A Systematic Evaluation of Ensemble Learning Methods for Fine-Grained Semantic Segmentation of Tuberculosis-Consistent Lesions in Chest Radiographs
Abstract
:1. Introduction
1.1. Related Literature
1.2. Contributions of the Study
- This study proposes to use fine-grained annotations of TB-consistent lesions to train and evaluate the performance of variants of U-Net-based segmentation models.
- The gains achieved through constructing an ensemble of the trained models were evaluated to demonstrate further improvement in the robustness and performance of the segmentation algorithms.
2. Materials and Methods
2.1. Datasets
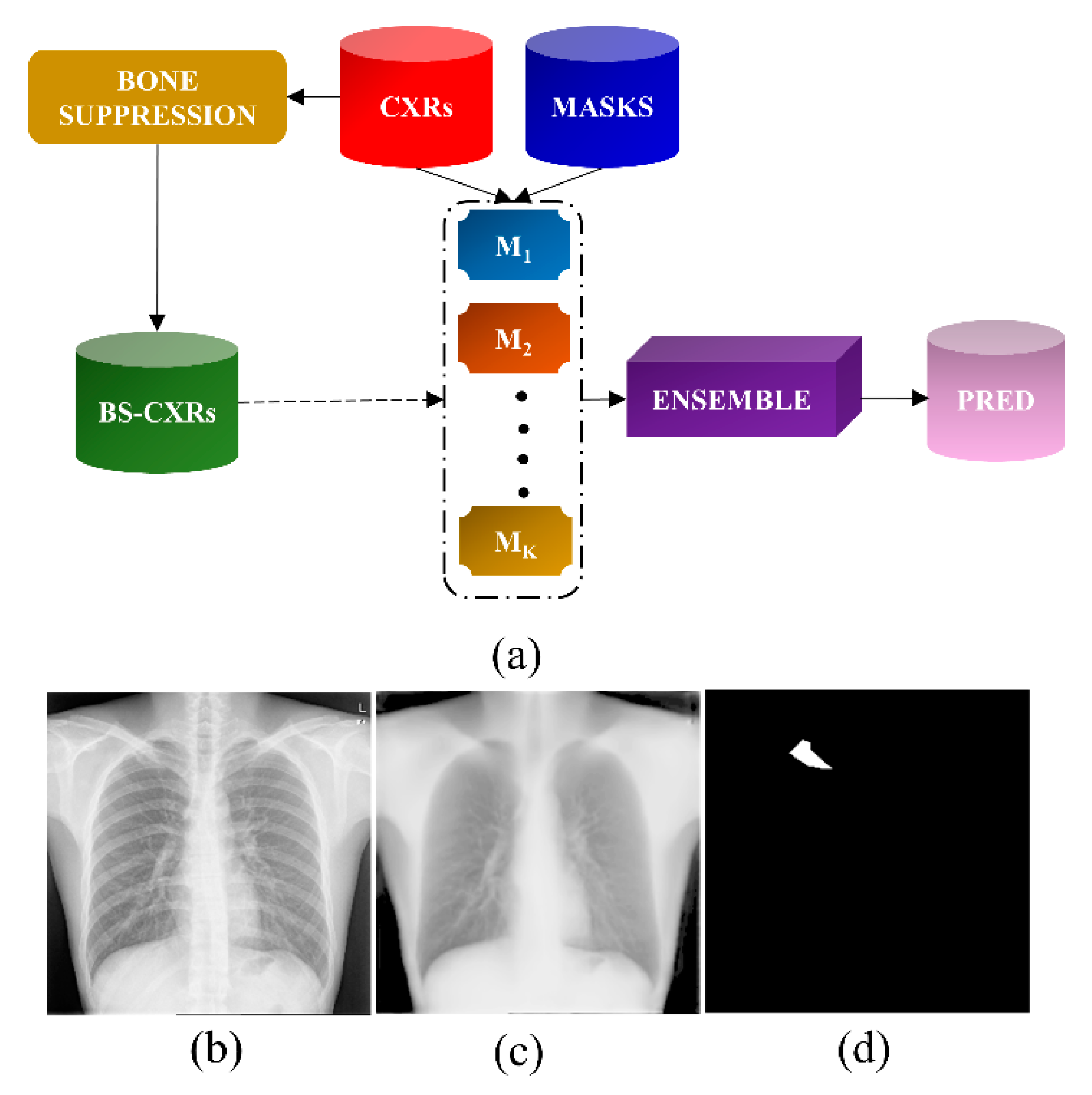
2.2. Model Architecture
2.2.1. Bone Suppression
2.2.2. Segmentation of TB-Consistent Lesions
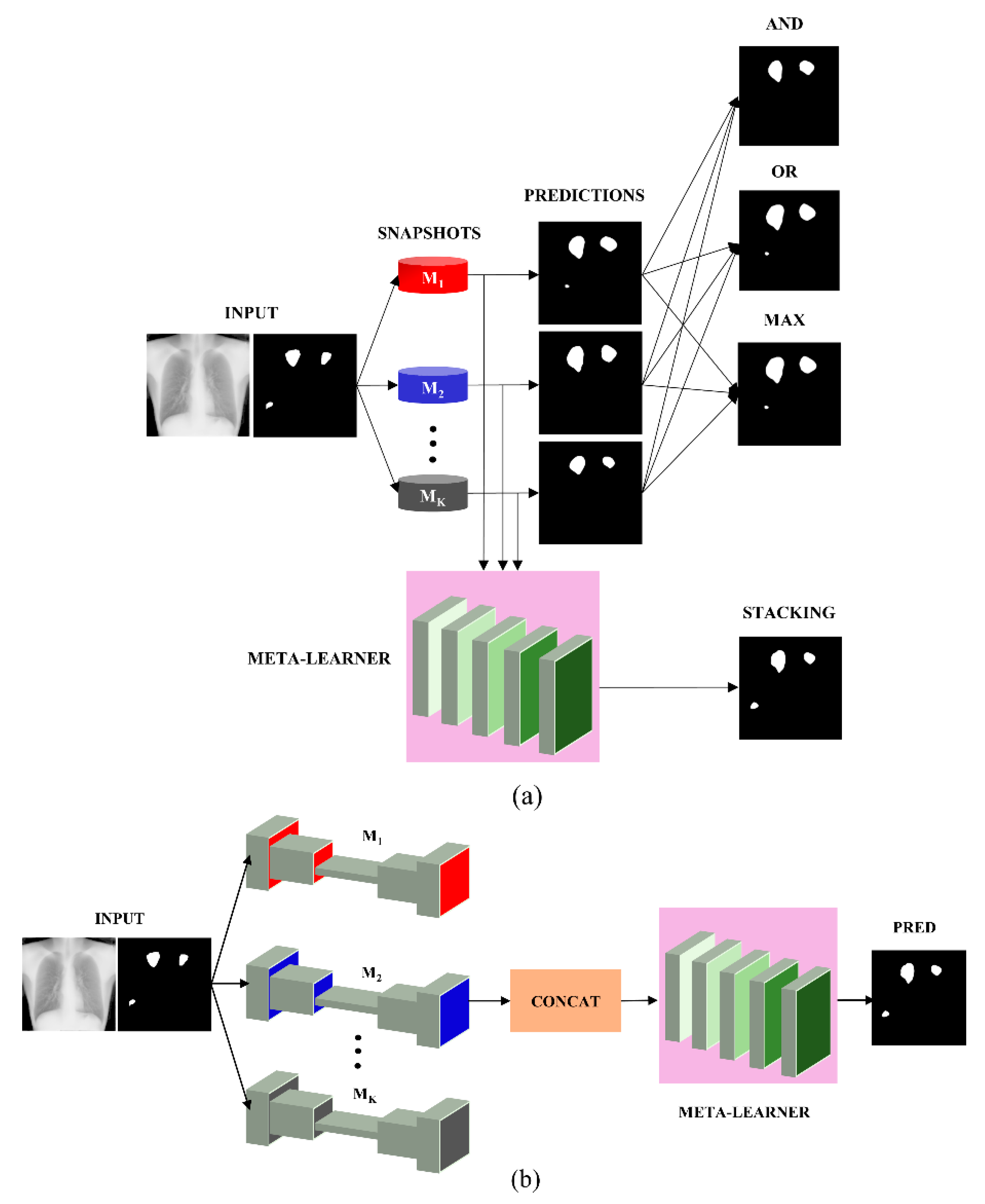
2.2.3. Ensemble Learning
2.2.4. Loss Functions and Evaluation Metrics
2.2.5. Statistical Analysis
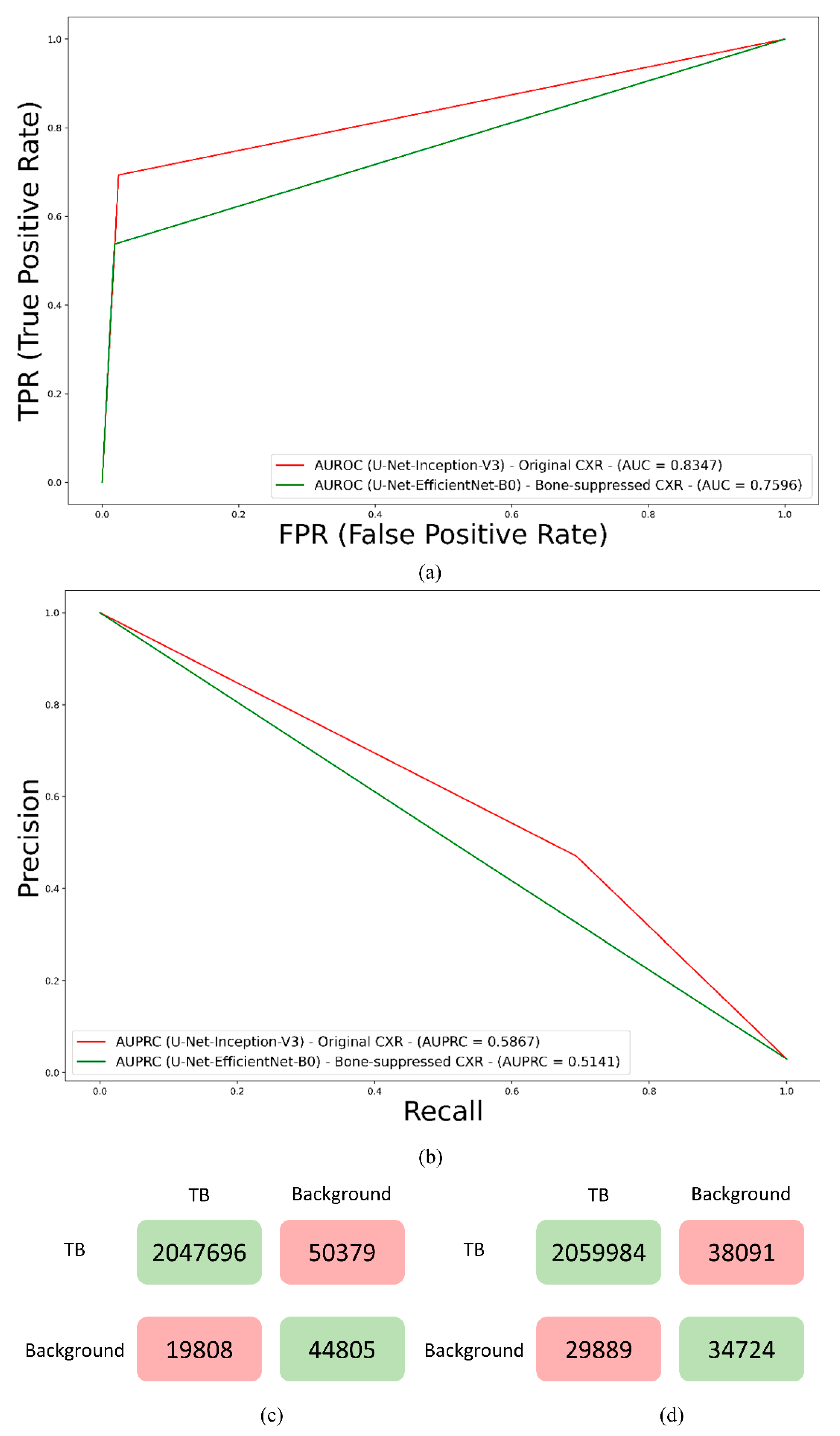
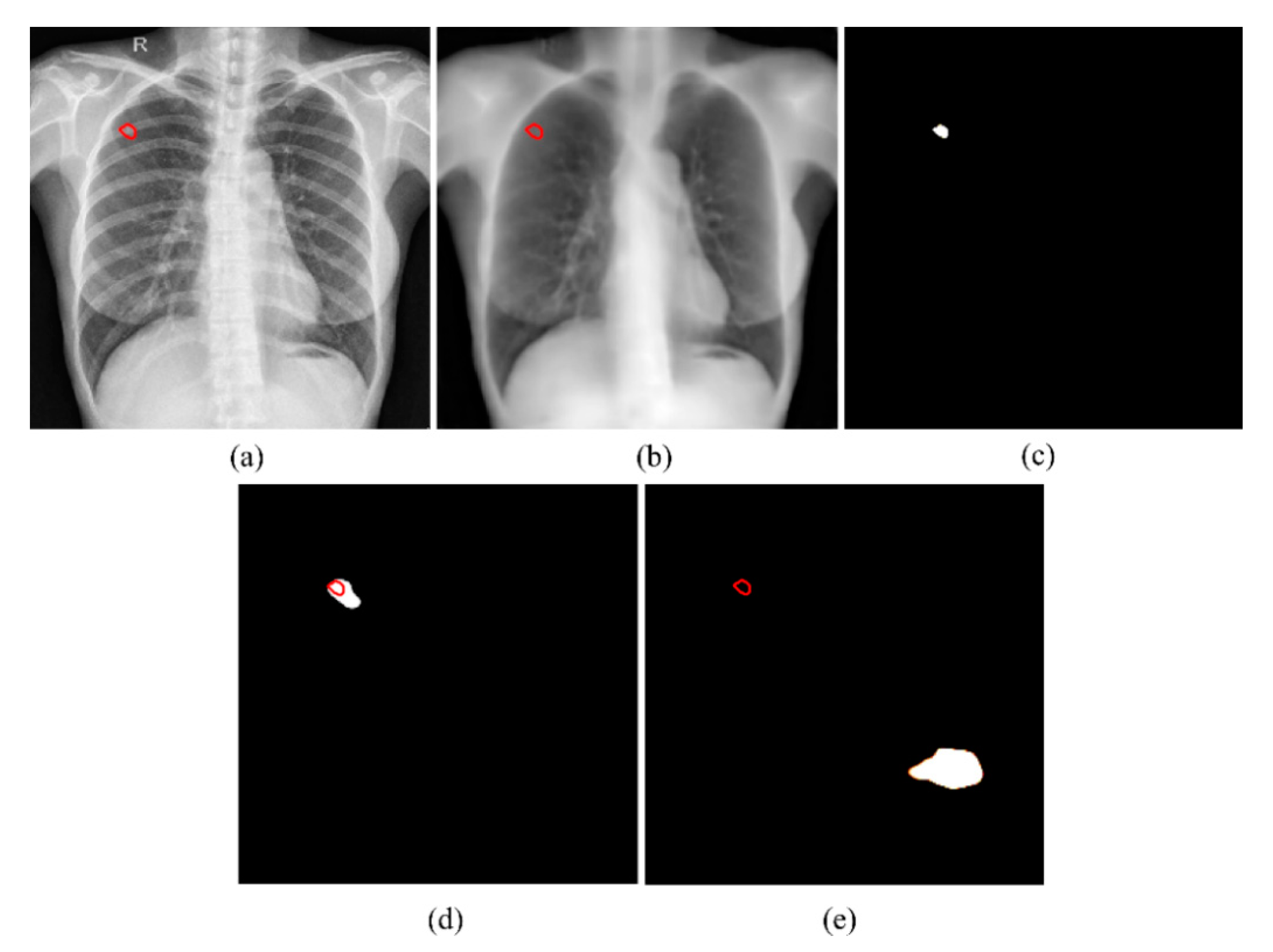
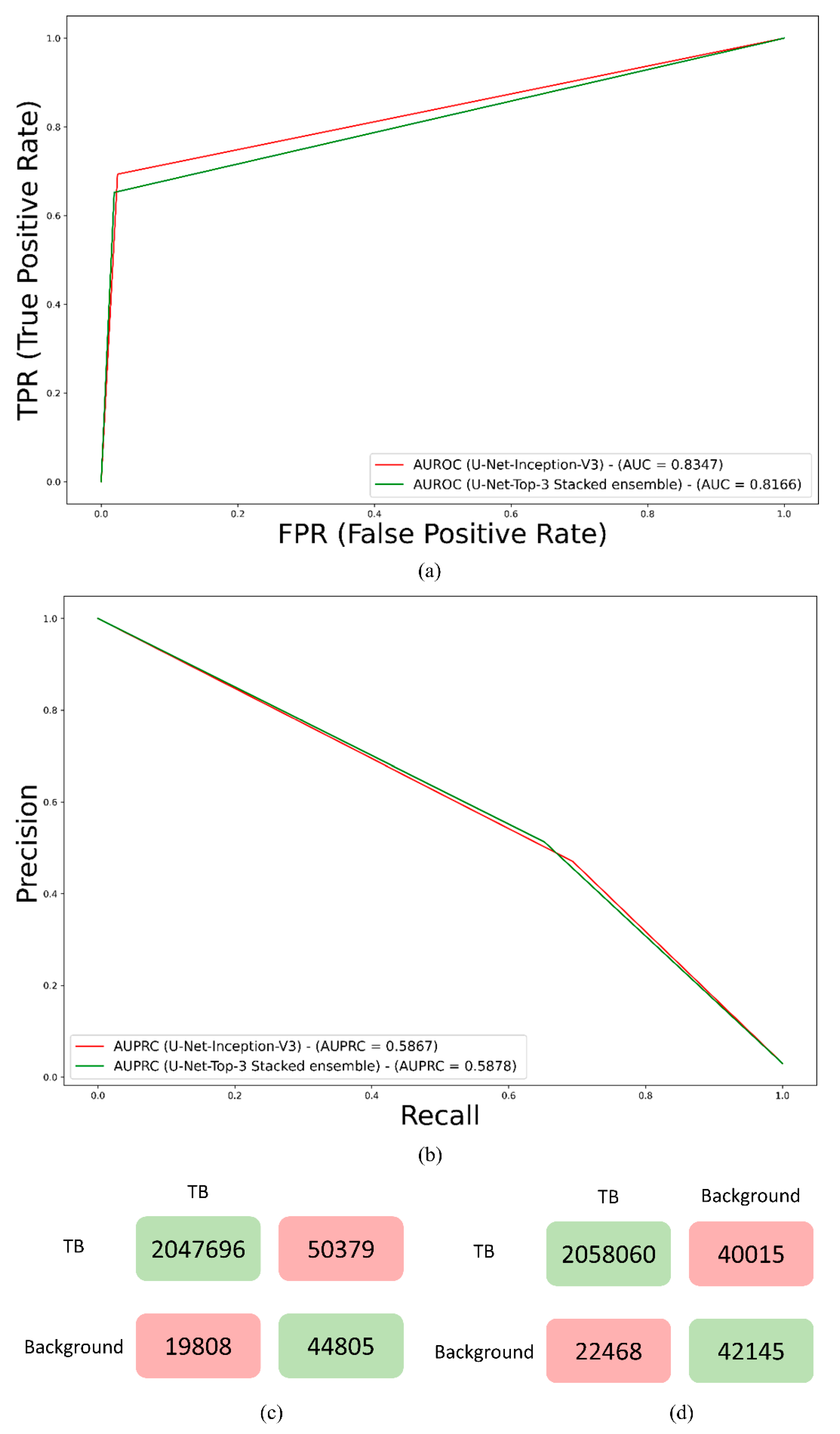
