiang, Z.; Kohlhaas, S.; Das, P.P.; Miska, E.A.; Rodriguez, A.; Bradley, A.; et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity 2007, 27, 847–859. [Google Scholar] [CrossRef] [Green Version]
- Eis, P.S.; Tam, W.; Sun, L.; Chadburn, A.; Li, Z.; Gomez, M.F.; Lund, E.; Dahlberg, J.E. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. USA 2005, 102, 3627–3632. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Ferracin, M.; Cimmino, A.; Di Leva, G.; Shimizu, M.; Wojcik, S.E.; Iorio, M.V.; Visone, R.; Sever, N.I.; Fabbri, M.; et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005, 353, 1793–1801. [Google Scholar] [CrossRef] [PubMed]
- Pekarsky, Y.; Santanam, U.; Cimmino, A.; Palamarchuk, A.; Efanov, A.; Maximov, V.; Volinia, S.; Alder, H.; Liu, C.G.; Rassenti, L.; et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006, 66, 11590–11593. [Google Scholar] [CrossRef] [PubMed]
- Medina, P.P.; Nolde, M.; Slack, F.J. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 2010, 467, 86–90. [Google Scholar] [CrossRef]
- Valeri, N.; Gasparini, P.; Braconi, C.; Paone, A.; Lovat, F.; Fabbri, M.; Sumani, K.M.; Alder, H.; Amadori, D.; Patel, T.; et al. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2). Proc. Natl. Acad. Sci. USA 2010, 107, 21098–21103. [Google Scholar] [CrossRef]
- Zhao, J.J.; Lin, J.; Lwin, T.; Yang, H.; Guo, J.; Kong, W.; Dessureault, S.; Moscinski, L.C.; Rezania, D.; Dalton, W.S.; et al. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood 2010, 115, 2630–2639. [Google Scholar] [CrossRef]
- Basso, J.; Paggi, M.G.; Fortuna, A.; Vitorino, C.; Vitorino, R. Deciphering specific miRNAs in brain tumors: A 5-miRNA signature in glioblastoma. Mol. Genet. Genomics 2022, 297, 507–521. [Google Scholar] [CrossRef]
- Kefas, B.; Godlewski, J.; Comeau, L.; Li, Y.; Abounader, R.; Hawkinson, M.; Lee, J.; Fine, H.; Chiocca, E.A.; Lawler, S.; et al. microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res. 2008, 68, 3566–3572. [Google Scholar] [CrossRef]
- Papagiannakopoulos, T.; Shapiro, A.; Kosik, K.S. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008, 68, 8164–8172. [Google Scholar] [CrossRef]
- Chan, J.A.; Krichevsky, A.M.; Kosik, K.S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005, 65, 6029–6033. [Google Scholar] [CrossRef] [Green Version]
- Costa, P.M.; Cardoso, A.L.; Pereira de Almeida, L.F.; Bruce, J.N.; Canoll, P.; Pedroso de Lima, M.C. PDGF-B-mediated downregulation of miR-21: New insights into PDGF signaling in glioblastoma. Hum. Mol. Genet. 2012, 21, 5118–5130. [Google Scholar] [CrossRef] [PubMed]
- Godlewski, J.; Nowicki, M.O.; Bronisz, A.; Nuovo, G.; Palatini, J.; De Lay, M.; Van Brocklyn, J.; Ostrowski, M.C.; Chiocca, E.A.; Lawler, S.E. MicroRNA-451 regulates LKB1/AMPK signaling and allows adaptation to metabolic stress in glioma cells. Mol. Cell 2010, 37, 620–632. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, D.; Ansari, K.; Nowicki, M.O.; Salińska, E.; Bronisz, A.; Godlewski, J. MicroRNA-451 Inhibits Migration of Glioblastoma while Making It More Susceptible to Conventional Therapy. Noncoding RNA 2019, 5, 25. [Google Scholar] [CrossRef] [PubMed]
- Kefas, B.; Comeau, L.; Erdle, N.; Montgomery, E.; Amos, S.; Purow, B. Pyruvate kinase M2 is a target of the tumor-suppressive microRNA-326 and regulates the survival of glioma cells. Neuro Oncol. 2010, 12, 1102–1112. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guessous, F.; Zhang, Y.; Dipierro, C.; Kefas, B.; Johnson, E.; Marcinkiewicz, L.; Jiang, J.; Yang, Y.; Schmittgen, T.D.; et al. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009, 69, 7569–7576. [Google Scholar] [CrossRef]
- Ciafrè, S.A.; Galardi, S.; Mangiola, A.; Ferracin, M.; Liu, C.G.; Sabatino, G.; Negrini, M.; Maira, G.; Croce, C.M.; Farace, M.G. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem. Biophys. Res. Commun. 2005, 334, 1351–1358. [Google Scholar] [CrossRef]
- Godlewski, J.; Nowicki, M.O.; Bronisz, A.; Williams, S.; Otsuki, A.; Nuovo, G.; Raychaudhury, A.; Newton, H.B.; Chiocca, E.A.; Lawler, S. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008, 68, 9125–9130. [Google Scholar] [CrossRef]
- Peruzzi, P.; Bronisz, A.; Nowicki, M.O.; Wang, Y.; Ogawa, D.; Price, R.; Nakano, I.; Kwon, C.H.; Hayes, J.; Lawler, S.E.; et al. MicroRNA-128 coordinately targets Polycomb Repressor Complexes in glioma stem cells. Neuro Oncol. 2013, 15, 1212–1224. [Google Scholar] [CrossRef]
- Conti, A.; Aguennouz, M.; La Torre, D.; Tomasello, C.; Cardali, S.; Angileri, F.F.; Maio, F.; Cama, A.; Germanò, A.; Vita, G.; et al. miR-21 and 221 upregulation and miR-181b downregulation in human grade II-IV astrocytic tumors. J. Neurooncol. 2009, 93, 325–332. [Google Scholar] [CrossRef]
- Chen, G.; Zhu, W.; Shi, D.; Lv, L.; Zhang, C.; Liu, P.; Hu, W. MicroRNA-181a sensitizes human malignant glioma U87MG cells to radiation by targeting Bcl-2. Oncol. Rep. 2010, 23, 997–1003. [Google Scholar]
- Wang, H.; Tao, T.; Yan, W.; Feng, Y.; Wang, Y.; Cai, J.; You, Y.; Jiang, T.; Jiang, C. Upregulation of miR-181s reverses mesenchymal transition by targeting KPNA4 in glioblastoma. Sci. Rep. 2015, 5, 13072. [Google Scholar] [CrossRef] [PubMed]
- Medina, R.; Zaidi, S.K.; Liu, C.G.; Stein, J.L.; van Wijnen, A.J.; Croce, C.M.; Stein, G.S. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res. 2008, 68, 2773–2780. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Tu, H.B.; Wu, L.; Liu, M.; Jiang, G.N. MicroRNA-21 Regulates Non-Small Cell Lung Cancer Cell Invasion and Chemo-Sensitivity through SMAD7. Cell. Physiol. Biochem. 2016, 38, 2152–2162. [Google Scholar] [CrossRef] [PubMed]
- Seike, M.; Goto, A.; Okano, T.; Bowman, E.D.; Schetter, A.J.; Horikawa, I.; Mathe, E.A.; Jen, J.; Yang, P.; Sugimura, H.; et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc. Natl. Acad. Sci. USA 2009, 106, 12085–12090. [Google Scholar] [CrossRef] [PubMed]
- Markou, A.; Tsaroucha, E.G.; Kaklamanis, L.; Fotinou, M.; Georgoulias, V.; Lianidou, E.S. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin. Chem. 2008, 54, 1696–1704. [Google Scholar] [CrossRef]
- Cai, J.; Fang, L.; Huang, Y.; Li, R.; Yuan, J.; Yang, Y.; Zhu, X.; Chen, B.; Wu, J.; Li, M. miR-205 targets PTEN and PHLPP2 to augment AKT signaling and drive malignant phenotypes in non-small cell lung cancer. Cancer Res. 2013, 73, 5402–5415. [Google Scholar] [CrossRef]
- Larzabal, L.; de Aberasturi, A.L.; Redrado, M.; Rueda, P.; Rodriguez, M.J.; Bodegas, M.E.; Montuenga, L.M.; Calvo, A. TMPRSS4 regulates levels of integrin α5 in NSCLC through miR-205 activity to promote metastasis. Br. J. Cancer 2014, 110, 764–774. [Google Scholar] [CrossRef]
- Kumar, M.S.; Erkeland, S.J.; Pester, R.E.; Chen, C.Y.; Ebert, M.S.; Sharp, P.A.; Jacks, T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc. Natl. Acad. Sci. USA 2008, 105, 3903–3908. [Google Scholar] [CrossRef]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS is regulated by the let-7 microRNA family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef]
- Kanthaje, S.; Baikunje, N.; Kandal, I.; Ratnacaram, C.K. Repertoires of MicroRNA-30 family as gate-keepers in lung cancer. Front. Biosci. 2021, 13, 141–156. [Google Scholar]
- Song, K.; Jiang, Y.; Zhao, Y.; Xie, Y.; Zhou, J.; Yu, W.; Wang, Q. Members of the miR-30 family inhibit the epithelial-to-mesenchymal transition of non-small-cell lung cancer cells by suppressing XB130 expression levels. Oncol. Lett. 2020, 20, 68. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Liu, H.; Jiang, L.; Rui, B.; Mei, J.; Xiao, H. miR-26 Induces Apoptosis and Inhibits Autophagy in Non-small Cell Lung Cancer Cells by Suppressing TGF-β1-JNK Signaling Pathway. Front. Pharmacol. 2019, 9, 1509. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Qiu, M.; Jiang, F.; Zhang, S.; Yang, X.; Wang, J.; Xu, L.; Yin, R. MiR-145 regulates cancer stem-like properties and epithelial-to-mesenchymal transition in lung adenocarcinoma-initiating cells. Tumour Biol. 2014, 35, 8953–8961. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.J.; Wei, R.S.; Li, X.H.; Li, Q.; Yu, J.R.; Zhuang, X.F. MiR-421 promotes lipid metabolism by targeting PTEN via activating PI3K/AKT/mTOR pathway in non-small cell lung cancer. Epigenomics, 2022; Online ahead of print. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, L.; Qi, Y.; Yang, X. MicroRNA-214 upregulates HIF-1α and VEGF by targeting ING4 in lung cancer cells. Mol. Med. Rep. 2019, 19, 4935–4945. [Google Scholar] [CrossRef]
- Liu, C.; Luo, J.; Zhao, Y.T.; Wang, Z.Y.; Zhou, J.; Huang, S.; Huang, J.N.; Long, H.X.; Zhu, B. TWIST1 upregulates miR-214 to promote epithelial-to-mesenchymal transition and metastasis in lung adenocarcinoma. Int. J. Mol. Med. 2018, 42, 461–470. [Google Scholar] [CrossRef]
- Joshi, P.; Jeon, Y.J.; Laganà, A.; Middleton, J.; Secchiero, P.; Garofalo, M.; Croce, C.M. MicroRNA-148a reduces tumorigenesis and increases TRAIL-induced apoptosis in NSCLC. Proc. Natl. Acad. Sci. USA 2015, 112, 8650–8655. [Google Scholar] [CrossRef]
- Lin, H.Y.; Chiang, C.H.; Hung, W.C. STAT3 upregulates miR-92a to inhibit RECK expression and to promote invasiveness of lung cancer cells. Br. J. Cancer 2013, 109, 731–738. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Y.; Qi, P.; Ma, Z. Biology of MiR-17-92 Cluster and Its Progress in Lung Cancer. Int. J. Med. Sci. 2018, 15, 1443–1448. [Google Scholar] [CrossRef]
- Ninio-Many, L.; Hikri, E.; Burg-Golani, T.; Stemmer, S.M.; Shalgi, R.; Ben-Aharon, I. miR-125a Induces HER2 Expression and Sensitivity to Trastuzumab in Triple-Negative Breast Cancer Lines. Front. Oncol. 2020, 10, 191. [Google Scholar] [CrossRef]
- Nandy, S.B.; Arumugam, A.; Subramani, R.; Pedroza, D.; Hernandez, K.; Saltzstein, E.; Lakshmanaswamy, R. MicroRNA-125a influences breast cancer stem cells by targeting leukemia inhibitory factor receptor which regulates the Hippo signaling pathway. Oncotarget 2015, 6, 17366–17378. [Google Scholar] [CrossRef]
- Iorio, M.V.; Ferracin, M.; Liu, C.G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adachi, R.; Horiuchi, S.; Sakurazawa, Y.; Hasegawa, T.; Sato, K.; Sakamaki, T. ErbB2 down-regulates microRNA-205 in breast cancer. Biochem. Biophys. Res. Commun. 2011, 411, 804–808. [Google Scholar] [CrossRef] [PubMed]
- Elgamal, O.A.; Park, J.K.; Gusev, Y.; Azevedo-Pouly, A.C.; Jiang, J.; Roopra, A.; Schmittgen, T.D. Tumor suppressive function of mir-205 in breast cancer is linked to HMGB3 regulation. PLoS ONE 2013, 8, e76402. [Google Scholar] [CrossRef]
- Liang, H.; Xiao, J.; Zhou, Z.; Wu, J.; Ge, F.; Li, Z.; Zhang, H.; Sun, J.; Li, F.; Liu, R.; et al. Hypoxia induces miR-153 through the IRE1α-XBP1 pathway to fine tune the HIF1α/VEGFA axis in breast cancer angiogenesis. Oncogene 2018, 37, 1961–1975. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Luo, A.; Liu, Y.; Wang, S.; Li, Y.; Shi, W.; Liu, Z.; Qu, X. MiR-214 increases the sensitivity of breast cancer cells to tamoxifen and fulvestrant through inhibition of autophagy. Mol. Cancer 2015, 14, 208. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Lv, P.; Liu, X.; Zhu, M.; Qiu, X. microRNA-214 enhances the invasion ability of breast cancer cells by targeting p53. Int. J. Mol. Med. 2015, 35, 1395–1402. [Google Scholar] [CrossRef] [PubMed]
- Frankel, L.B.; Christoffersen, N.R.; Jacobsen, A.; Lindow, M.; Krogh, A.; Lund, A.H. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J. Biol. Chem. 2008, 283, 1026–1033. [Google Scholar] [CrossRef]
- Yu, F.; Yao, H.; Zhu, P.; Zhang, X.; Pan, Q.; Gong, C.; Huang, Y.; Hu, X.; Su, F.; Lieberman, J.; et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 2007, 131, 1109–1123. [Google Scholar] [CrossRef]
- Ma, L.; Teruya-Feldstein, J.; Weinberg, R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007, 449, 682–688. [Google Scholar] [CrossRef]
- Camps, C.; Buffa, F.M.; Colella, S.; Moore, J.; Sotiriou, C.; Sheldon, H.; Harris, A.L.; Gleadle, J.M.; Ragoussis, J. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin. Cancer Res. 2008, 14, 1340–1348. [Google Scholar] [CrossRef]
- Hu, Y.; Qiu, Y.; Yagüe, E.; Ji, W.; Liu, J.; Zhang, J. miRNA-205 targets VEGFA and FGF2 and regulates resistance to chemotherapeutics in breast cancer. Cell Death Dis. 2016, 7, e2291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddy, S.D.; Ohshiro, K.; Rayala, S.K.; Kumar, R. MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions. Cancer Res. 2008, 68, 8195–8200. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, C.; Miao, S.; Li, C.; Chen, Z.; Li, F. Enhancing E-cadherin expression via promoter-targeted miR-373 suppresses bladder cancer cells growth and metastasis. Oncotarget 2017, 8, 93969–93983. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, Z.; Wang, X. miRNA-373 promotes urinary bladder cancer cell proliferation, migration and invasion through upregulating epidermal growth factor receptor. Exp. Ther. Med. 2019, 17, 1190–1195. [Google Scholar] [CrossRef]
- Ohno, R.; Uozaki, H.; Kikuchi, Y.; Kumagai, A.; Aso, T.; Watanabe, M.; Watabe, S.; Muto, S.; Yamaguchi, R. Both cancerous miR-21 and stromal miR-21 in urothelial carcinoma are related to tumour progression. Histopathology 2016, 69, 993–999. [Google Scholar] [CrossRef]
- Andrew, A.S.; Marsit, C.J.; Schned, A.R.; Seigne, J.D.; Kelsey, K.T.; Moore, J.H.; Perreard, L.; Karagas, M.R.; Sempere, L.F. Expression of tumor suppressive microRNA-34a is associated with a reduced risk of bladder cancer recurrence. Int. J. Cancer 2015, 137, 1158–1166. [Google Scholar] [CrossRef]
- Wszolek, M.F.; Rieger-Christ, K.M.; Kenney, P.A.; Gould, J.J.; Silva Neto, B.; Lavoie, A.K.; Logvinenko, T.; Libertino, J.A.; Summerhayes, I.C. A MicroRNA expression profile defining the invasive bladder tumor phenotype. Urol. Oncol. 2011, 29, 794–801. [Google Scholar] [CrossRef]
- Gottardo, F.; Liu, C.G.; Ferracin, M.; Calin, G.A.; Fassan, M.; Bassi, P.; Sevignani, C.; Byrne, D.; Negrini, M.; Pagano, F.; et al. Micro-RNA profiling in kidney and bladder cancers. Urol. Oncol. 2007, 25, 387–392. [Google Scholar] [CrossRef]
- Mao, X.W.; Xiao, J.Q.; Li, Z.Y.; Zheng, Y.C.; Zhang, N. Effects of microRNA-135a on the epithelial-mesenchymal transition, migration and invasion of bladder cancer cells by targeting GSK3β through the Wnt/β-catenin signaling pathway. Exp. Mol. Med. 2018, 50, e429. [Google Scholar] [CrossRef]
- Xu, T.; Qin, L.; Zhu, Z.; Wang, X.; Liu, Y.; Fan, Y.; Zhong, S.; Wang, X.; Zhang, X.; Xia, L.; et al. MicroRNA-31 functions as a tumor suppressor and increases sensitivity to mitomycin-C in urothelial bladder cancer by targeting integrin α5. Oncotarget 2016, 7, 27445–27457. [Google Scholar] [CrossRef]
- Dey, N.; Das, F.; Ghosh-Choudhury, N.; Mandal, C.C.; Parekh, D.J.; Block, K.; Kasinath, B.S.; Abboud, H.E.; Choudhury, G.G. microRNA-21 governs TORC1 activation in renal cancer cell proliferation and invasion. PLoS ONE 2012, 7, e37366. [Google Scholar]
- Xu, X.; Wu, J.; Li, S.; Hu, Z.; Xu, X.; Zhu, Y.; Liang, Z.; Wang, X.; Lin, Y.; Mao, Y.; et al. Downregulation of microRNA-182-5p contributes to renal cell carcinoma proliferation via activating the AKT/FOXO3a signaling pathway. Mol. Cancer 2014, 13, 109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, P.; Wang, L.; Lu, Y.; Liu, Y.; Li, L.; Yin, L.; Zhang, C.; Zhao, W.; Shen, B.; Xu, W. MicroRNA-195 targets VEGFR2 and has a tumor suppressive role in ACHN cells via PI3K/Akt and Raf/MEK/ERK signaling pathways. Int. J. Oncol. 2016, 49, 1155–1163. [Google Scholar] [CrossRef]
- Liu, F.; Wu, L.; Wang, A.; Xu, Y.; Luo, X.; Liu, X.; Hua, Y.; Zhang, D.; Wu, S.; Lin, T.; et al. MicroRNA-138 attenuates epithelial-to-mesenchymal transition by targeting SOX4 in clear cell renal cell carcinoma. Am. J. Transl. Res. 2017, 9, 3611–3622. [Google Scholar]
- Lu, J.; Wei, J.H.; Feng, Z.H.; Chen, Z.H.; Wang, Y.Q.; Huang, Y.; Fang, Y.; Liang, Y.P.; Cen, J.J.; Pan, Y.H.; et al. miR-106b-5p promotes renal cell carcinoma aggressiveness and stem-cell-like phenotype by activating Wnt/β-catenin signalling. Oncotarget 2017, 8, 21461–21471. [Google Scholar] [CrossRef]