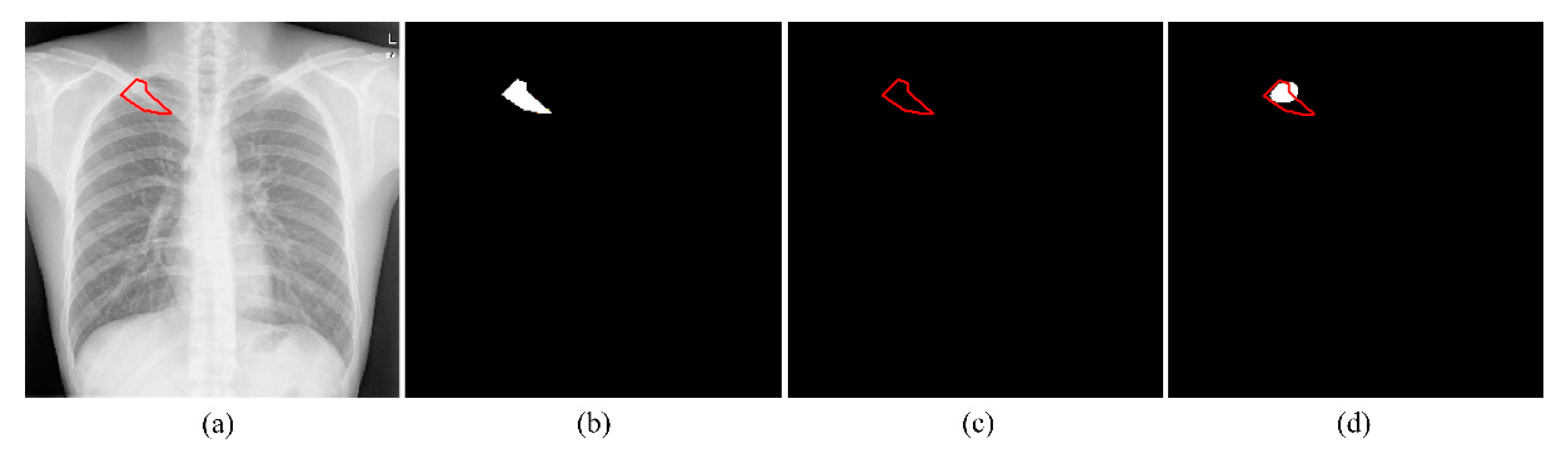
3. Results and Discussion
4. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. World Health Organization Global Tuberculosis Report; WHO: Geneve, Switzerland, 2021.
- Mettler, F.A.; Huda, W.; Yoshizumi, T.T.; Mahesh, M. Effective Doses in Radiology and Diagnostic Nuclear Medicine: A Catalog. Radiology 2008, 248, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, S.; Karargyris, A.; Candemir, S.; Siegelman, J.; Folio, L.; Antani, S.; Thoma, G. Automatic Screening for