- Hu, G.; Lai, P.; Liu, M.; Xu, L.; Guo, Z.; Liu, H.; Li, W.; Wang, G.; Yao, X.; Zheng, J.; et al. miR-203a regulates proliferation, migration, and apoptosis by targeting glycogen synthase kinase-3β in human renal cell carcinoma. Tumour Biol. 2014, 35, 11443–11453. [Google Scholar] [CrossRef] [PubMed]
- Strillacci, A.; Griffoni, C.; Sansone, P.; Paterini, P.; Piazzi, G.; Lazzarini, G.; Spisni, E.; Pantaleo, M.A.; Biasco, G.; Tomasi, V. MiR-101 downregulation is involved in cyclooxygenase-2 overexpression in human colon cancer cells. Exp. Cell Res. 2009, 315, 1439–1447. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Yu, L.L.; Han, N.; Zhang, B.T. miR-141 promotes colon cancer cell proliferation by inhibiting MAP2K4. Oncol. Lett. 2017, 13, 1665–1671. [Google Scholar] [CrossRef]
- Tian, Y.; Pan, Q.; Shang, Y.; Zhu, R.; Ye, J.; Liu, Y.; Zhong, X.; Li, S.; He, Y.; Chen, L.; et al. MicroRNA-200 (miR-200) cluster regulation by achaete scute-like 2 (Ascl2): Impact on the epithelial-mesenchymal transition in colon cancer cells. J. Biol. Chem. 2014, 289, 36101–36115. [Google Scholar] [CrossRef]
- Wang, P.; Zou, F.; Zhang, X.; Li, H.; Dulak, A.; Tomko, R.J., Jr.; Lazo, J.S.; Wang, Z.; Zhang, L.; Yu, J. microRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer Res. 2009, 69, 8157–8165. [Google Scholar] [CrossRef]
- Yu, Y.; Kanwar, S.S.; Patel, B.B.; Oh, P.S.; Nautiyal, J.; Sarkar, F.H.; Majumdar, A.P. MicroRNA-21 induces stemness by downregulating transforming growth factor beta receptor 2 (TGFβR2) in colon cancer cells. Carcinogenesis 2012, 33, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Christensen, L.L.; Tobiasen, H.; Holm, A.; Schepeler, T.; Ostenfeld, M.S.; Thorsen, K.; Rasmussen, M.H.; Birkenkamp-Demtroeder, K.; Sieber, O.M.; Gibbs, P.; et al. MiRNA-362-3p induces cell cycle arrest through targeting of E2F1, USF2 and PTPN1 and is associated with recurrence of colorectal cancer. Int. J. Cancer 2013, 133, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, X.; Yang, Z.; Du, H.; Wu, Z.; Gong, J.; Yan, J.; Zheng, Q. MiR-145 regulates PAK4 via the MAPK pathway and exhibits an antitumor effect in human colon cells. Biochem. Biophys. Res. Commun. 2012, 427, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Sun, Y.; Zhang, Y.; Han, S. Downregulation of miR-181b inhibits human colon cancer cell proliferation by targeting CYLD and inhibiting the NF-κB signaling pathway. Int. J. Mol. Med. 2020, 46, 1755–1764. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Li, Z.; Zhou, H. Identification of prognostic miRNA biomarkers for predicting overall survival of colon adenocarcinoma and bioinformatics analysis: A study based on the Cancer Genome Atlas database. J. Cell. Biochem. 2019, 120, 9839–9849. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, Y.; Fu, Y.; Han, W.; Xu, H.; Wen, L.; Deng, Y.; Liu, K. Long non-coding RNA LINC00160 functions as a decoy of microRNA-132 to mediate autophagy and drug resistance in hepatocellular carcinoma via inhibition of PIK3R3. Cancer Lett 2020, 478, 22–33. [Google Scholar] [CrossRef]
- Bao, C.; Li, Y.; Huan, L.; Zhang, Y.; Zhao, F.; Wang, Q.; Liang, L.; Ding, J.; Liu, L.; Chen, T.; et al. NF-κB signaling relieves negative regulation by miR-194 in hepatocellular carcinoma by suppressing the transcription factor HNF-1α. Sci. Signal. 2015, 8, ra75. [Google Scholar] [CrossRef] [PubMed]
- Cui, G.; Wang, H.; Liu, W.; Xing, J.; Song, W.; Zeng, Z.; Liu, L.; Wang, H.; Wang, X.; Luo, H.; et al. Glycogen Phosphorylase B Is Regulated by miR101-3p and Promotes Hepatocellular Carcinoma Tumorigenesis. Front. Cell Dev. Biol. 2020, 8, 566494. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.N.; Zhou, J.; Lu, X.J. The long noncoding RNA NEAT1 contributes to hepatocellular carcinoma development by sponging miR-485 and enhancing the expression of the STAT3. J. Cell. Physiol. 2018, 233, 6733–6741. [Google Scholar] [CrossRef]
- Wang, Y.; Lee, A.T.; Ma, J.Z.; Wang, J.; Ren, J.; Yang, Y.; Tantoso, E.; Li, K.B.; Ooi, L.L.; Tan, P.; et al. Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J. Biol. Chem. 2008, 283, 13205–13215. [Google Scholar] [CrossRef]
- Connolly, E.; Melegari, M.; Landgraf, P.; Tchaikovskaya, T.; Tennant, B.C.; Slagle, B.L.; Rogler, L.E.; Zavolan, M.; Tuschl, T.; Rogler, C.E. Elevated expression of the miR-17-92 polycistron and miR-21 in hepadnavirus-associated hepatocellular carcinoma contributes to the malignant phenotype. Am. J. Pathol. 2008, 173, 856–864. [Google Scholar] [CrossRef] [PubMed]
- Kong, J.; Liu, X.; Li, X.; Wu, J.; Wu, N.; Chen, J.; Fang, F. miR-125/Pokemon auto-circuit contributes to the progression of hepatocellular carcinoma. Tumour Biol. 2016, 37, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xie, L.; He, X.; Li, J.; Tu, K.; Wei, L.; Wu, J.; Guo, Y.; Ma, X.; Zhang, P.; et al. Diagnostic and prognostic implications of microRNAs in human hepatocellular carcinoma. Int. J. Cancer 2008, 123, 1616–1622. [Google Scholar] [CrossRef] [PubMed]
- Murakami, Y.; Yasuda, T.; Saigo, K.; Urashima, T.; Toyoda, H.; Okanoue, T.; Shimotohno, K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene 2006, 25, 2537–2545. [Google Scholar] [CrossRef]
- Yang, T.S.; Yang, X.H.; Wang, X.D.; Wang, Y.L.; Zhou, B.; Song, Z.S. MiR-214 regulate gastric cancer cell proliferation, migration and invasion by targeting PTEN. Cancer Cell Int. 2013, 13, 68. [Google Scholar] [CrossRef]
- Carvalho, J.; van Grieken, N.C.; Pereira, P.M.; Sousa, S.; Tijssen, M.; Buffart, T.E.; Diosdado, B.; Grabsch, H.; Santos, M.A.; Meijer, G.; et al. Lack of microRNA-101 causes E-cadherin functional deregulation through EZH2 up-regulation in intestinal gastric cancer. J. Pathol. 2012, 228, 31–44. [Google Scholar] [CrossRef]
- Ning, T.; Zhang, H.; Wang, X.; Li, S.; Zhang, L.; Deng, T.; Zhou, L.; Liu, R.; Wang, X.; Bai, M.; et al. miR-370 regulates cell proliferation and migration by targeting EGFR in gastric cancer. Oncol. Rep. 2017, 38, 384–392. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, H.; Ge, S.; Fan, Q.; Zhou, L.; Li, H.; Bai, M.; Ning, T.; Liu, R.; Wang, X.; et al. Effects of miR-138-5p and miR-204-5p on the migration and proliferation of gastric cancer cells by targeting EGFR. Oncol. Rep. 2018, 39, 2624–2634. [Google Scholar] [CrossRef]
- Kogo, R.; Mimori, K.; Tanaka, F.; Komune, S.; Mori, M. Clinical significance of miR-146a in gastric cancer cases. Clin. Cancer Res. 2011, 17, 4277–4284. [Google Scholar] [CrossRef]
- Yao, Y.; Suo, A.L.; Li, Z.F.; Liu, L.Y.; Tian, T.; Ni, L.; Zhang, W.G.; Nan, K.J.; Song, T.S.; Huang, C. MicroRNA profiling of human gastric cancer. Mol. Med. Rep. 2009, 2, 963–970. [Google Scholar] [PubMed]
- Lu, Z.; Liu, M.; Stribinskis, V.; Klinge, C.M.; Ramos, K.S.; Colburn, N.H.; Li, Y. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene 2008, 27, 4373–4379. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.; Gu, J.; Wang, Q.; Zheng, L. Translational control of Bcl-2 promotes apoptosis of gastric carcinoma cells. BMC Cancer 2021, 21, 12. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Chen, Q. Poor expression of microRNA-135b results in the inhibition of cisplatin resistance and proliferation and induces the apoptosis of gastric cancer cells through MST1-mediated MAPK signaling pathway. FASEB J. 2019, 33, 3420–3436. [Google Scholar] [CrossRef] [PubMed]
- Mody, H.R.; Hung, S.W.; Pathak, R.K.; Griffin, J.; Cruz-Monserrate, Z.; Govindarajan, R. miR-202 Diminishes TGFβ Receptors and Attenuates TGFβ1-Induced EMT in Pancreatic Cancer. Mol. Cancer Res. 2017, 15, 1029–1039. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Xu, Y.; Zhao, J.; Liu, Q.; Feng, W.; Fan, J.; Wang, P. miR-367 promotes epithelial-to-mesenchymal transition and invasion of pancreatic ductal adenocarcinoma cells by targeting the Smad7-TGF-β signalling pathway. Br. J. Cancer 2015, 112, 1367–1375. [Google Scholar] [CrossRef]
- Morgan, E.L.; Patterson, M.R.; Ryder, E.L.; Lee, S.Y.; Wasson, C.W.; Harper, K.L.; Li, Y.; Griffin, S.; Blair, G.E.; Whitehouse, A.; et al. MicroRNA-18a targeting of the STK4/MST1 tumour suppressor is necessary for transformation in HPV positive cervical cancer. PLoS Pathog. 2020, 16, e1008624. [Google Scholar] [CrossRef]
- Shen, Y.; Zhou, J.; Li, Y.; Ye, F.; Wan, X.; Lu, W.; Xie, X.; Cheng, X. miR-375 mediated acquired chemo-resistance in cervical cancer by facilitating EMT. PLoS ONE 2014, 9, e109299. [Google Scholar] [CrossRef]
- Babion, I.; Miok, V.; Jaspers, A.; Huseinovic, A.; Steenbergen, R.D.M.; van Wieringen, W.N.; Wilting, S.M. Identification of Deregulated Pathways.; Key Regulators.; and Novel miRNA-mRNA Interactions in HPV-Mediated Transformation. Cancers 2020, 12, 700. [Google Scholar] [CrossRef]
- Song, L.; Liu, S.; Zhang, L.; Yao, H.; Gao, F.; Xu, D.; Li, Q. MiR-21 modulates radiosensitivity of cervical cancer through inhibiting autophagy via the PTEN/Akt/HIF-1α feedback loop and the Akt-mTOR signaling pathway. Tumour Biol. 2016, 37, 12161–12168. [Google Scholar] [CrossRef]
- Chen, R.; Gan, Q.; Zhao, S.; Zhang, D.; Wang, S.; Yao, L.; Yuan, M.; Cheng, J. DNA methylation of miR-138 regulates cell proliferation and EMT in cervical cancer by targeting EZH2. BMC Cancer 2022, 22, 488. [Google Scholar] [CrossRef]
- Zheng, Q.; Peskoe, S.B.; Ribas, J.; Rafiqi, F.; Kudrolli, T.; Meeker, A.K.; De Marzo, A.M.; Platz, E.A.; Lupold, S.E. Investigation of miR-21, miR-141, and miR-221 expression levels in prostate adenocarcinoma for associated risk of recurrence after radical prostatectomy. Prostate 2014, 74, 1655–1662. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Ge, J.; Zhang, Z.; Zhou, W. miR-141 inhibits prostatic cancer cell proliferation and migration, and induces cell apoptosis via targeting of RUNX1. Oncol. Rep. 2018, 39, 1454–1460. [Google Scholar] [CrossRef]
- Hirata, H.; Ueno, K.; Shahryari, V.; Deng, G.; Tanaka, Y.; Tabatabai, Z.L.; Hinoda, Y.; Dahiya, R. MicroRNA-182-5p promotes cell invasion and proliferation by down regulating FOXF2.; RECK and MTSS1 genes in human prostate cancer. PLoS ONE 2013, 8, e55502. [Google Scholar] [CrossRef] [Green Version]
- Peng, X.; Guo, W.; Liu, T.; Wang, X.; Tu, X.; Xiong, D.; Chen, S.; Lai, Y.; Du, H.; Chen, G.; et al. Identification of miRs-143 and -145 that is associated with bone metastasis of prostate cancer and involved in the regulation of EMT. PLoS ONE 2011, 6, e20341. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, J.; Ma, Y.; Xu, C.; Wang, Y.; He, Y. MicroRNA miR-145-5p inhibits Phospholipase D 5 (PLD5) to downregulate cell proliferation and metastasis to mitigate prostate cancer. Bioengineered 2021, 12, 3240–3251. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, N.; Li, X.; Padi, S.K.; Zhang, Q.; Tang, M.S.; Guo, B. Downregulation of miR-205 and miR-31 confers resistance to chemotherapy-induced apoptosis in prostate cancer cells. Cell Death Dis. 2010, 1, e105. [Google Scholar] [CrossRef] [PubMed]
- Galardi, S.; Mercatelli, N.; Giorda, E.; Massalini, S.; Frajese, G.V.; Ciafrè, S.A.; Farace, M.G. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J. Biol. Chem. 2007, 282, 23716–23724. [Google Scholar] [CrossRef]
- Schubert, M.; Spahn, M.; Kneitz, S.; Scholz, C.J.; Joniau, S.; Stroebel, P.; Riedmiller, H.; Kneitz, B. Distinct microRNA expression profile in prostate cancer patients with early clinical failure and the impact of let-7 as prognostic marker in high-risk prostate cancer. PLoS ONE 2013, 8, e65064. [Google Scholar]
- Li, T.; Li, D.; Sha, J.; Sun, P.; Huang, Y. MicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cells. Biochem. Biophys. Res. Commun. 2009, 383, 280–285. [Google Scholar] [CrossRef]
- Liu, L.Z.; Li, C.; Chen, Q.; Jing, Y.; Carpenter, R.; Jiang, Y.; Kung, H.F.; Lai, L.; Jiang, B.H. MiR-21 induced angiogenesis through AKT and ERK activation and HIF-1α expression. PLoS ONE 2011, 6, e19139. [Google Scholar] [CrossRef]
- Palmer, R.D.; Murray, M.J.; Saini, H.K.; van Dongen, S.; Abreu-Goodger, C.; Muralidhar, B.; Pett, M.R.; Thornton, C.M.; Nicholson, J.C.; Enright, A.J.; et al. Malignant germ cell tumors display common microRNA profiles resulting in global changes in expression of messenger RNA targets. Cancer Res. 2010, 70, 2911–2923. [Google Scholar] [CrossRef] [PubMed]
- Looijenga, L.H.; Gillis, A.J.; Stoop, H.J.; Hersmus, R.; Oosterhuis, J.W. Chromosomes and expression in human testicular germ-cell tumors: Insight into their cell of origin and pathogenesis. Ann. N. Y. Acad. Sci. 2007, 1120, 187–214. [Google Scholar] [CrossRef] [PubMed]
- Voorhoeve, P.M.; le Sage, C.; Schrier, M.; Gillis, A.J.; Stoop, H.; Nagel, R.; Liu, Y.P.; van Duijse, J.; Drost, J.; Griekspoor, A.; et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 2006, 124, 1169–1181. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Shi, H.; Li, X.; Chen, G.; Larsson, C.; Lui, W.O. miR-223-3p regulates cell growth and apoptosis via FBXW7 suggesting an oncogenic role in human testicular germ cell tumors. Int. J. Oncol. 2017, 50, 356–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.; Tian, H.; Duan, Z.; Cao, Y.; Zhang, X.S.; Sun, F. microRNA-383 impairs phosphorylation of H2AX by targeting PNUTS and inducing cell cycle arrest in testicular embryonal carcinoma cells. Cell Signal. 2014, 26, 903–911. [Google Scholar] [CrossRef]
- Das, M.K.; Evensen, H.S.F.; Furu, K.; Haugen, T.B. miRNA-302s may act as oncogenes in human testicular germ cell tumours. Sci. Rep. 2019, 9, 9189. [Google Scholar] [CrossRef]
- Liu, L.; Lian, J.; Zhang, H.; Tian, H.; Liang, M.; Yin, M.; Sun, F. MicroRNA-302a sensitizes testicular embryonal carcinoma cells to cisplatin-induced cell death. J. Cell. Physiol. 2013, 228, 2294–2304. [Google Scholar] [CrossRef]
- Özata, D.M.; Li, X.; Lee, L.; Liu, J.; Warsito, D.; Hajeri, P.; Hultman, I.; Fotouhi, O.; Marklund, S.; Ährlund-Richter, L.; et al. Loss of miR-514a-3p regulation of PEG3 activates the NF-kappa B pathway in human testicular germ cell tumors. Cell Death Dis. 2017, 8, e2759. [Google Scholar] [CrossRef]
- Chen, B.F.; Gu, S.; Suen, Y.K.; Li, L.; Chan, W.Y. microRNA-199a-3p, DNMT3A, and aberrant DNA methylation in testicular cancer. Epigenetics 2014, 9, 119–128. [Google Scholar] [CrossRef]
- Zhao, G.; Yin, Y.; Zhao, B. miR-140-5p is negatively correlated with proliferation.; invasion.; and tumorigenesis in malignant melanoma by targeting SOX4 via the Wnt/β-catenin and NF-κB cascades. J. Cell. Physiol. 2020, 235, 2161–2170. [Google Scholar] [CrossRef]
- Bemis, L.T.; Chen, R.; Amato, C.M.; Classen, E.H.; Robinson, S.E.; Coffey, D.G.; Erickson, P.F.; Shellman, Y.G.; Robinson, W.A. MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer Res. 2008, 68, 1362–1368. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Dong, X.D.; Chen, X.; Yao, S.; Wang, L.; Wang, J.; Wang, C.; Hu, D.N.; Qu, J.; Tu, L. Role of microRNA-182 in posterior uveal melanoma: Regulation of tumor development through MITF, BCL2 and cyclin D2. PLoS ONE 2012, 7, e40967. [Google Scholar]
- Haflidadóttir, B.S.; Bergsteinsdóttir, K.; Praetorius, C.; Steingrímsson, E. miR-148 regulates Mitf in melanoma cells. PLoS ONE 2010, 5, e11574. [Google Scholar] [CrossRef]
- Margue, C.; Philippidou, D.; Reinsbach, S.E.; Schmitt, M.; Behrmann, I.; Kreis, S. New target genes of MITF-induced microRNA-211 contribute to melanoma cell invasion. PLoS ONE 2013, 8, e73473. [Google Scholar]
- Sun, X.; Li, J.; Sun, Y.; Zhang, Y.; Dong, L.; Shen, C.; Yang, L.; Yang, M.; Li, Y.; Shen, G.; et al. miR-7 reverses the resistance to BRAFi in melanoma by targeting EGFR/IGF-1R/CRAF and inhibiting the MAPK and PI3K/AKT signaling pathways. Oncotarget 2016, 7, 53558–53570. [Google Scholar] [CrossRef] [PubMed]
- Rambow, F.; Job, B.; Petit, V.; Gesbert, F.; Delmas, V.; Seberg, H.; Meurice, G.; Van Otterloo, E.; Dessen, P.; Robert, C.; et al. New Functional Signatures for Understanding Melanoma Biology from Tumor Cell Lineage-Specific Analysis. Cell Rep. 2015, 13, 840–853. [Google Scholar] [CrossRef]
- Arts, N.; Cané, S.; Hennequart, M.; Lamy, J.; Bommer, G.; Van den Eynde, B.; De Plaen, E. microRNA-155, induced by interleukin-1ß, Represses the expression of microphthalmia-associated transcription factor (MITF-M) in melanoma cells. PLoS ONE 2015, 10, e0122517. [Google Scholar] [CrossRef]
- Philippidou, D.; Schmitt, M.; Moser, D.; Margue, C.; Nazarov, P.V.; Muller, A.; Vallar, L.; Nashan, D.; Behrmann, I.; Kreis, S. Signatures of microRNAs and selected microRNA target genes in human melanoma. Cancer Res. 2010, 70, 4163–4173. [Google Scholar] [CrossRef]
- Xu, N.; Zhang, L.; Meisgen, F.; Harada, M.; Heilborn, J.; Homey, B.; Grandér, D.; Ståhle, M.; Sonkoly, E.; Pivarcsi, A. MicroRNA-125b down-regulates matrix metallopeptidase 13 and inhibits cutaneous squamous cell carcinoma cell proliferation, migration, and invasion. J. Biol. Chem. 2012, 287, 29899–29908. [Google Scholar] [CrossRef]
- Toll, A.; Salgado, R.; Espinet, B.; Díaz-Lagares, A.; Hernández-Ruiz, E.; Andrades, E.; Sandoval, J.; Esteller, M.; Pujol, R.M.; Hernández-Muñoz, I. MiR-204 silencing in intraepithelial to invasive cutaneous squamous cell carcinoma progression. Mol. Cancer 2016, 15, 53. [Google Scholar] [CrossRef]
- Neu, J.; Dziunycz, P.J.; Dzung, A.; Lefort, K.; Falke, M.; Denzler, R.; Freiberger, S.N.; Iotzova-Weiss, G.; Kuzmanov, A.; Levesque, M.P.; et al. miR-181a decelerates proliferation in cutaneous squamous cell carcinoma by targeting the proto-oncogene KRAS. PLoS ONE 2017, 12, e0185028. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Zhou, Y.; Chen, P.; Yang, M.; Xu, J. MicroRNA-142-5p induces cancer stem cell-like properties of cutaneous squamous cell carcinoma via inhibiting PTEN. J. Cell. Biochem. 2018, 119, 2179–2188. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.H.; Zhou, F.; Shi, C.; Xiang, T.; Zhou, C.K.; Wang, Q.Q.; Jiang, Y.S.; Gao, S.F. miRNA-221 promotes cutaneous squamous cell carcinoma progression by targeting PTEN. Cell. Mol. Biol. Lett. 2019, 24, 9. [Google Scholar] [CrossRef]
- Lohcharoenkal, W.; Harada, M.; Lovén, J.; Meisgen, F.; Landén, N.X.; Zhang, L.; Lapins, J.; Mahapatra, K.D.; Shi, H.; Nissinen, L.; et al. MicroRNA-203 Inversely Correlates with Differentiation Grade, Targets c-MYC, and Functions as a Tumor Suppressor in cSCC. J. Investig. Dermatol. 2016, 136, 2485–2494. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Li, W.; Zhao, T.; Tian, X.; Liu, Y.; Zhang, X. Role of miR-148a in cutaneous squamous cell carcinoma by repression of MAPK pathway. Arch. Biochem. Biophys. 2015, 583, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.H.; Zhou, J.D.; He, Q.Y.; Yin, Z.Q.; Cao, K.; Luo, C.Q. MiR-199a inhibits the ability of proliferation and migration by regulating CD44-Ezrin signaling in cutaneous squamous cell carcinoma cells. Int. J. Clin. Exp. Pathol. 2014, 7, 7131–7141. [Google Scholar] [PubMed]
- Yamane, K.; Jinnin, M.; Etoh, T.; Kobayashi, Y.; Shimozono, N.; Fukushima, S.; Masuguchi, S.; Maruo, K.; Inoue, Y.; Ishihara, T.; et al. Down-regulation of miR-124/-214 in cutaneous squamous cell carcinoma mediates abnormal cell proliferation via the induction of ERK. J. Mol. Med. 2013, 91, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Pallante, P.; Visone, R.; Ferracin, M.; Ferraro, A.; Berlingieri, M.T.; Troncone, G.; Chiappetta, G.; Liu, C.G.; Santoro, M.; Negrini, M.; et al. MicroRNA deregulation in human thyroid papillary carcinomas. Endocr. Relat. Cancer 2006, 13, 497–508. [Google Scholar] [CrossRef]
- He, H.; Jazdzewski, K.; Li, W.; Liyanarachchi, S.; Nagy, R.; Volinia, S.; Calin, G.A.; Liu, C.G.; Franssila, K.; Suster, S.; et al. The role of microRNA genes in papillary thyroid carcinoma. Proc. Natl. Acad. Sci. USA 2005, 102, 19075–19080. [Google Scholar] [CrossRef] [PubMed]
- Visone, R.; Russo, L.; Pallante, P.; De Martino, I.; Ferraro, A.; Leone, V.; Borbone, E.; Petrocca, F.; Alder, H.; Croce, C.M.; et al. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr. Relat. Cancer 2007, 14, 791–798. [Google Scholar] [CrossRef]
- Nikiforova, M.N.; Tseng, G.C.; Steward, D.; Diorio, D.; Nikiforov, Y.E. MicroRNA expression profiling of thyroid tumors: Biological significance and diagnostic utility. J. Clin. Endocrinol. Metab. 2008, 93, 1600–1608. [Google Scholar] [CrossRef]
- Liao, B.; Liu, S.; Liu, J.; Reddy, P.A.K.; Ying, Y.; Xie, Y.; Wang, J.; Zeng, X. Long Noncoding RNA CTC Inhibits Proliferation and Invasion by Targeting miR-146 to Regulate KIT in Papillary Thyroid Carcinoma. Sci. Rep. 2020, 10, 4616. [Google Scholar] [CrossRef] [PubMed]
- Czajka, A.A.; Wójcicka, A.; Kubiak, A.; Kotlarek, M.; Bakuła-Zalewska, E.; Koperski, Ł.; Wiechno, W.; Jażdżewski, K. Family of microRNA-146 Regulates RARβ in Papillary Thyroid Carcinoma. PLoS ONE 2016, 11, e0151968. [Google Scholar] [CrossRef] [PubMed]
- Braun, J.; Hoang-Vu, C.; Dralle, H.; Hüttelmaier, S. Downregulation of microRNAs directs the EMT and invasive potential of anaplastic thyroid carcinomas. Oncogene 2010, 29, 4237–4244. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, Z.B.; Ren, W.M.; Ye, X.G.; Zhang, Y.Y. The miR-200 family regulates the epithelial-mesenchymal transition induced by EGF/EGFR in anaplastic thyroid cancer cells. Int. J. Mol. Med. 2012, 30, 856–862. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Gao, Y.; Wang, G.; Dai, G.; Tong, L. MiR-145 inhibits the migration and invasion of papillary thyroid carcinoma cells through NF-κB pathway regulation. J. Cell. Biochem. 2020, 121, 3325–3332. [Google Scholar] [CrossRef]
- Hudson, J.; Duncavage, E.; Tamburrino, A.; Salerno, P.; Xi, L.; Raffeld, M.; Moley, J.; Chernock, R.D. Overexpression of miR-10a and miR-375 and downregulation of YAP1 in medullary thyroid carcinoma. Exp. Mol. Pathol. 2013, 95, 62–67. [Google Scholar] [CrossRef]
- Abraham, D.; Jackson, N.; Gundara, J.S.; Zhao, J.; Gill, A.J.; Delbridge, L.; Robinson, B.G.; Sidhu, S.B. MicroRNA profiling of sporadic and hereditary medullary thyroid cancer identifies predictors of nodal metastasis, prognosis, and potential therapeutic targets. Clin. Cancer Res. 2011, 17, 4772–4781. [Google Scholar] [CrossRef]
- Pennelli, G.; Galuppini, F.; Barollo, S.; Cavedon, E.; Bertazza, L.; Fassan, M.; Guzzardo, V.; Pelizzo, M.R.; Rugge, M.; Mian, C. The PDCD4/miR-21 pathway in medullary thyroid carcinoma. Hum. Pathol. 2015, 46, 50–57. [Google Scholar] [CrossRef]
- Gao, Y.; Luo, L.H.; Li, S.; Yang, C. miR-17 inhibitor suppressed osteosarcoma tumor growth and metastasis via increasing PTEN expression. Biochem. Biophys. Res. Commun. 2014, 444, 230–234. [Google Scholar] [CrossRef] [PubMed]
- McKinsey, E.L.; Parrish, J.K.; Irwin, A.E.; Niemeyer, B.F.; Kern, H.B.; Birks, D.K.; Jedlicka, P. A novel oncogenic mechanism in Ewing sarcoma involving IGF pathway targeting by EWS/Fli1-regulated microRNAs. Oncogene 2011, 30, 4910–4920. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, S.; Xu, Y.; Zhang, Y.; Guan, H.; Li, X.; Li, Y.; Wang, Y. Upregulation of miR-192 inhibits cell growth and invasion and induces cell apoptosis by targeting TCF7 in human osteosarcoma. Tumour Biol. 2016, 37, 15211–15220. [Google Scholar] [CrossRef] [PubMed]
- Gindin, Y.; Jiang, Y.; Francis, P.; Walker, R.L.; Abaan, O.D.; Zhu, Y.J.; Meltzer, P.S. miR-23a impairs bone differentiation in osteosarcoma via down-regulation of GJA1. Front. Genet. 2015, 6, 233. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Meng, C.; Shao, Z.; Wang, H.; Yang, S. MiR-23a functions as a tumor suppressor in osteosarcoma. Cell. Physiol. Biochem. 2014, 34, 1485–1496. [Google Scholar] [CrossRef] [PubMed]
- Llobat, L.; Gourbault, O. Role of MicroRNAs in Human Osteosarcoma: Future Perspectives. Biomedicines 2021, 9, 463. [Google Scholar] [CrossRef]
- Sharma, P.C.; Gupta, A. MicroRNAs: Potential biomarkers for diagnosis and prognosis of different cancers. Transl. Cancer Res. 2020, 9, 5798–5818. [Google Scholar] [CrossRef]
- Olive, V.; Li, Q.; He, L. mir-17-92: A polycistronic oncomir with pleiotropic functions. Immunol. Rev. 2013, 253, 158–166. [Google Scholar] [CrossRef]
- O’Donnell, K.A.; Wentzel, E.A.; Zeller, K.I.; Dang, C.V.; Mendell, J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005, 435, 839–843. [Google Scholar] [CrossRef]
- Korać, P.; Antica, M.; Matulić, M. MiR-7 in Cancer Development. Biomedicines 2021, 9, 325. [Google Scholar] [CrossRef]
- Masoudi, M.S.; Mehrabian, E.; Mirzaei, H. MiR-21: A key player in glioblastoma pathogenesis. J. Cell. Biochem. 2018, 119, 1285–1290. [Google Scholar] [CrossRef]
- Hamamoto, J.; Soejima, K.; Yoda, S.; Naoki, K.; Nakayama, S.; Satomi, R.; Terai, H.; Ikemura, S.; Sato, T.; Yasuda, H.; et al. Identification of microRNAs differentially expressed between lung squamous cell carcinoma and lung adenocarcinoma. Mol. Med. Rep. 2013, 8, 456–462. [Google Scholar] [CrossRef] [PubMed]
- Landi, M.T.; Zhao, Y.; Rotunno, M.; Koshiol, J.; Liu, H.; Bergen, A.W.; Rubagotti, M.; Goldstein, A.M.; Linnoila, I.; Marincola, F.M.; et al. MicroRNA expression differentiates histology and predicts survival of lung cancer. Clin. Cancer Res. 2010, 16, 430–441. [Google Scholar] [CrossRef] [PubMed]
- Lebanony, D.; Benjamin, H.; Gilad, S.; Ezagouri, M.; Dov, A.; Ashkenazi, K.; Gefen, N.; Izraeli, S.; Rechavi, G.; Pass, H.; et al. Diagnostic assay based on hsa-miR-205 expression distinguishes squamous from nonsquamous non-small-cell lung carcinoma. J. Clin. Oncol. 2009, 27, 2030–2037. [Google Scholar] [CrossRef] [PubMed]
- Bishop, J.A.; Benjamin, H.; Cholakh, H.; Chajut, A.; Clark, D.P.; Westra, W.H. Accurate classification of non-small cell lung carcinoma using a novel microRNA-based approach. Clin. Cancer Res. 2010, 16, 610–619. [Google Scholar] [CrossRef]
- El Founini, Y.; Chaoui, I.; Dehbi, H.; El Mzibri, M.; Abounader, R.; Guessous, F. MicroRNAs: Key Regulators in Lung Cancer. Microrna 2021, 10, 109–122. [Google Scholar] [CrossRef]
- Fang, R.; Xiao, T.; Fang, Z.; Sun, Y.; Li, F.; Gao, Y.; Feng, Y.; Li, L.; Wang, Y.; Liu, X.; et al. MicroRNA-143 (miR-143) regulates cancer glycolysis via targeting hexokinase 2 gene. J. Biol. Chem. 2012, 287, 23227–23235. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Mao, W.; Zheng, S. MicroRNA-183 regulates Ezrin expression in lung cancer cells. FEBS Lett. 2008, 582, 3663–3668. [Google Scholar] [CrossRef]
- Xie, X.; Tan, W.; Chen, B.; Huang, X.; Peng, C.; Yan, S.; Yang, L.; Song, C.; Wang, J.; Zheng, W.; et al. Preoperative prediction nomogram based on primary tumor miRNAs signature and clinical-related features for axillary lymph node metastasis in early-stage invasive breast cancer. Int. J. Cancer 2018, 142, 1901–1910. [Google Scholar] [CrossRef]
- Quesne, J.L.; Jones, J.; Warren, J.; Dawson, S.J.; Ali, H.R.; Bardwell, H.; Blows, F.; Pharoah, P.; Caldas, C. Biological and prognostic associations of miR-205 and let-7b in breast cancer revealed by in situ hybridization analysis of micro-RNA expression in arrays of archival tumour tissue. J. Pathol. 2012, 227, 306–314. [Google Scholar] [CrossRef]
- Kalinkova, L.; Nikolaieva, N.; Smolkova, B.; Ciernikova, S.; Kajo, K.; Bella, V.; Kajabova, V.H.; Kosnacova, H.; Minarik, G.; Fridrichova, I. miR-205-5p Downregulation and ZEB1 Upregulation Characterize the Disseminated Tumor Cells in Patients with Invasive Ductal Breast Cancer. Int. J. Mol. Sci. 2021, 23, 103. [Google Scholar] [CrossRef]
- Abolghasemi, M.; Tehrani, S.S.; Yousefi, T.; Karimian, A.; Mahmoodpoor, A.; Ghamari, A.; Jadidi-Niaragh, F.; Yousefi, M.; Kafil, H.S.; Bastami, M.; et al. MicroRNAs in breast cancer: Roles, functions, and mechanism of actions. J. Cell. Physiol. 2020, 235, 5008–5029. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Shirvani-Farsani, Z.; Branicki, W.; Taheri, M. MicroRNA Signature in Renal Cell Carcinoma. Front. Oncol. 2020, 10, 596359. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, L.; Qu, J.; Guan, N.N.; Li, J.Q. Predicting miRNA–disease association based on inductive matrix completion. Bioinformatics 2018, 34, 4256–4265. [Google Scholar] [CrossRef] [PubMed]
- Khashkhashi Moghadam, S.; Bakhshinejad, B.; Khalafizadeh, A.; Mahmud Hussen, B.; Babashah, S. Non-coding RNA-associated competitive endogenous RNA regulatory networks: Novel diagnostic and therapeutic opportunities for hepatocellular carcinoma. J. Cell. Mol. Med. 2022, 26, 287–305. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, Y.; Bai, R.; Yang, K.; Tian, Z. MiR-186 inhibited aerobic glycolysis in gastric cancer via HIF-1α regulation. Oncogenesis 2016, 5, e224. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Lin, X.; Qiu, X.; Yang, L.; Wang, R. The Molecular Roles and Clinical Implications of Non-Coding RNAs in Gastric Cancer. Front. Cell Dev. Biol. 2021, 9, 802745. [Google Scholar] [CrossRef]
- Bañuelos-Villegas, E.G.; Pérez-y Pérez, M.F.; Alvarez-Salas, L.M. Cervical Cancer. Papillomavirus. and miRNA Dysfunction. Front. Mol. Biosci. 2021, 8, 758337. [Google Scholar] [CrossRef]
- Gillis, A.J.; Stoop, H.J.; Hersmus, R.; Oosterhuis, J.W.; Sun, Y.; Chen, C.; Guenther, S.; Sherlock, J.; Veltman, I.; Baeten, J.; et al. High-throughput microRNAome analysis in human germ cell tumours. J. Pathol. 2007, 213, 319–328. [Google Scholar] [CrossRef]
- Das, M.K.; Haugen, Ø.P.; Haugen, T.B. Diverse Roles and Targets of miRNA in the Pathogenesis of Testicular Germ Cell Tumour. Cancers 2022, 14, 1190. [Google Scholar] [CrossRef]
- Varrone, F.; Caputo, E. The miRNAs Role in Melanoma and in Its Resistance to Therapy. Int. J. Mol. Sci. 2020, 21, 878. [Google Scholar] [CrossRef]
- Simmons, J.L.; Pierce, C.J.; Al-Ejeh, F.; Boyle, G.M. MITF and BRN2 contribute to metastatic growth after dissemination of melanoma. Sci. Rep. 2017, 7, 10909. [Google Scholar] [CrossRef] [PubMed]
- Fuziwara, C.S.; Kimura, E.T. MicroRNA Deregulation in Anaplastic Thyroid Cancer Biology. Int. J. Endocrinol. 2014, 2014, 743450. [Google Scholar] [CrossRef] [PubMed]
- Bernier, A.; Sagan, S. The Diverse Roles of microRNAs at the Host–Virus Interface. Viruses 2018, 10, 440. [Google Scholar] [CrossRef]
- Girardi, E.; López, P.; Pfeffer, S. On the Importance of Host Micrornas during Viral Infection. Front. Genet. 2018, 9, 439. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Kumar, A.; Ingle, H.; Kumar, H. The Interplay between Viral-Derived Mirnas and Host Immunity during Infection. Front. Immunol. 2019, 10, 3079. [Google Scholar] [CrossRef]
- Ahmad, I.; Valverde, A.; Siddiqui, H.; Schaller, S.; Naqvi, A. Viral Micrornas: Interfering the Interferon Signaling. Curr. Pharm. Des. 2020, 26, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Boss, I.; Renne, R. Viral Mirnas and Immune Evasion. Biochim. Biophys. Acta 2011, 1809, 708–714. [Google Scholar] [CrossRef]
- Lagos, D.; Pollara, G.; Henderson, S.; Gratrix, F.; Fabani, M.; Milne, R.; Gotch, F.; Boshoff, C. Mir-132 Regulates Antiviral Innate Immunity through Suppression of the P300 Transcriptional Co-Activator. Nat. Cell. Biol. 2010, 12, 513–519. [Google Scholar] [CrossRef]
- O’Connor, C.; Vanicek, J.; Murphy, E. Host Microrna Regulation of Human Cytomegalovirus Immediate Early Protein Translation Promotes Viral Latency. J. Virol. 2014, 88, 5524–5532. [Google Scholar] [CrossRef]
- Mansouri, S.; Pan, Q.; Blencowe, B.; Claycomb, J.; Frappier, L. Epstein-Barr Virus EBNA1 Protein Regulates Viral Latency Through Effects on Let-7 Microrna and Dicer. J. Virol. 2014, 88, 11166–11177. [Google Scholar] [CrossRef]
- Imig, J.; Motsch, N.; Zhu, J.; Barth, S.; Okoniewski, M.; Reineke, T.; Tinguely, M.; Faggioni, A.; Trivedi, P.; Meister, G.; et al. Microrna Profiling in Epstein–Barr Virus-Associated B-Cell Lymphoma. Nucleic Acids Res. 2010, 39, 1880–1893. [Google Scholar] [CrossRef]
- Iizasa, H.; Kim, H.; Kartika, A.; Kanehiro, Y.; Yoshiyama, H. Role of Viral and Host Micrornas in Immune Regulation of Epstein-Barr Virus-Associated Diseases. Front. Immunol. 2020, 11, 367. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Majerciak, V.; Uldrick, T.; Wang, X.; Kruhlak, M.; Yarchoan, R.; Zheng, Z. Kaposi’s Sarcoma-Associated Herpesviral IL-6 and Human IL-6 Open Reading Frames Contain Mirna Binding Sites and Are Subject to Cellular Mirna Regulation. J. Pathol. 2011, 225, 378–389. [Google Scholar] [CrossRef]
- Qin, Z.; Peruzzi, F.; Reiss, K.; Dai, L. Role of Host Micrornas in Kaposi’S Sarcoma-Associated Herpesvirus Pathogenesis. Viruses 2014, 6, 4571–4580. [Google Scholar] [CrossRef]
- Duan, F.; Liao, J.; Huang, Q.; Nie, Y.; Wu, K. HSV-1 Mir-H6 Inhibits HSV-1 Replication and IL-6 Expression in Human Corneal Epithelial Cells In Vitro. Clin. Dev. Immunol. 2012, 2012, 192791. [Google Scholar] [CrossRef]
- Kawamura, Y.; Bosch-Marce, M.; Tang, S.; Patel, A.; Krause, P. Herpes Simplex Virus 2 Latency-Associated Transcript (LAT) Region Mutations Do Not Identify A Role For LAT-Associated Micrornas in Viral Reactivation in Guinea Pig Genital Models. J. Virol. 2018, 92, e00642-18. [Google Scholar] [CrossRef] [Green Version]
- Naqvi, A. Immunomodulatory Roles of Human Herpesvirus-Encoded Microrna in Host-Virus Interaction. Rev. Med. Virol. 2019, 30, e2081. [Google Scholar] [CrossRef]
- Zeng, G.; Wang, Z.; Huang, Y.; Abedin, Z.; Liu, Y.; Randhawa, P. Cellular and Viral Mirna Expression in Polyomavirus BK Infection. Transpl. Inf. Dis. 2019, 21, e13159. [Google Scholar] [CrossRef]
- Sadri Nahand, J.; Moghoofei, M.; Salmaninejad, A.; Bahmanpour, Z.; Karimzadeh, M.; Nasiri, M.; Mirzaei, H.; Pourhanifeh, M.; Bokharaei-Salim, F.; Mirzaei, H.; et al. Pathogenic Role of Exosomes and Micrornas in HPV-Mediated Inflammation and Cervical Cancer: A Review. Int. J. Cancer 2019, 146, 305–320. [Google Scholar] [CrossRef]
- Piedade, D.; Azevedo-Pereira, J. Micrornas as Important Players in Host–Adenovirus Interactions. Front. Microbiol. 2017, 8, 1324. [Google Scholar] [CrossRef]
- Loureiro, D.; Tout, I.; Narguet, S.; Benazzouz, S.; Mansouri, A.; Asselah, T. Mirnas as Potential Biomarkers for Viral Hepatitis B and C. Viruses 2020, 12, 1440. [Google Scholar] [CrossRef]
- Martinez-Espinoza, I.; Banos-Lara, M.D.R.; Guerrero-Plata, A. The Importance of Mirna Identification during Respiratory Viral Infections. J. Cell. Immunol. 2021, 3, 207–214. [Google Scholar]
- Haque, M.; Murale, D.; Lee, J. Role of Microrna and Oxidative Stress in Influenza A Virus Pathogenesis. Int. J. Mol. Sci. 2020, 21, 8962. [Google Scholar] [CrossRef]
- Nanbo, A.; Furuyama, W.; Lin, Z. RNA Virus-Encoded Mirnas: Current Insights and Future Challenges. Front. Microbiol. 2021, 12, 679210. [Google Scholar] [CrossRef]
- Grey, F.; Hook, L.; Nelson, J. The Functions of Herpesvirus-Encoded Micrornas. Med. Microbiol. Immunol. 2007, 197, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Imperiale, M. Biology of Polyomavirus Mirna. Front. Microbiol. 2021, 12, 662829. [Google Scholar] [CrossRef]
- Piedade, D.; Azevedo-Pereira, J. Micrornas, HIV and HCV: A Complex Relation towards Pathology. Rev. Med. Virol. 2016, 26, 197–215. [Google Scholar] [CrossRef]
- Siddika, T.; Heinemann, I.U. Bringing MicroRNAs to Light: Methods for MicroRNA Quantification and Visualization in Live Cells. Front. Bioeng. Biotechnol. 2021, 8, 619583. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.Q.; Zhao, R.C.; Morris, K.V. Profiling microRNA expression with microarrays. Trends Biotechnol. 2008, 26, 70–76. [Google Scholar] [CrossRef]
- Chang-Gong, L.; Calin, G.A.; Volinia, S.; Croce, C.M. MicroRNA expression profiling using microarrays. Nat. Protoc. 2008, 3, 563–578. [Google Scholar]
- Beuvink, I.; Kolb, F.A.; Budach, W.; Garnier, A.; Lange, J.; Natt, F.; Dengler, U.; Hall, J.; Filipowicz, W.; Weiler, J. A novel microarray approach reveals new tissue-specific signatures of known and predicted mammalian microRNAs. Nucleic Acids Res. 2007, 35, e52. [Google Scholar] [CrossRef] [PubMed]
- Duan, D.; Zheng, K.; Shen, Y.; Cao, R.; Jiang, L.; Lu, Z.; Yan, X.; Li, J. Label-free high-throughput microRNA expression profiling from total RNA. Nucleic Acids Res. 2011, 39, e154. [Google Scholar] [CrossRef]
- Fan, J.B. Next-Generation MicroRNA Expression Profiling Technology: Methods and Protocols; Humana Press: New York, NY, USA, 2012. [Google Scholar]
- Ge, Q.; Tian, F.; Zhou, Y.; Zhu, Y.; Lu, J.; Baib, Y.; Lu, Z. A universal linker-RT PCR based quantitative method for the detection of circulating miRNAs. Anal Methods 2014, 6, 9101–9107. [Google Scholar] [CrossRef]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef]
- Raymond, C.K.; Roberts, B.S.; Garrett-Engele, P.; Lim, L.P.; Johnson, J.M. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA 2005, 11, 1737–1744. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Yang, J.; Liu, G.; Jin, W.; Liu, Z.; Zhao, S.; Ya, M. Quantification of mature MicroRNAs using pincer probes and real-time PCR amplification. PLoS ONE 2015, 10, e0120160. [Google Scholar] [CrossRef] [PubMed]
- Androvic, P.; Valihrach, L.; Elling, J.; Sjoback, R.; Kubist, M. Two-tailed RT-qPCR: A novel method for highly accurate miRNA quantification. Nucleic Acids Res. 2017, 45, e144. [Google Scholar] [CrossRef]
- Hanna, J.A.; Wimberly, H.; Kumar, S.; Slack, F.; Agarwal, S.; Rimm, D.L. Quantitative analysis of microRNAs in tissue microarrays by in situ hybridization. Biotechniques 2012, 52, 235–245. [Google Scholar] [CrossRef]
- Hilario, E. End labeling procedures: An overview. Mol. Biotechnol. 2004, 28, 77–80. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Chen, W.; Adamidi, C.; Maaskola, J.; Einspanier, R.; Knespel, S.; Rajewsky, N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008, 26, 407–415. [Google Scholar] [CrossRef]
- Martinez-Gutierrez, A.D.; Catalan, O.M.; Vázquez-Romo, R.; Porras Reyes, F.I.; Alvarado-Miranda, A.; Lara Medina, F.; Bargallo-rocha, J.E.; Orozco Moreno, L.T.; Cantú De León, D.; Herrera, L.A.; et al. miRNA profile obtained by next-generation sequencing in metastatic breast cancer patients is able to predict the response to systemic treatments. Int. J. Mol. Med. 2019, 44, 1267–1280. [Google Scholar] [CrossRef] [PubMed]
- Alexiou, A.; Zisis, D.; Kavakiotis, I.; Miliotis, M.; Koussounadis, A.; Karagkouni, D.; Hatzigeorgiou, A.G. DIANA-mAP: Analyzing miRNA from Raw NGS Data to Quantification. Genes 2021, 12, 46. [Google Scholar] [CrossRef]
- Motameny, S.; Wolters, S.; Nürnberg, P.; Schumacher, B. Next Generation Sequencing of miRNAs—Strategies, Resources and Methods. Genes 2010, 1, 70–84. [Google Scholar] [CrossRef]
- Dave, V.P.; Ngo, T.A.; Pernestig, A.K.; Tilevik, D.; Kant, K.; Nguyen, T.; Wolff, A.; Bang, D.D. MicroRNA amplification and detection technologies: Opportunities and challenges for point of care diagnostics. Lab. Investig. 2019, 99, 452–469. [Google Scholar] [CrossRef]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, D.; Tyryshkin, K.; Renwick, N. microRNA-guided diagnostics in clinical samples. Best Pract. Res. Clin. Endocrinol. Metab. 2016, 30, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Sempere, L.F.; Azmi, A.S.; Moore, A. microRNA-based diagnostic and therapeutic applications in cancer medicine. Wiley Interdiscip. Rev. RNA 2021, 12, e1662. [Google Scholar] [CrossRef]
- Schulte, C.; Zeller, T. microRNA-based diagnostics and therapy in cardiovascular disease-Summing up the facts. Cardiovasc. Diagn. Ther. 2015, 5, 17–36. [Google Scholar]
- Kitano, Y.; Aoki, K.; Ohka, F.; Yamazaki, S.; Motomura, K.; Tanahashi, K.; Hirano, M.; Naganawa, T.; Iida, M.; Shiraki, Y.; et al. Urinary MicroRNA-Based Diagnostic Model for Central Nervous System Tumors Using Nanowire Scaffolds. ACS Appl. Mater. Interfaces 2021, 13, 17316–17329. [Google Scholar] [CrossRef]
- Enokida, H.; Yoshino, H.; Matsushita, R.; Nakagawa, M. The role of microRNAs in bladder cancer. Investig. Clin. Urol. 2016, 57, S60–S76. [Google Scholar] [CrossRef]
- Bonneau, E.; Neveu, B.; Kostantin, E.; Tsongalis, G.J.; De Guire, V. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. EJIFCC 2019, 30, 114–127. [Google Scholar]
- ThyGeNext-ThyraMIR. Available online: https://thygenext-thyramir.com (accessed on 25 March 2022).
- López, R.I.; Gómez, V.A.; Gómez, R.J. Current use of molecular profiling for indeterminate thyroid nodules. Cir. Esp. 2018, 96, 395–400. [Google Scholar] [CrossRef]
- Lithwick-Yanai, G.; Dromi, N.; Shtabsky, A.; Morgenstern, S.; Strenov, Y.; Feinmesser, M.; Kravtsov, V.; Leon, M.E.; Hajdúch, M.; Ali, S.Z.; et al. Multicentre validation of a microRNA-based assay for diagnosing indeterminate thyroid nodules utilising fine needle aspirate smears. J. Clin. Pathol. 2017, 70, 500–507. [Google Scholar] [CrossRef]
- Walts, A.E.; Sacks, W.L.; Wu, H.H.; Randolph, M.L.; Bose, S. A retrospective analysis of the performance of the RosettaGX® Reveal™ thyroid miRNA and the Afirma Gene Expression Classifiers in a cohort of cytologically indeterminate thyroid nodules. Diagn. Cytopathol. 2018, 46, 901–907. [Google Scholar] [CrossRef]
- Walter, E.; Dellago, H.; Grillari, J.; Dimai, H.P.; Hackl, M. Cost-utility analysis of fracture risk assessment using microRNAs compared with standard tools and no monitoring in the Austrian female population. Bone 2018, 108, 44–54. [Google Scholar] [CrossRef]
- Harnessing the Predictive Power of Blood-Borne miRNAs. Available online: https://www.hummingbird-diagnostics.com (accessed on 2 March 2022).
- Faruq, O.; Vecchione, A. microRNA: Diagnostic Perspective. Front. Med. 2015, 2, 51. [Google Scholar] [CrossRef]
- Tomeva, E.; Switzeny, O.J.; Heitzinger, C.; Hippe, B.; Haslberger, A.G. Comprehensive Approach to Distinguish Patients with Solid Tumors from Healthy Controls by Combining Androgen Receptor Mutation p.H875Y with Cell-Free DNA Methylation and Circulating miRNAs. Cancers 2022, 14, 462. [Google Scholar] [CrossRef]
- Singh, A.; Dwivedi, A. Circulating miRNA and cell-free DNA as a potential diagnostic tool in early detection of biliary tract cancer: A meta-analysis. Biomarkers 2022, 27, 399–406. [Google Scholar] [CrossRef]
- Yang, Z.; LaRiviere, M.J.; Ko, J.; Till, J.E.; Christensen, T.; Yee, S.S.; Black, T.A.; Tien, K.; Lin, A.; Shen, H.; et al. A Multianalyte Panel Consisting of Extracellular Vesicle miRNAs and mRNAs, cfDNA, and CA19-9 Shows Utility for Diagnosis and Staging of Pancreatic Ductal Adenocarcinoma. Clin. Cancer Res. 2020, 26, 3248–3258. [Google Scholar] [CrossRef]
- Pezzuto, F.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. The Role of Circulating Free DNA and MicroRNA in Non-Invasive Diagnosis of HBV- and HCV-Related Hepatocellular Carcinoma. Int. J. Mol. Sci. 2018, 19, 1007. [Google Scholar] [CrossRef] [Green Version]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Wu, Q.; Yu, J.; Rao, Y.; Kou, Z.; Fang, G.; Shi, X.; Liu, W.; Han, H. A Systematic Way to Infer the Regulation Relations of miRNAs on Target Genes and Critical miRNAs in Cancers. Front. Genet. 2020, 11, 278. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Qi, H.; Teng, J.; Su, B.; Chen, H.; Wang, C.; Xia, Q. Identification of serum miRNAs by nano-quantum dots microarray as diagnostic biomarkers for early detection of non-small cell lung cancer. Tumour Biol. 2016, 37, 7777–7784. [Google Scholar] [CrossRef]
- Macerola, E.; Poma, A.M.; Proietti, A.; Rago, T.; Romani, R.; Vignali, P.; Ugolini, C.; Torregrossa, L.; Basolo, A.; Santini, F.; et al. Down-regulation of miR-7-5p and miR-548ar-5p predicts malignancy in indeterminate thyroid nodules negative for BRAF and RAS mutations. Endocrine 2022, 76, 677–686. [Google Scholar] [CrossRef]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef]
- Shen, J.; Todd, N.W.; Zhang, H.; Yu, L.; Lingxiao, X.; Mei, Y.; Guarnera, M.; Liao, J.; Chou, A.; Lu, C.L.; et al. Plasma microRNAs as potential biomarkers for non-small-cell lung cancer. Lab. Investig. 2011, 91, 579–587. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.; Olkhov-Mitsel, E.; Xie, H.; Yao, C.Q.; Zhao, F.; Jahangiri, S.; Cuizon, C.; Scarcello, S.; Jeyapala, R.; Watson, J.D.; et al. Temporal Stability and Prognostic Biomarker Potential of the Prostate Cancer Urine miRNA Transcriptome. J. Natl. Cancer Inst. 2020, 112, 247–255. [Google Scholar] [CrossRef]
- Aftab, M.; Poojary, S.S.; Seshan, V.; Kumar, S.; Agarwal, P.; Tandon, S.; Zutshi, V.; Das, B.C. Urine miRNA signature as a potential non-invasive diagnostic and prognostic biomarker in cervical cancer. Sci. Rep. 2021, 11, 10323. [Google Scholar] [CrossRef]
- von Siebenthal, M.; Besic, M.; Gheinani, A.H.; Akshay, A.; Lizun-Platoni, S.; Kunz, N.; Burkhard, F.C.; Monastyrskaya, K. Urinary miRNA profiles discriminate between obstruction-induced bladder dysfunction and healthy controls. Sci. Rep. 2021, 11, 10204. [Google Scholar] [CrossRef]
- Bertoli, G.; Cava, C.; Castiglioni, I. MicroRNAs: New Biomarkers for Diagnosis, Prognosis, Therapy Prediction and Therapeutic Tools for Breast Cancer. Theranostics 2015, 5, 1122–1143. [Google Scholar] [CrossRef] [Green Version]
- Shea, A.; Harish, V.; Afzal, Z.; Chijioke, J.; Kedir, H.; Dusmatova, S.; Roy, A.; Ramalinga, M.; Harris, B.; Blancato, J.; et al. MicroRNAs in glioblastoma multiforme pathogenesis and therapeutics. Cancer Med. 2016, 5, 917–946. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhou, Y.; Richards, A.M. Effective tools for RNA-derived therapeutics: siRNA interference or miRNA mimicry. Theranostics 2021, 11, 8771–8796. [Google Scholar] [CrossRef] [PubMed]
- NIH US National Library of Medicine. Available online: https://www.clinicaltrials.gov/ (accessed on 15 March 2022).
- Ramamoorthy, A.; Skaar, T.C. In silico identification of microRNAs predicted to regulate the drug metabolizing cytochrome P450 genes. Drug. Metab. Lett. 2011, 5, 126–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matulić, M.; Gršković, P.; Petrović, A.; Begić, V.; Harabajsa, S.; Korać, P. miRNA in Molecular Diagnostics. Bioengineering 2022, 9, 459. https://doi.org/10.3390/bioengineering9090459
Matulić M, Gršković P, Petrović A, Begić V, Harabajsa S, Korać P. miRNA in Molecular Diagnostics. Bioengineering. 2022; 9(9):459. https://doi.org/10.3390/bioengineering9090459
Chicago/Turabian StyleMatulić, Maja, Paula Gršković, Andreja Petrović, Valerija Begić, Suzana Harabajsa, and Petra Korać. 2022. "miRNA in Molecular Diagnostics" Bioengineering 9, no. 9: 459. https://doi.org/10.3390/bioengineering9090459
APA StyleMatulić, M., Gršković, P., Petrović, A., Begić, V., Harabajsa, S., & Korać, P. (2022). miRNA in Molecular Diagnostics. Bioengineering, 9(9), 459. https://doi.org/10.3390/bioengineering9090459