Tuberculosis in Chest Radiographs: A Survey. Quant. Imaging Med. Surg. 2013, 3, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Zamzmi, G.; Rajaraman, S.; Antani, S. UMS-Rep: Unified Modality-Specific Representation for Efficient Medical Image Analysis. Inform. Med. Unlocked 2021, 24, 100571. [Google Scholar] [CrossRef]
- Minaee, S.; Boykov, Y.; Porikli, F.; Plaza, A.; Kehtarnavaz, N.; Terzopoulos, D. Image Segmentation Using Deep Learning: A Survey. IEEE Trans. Pattern Anal. Mach. Intell. 2022, 44, 3523–3542. [Google Scholar] [CrossRef]
- Tang, P.; Yang, P.; Nie, D.; Wu, X.; Zhou, J.; Wang, Y. Unified Medical Image Segmentation by Learning from Uncertainty in an End-to-End Manner. Knowl. -Based Syst. 2022, 241, 108215. [Google Scholar] [CrossRef]
- Rajaraman, S.; Folio, L.R.; Dimperio, J.; Alderson, P.O.; Antani, S.K. Improved Semantic Segmentation of Tuberculosis—Consistent Findings in Chest X-rays Using Augmented Training of Modality-Specific u-Net Models with Weak Localizations. Diagnostics 2021, 11, 616. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, Y.H.; Ban, Y.; Wang, H.; Cheng, M.M. Rethinking Computer-Aided Tuberculosis Diagnosis. In Proceedings of the Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020. [Google Scholar]
- LeCun, Y.; Bengio, Y. Convolutional Networks for Images, Speech, and Time Series. Handb. Brain Theory Neural Netw. 1995, 3361, 255–258. [Google Scholar] [CrossRef]
- Smolyakov, V. Ensemble Learning to Improve Machine Learning Results. Available online: https://www.kdnuggets.com/2017/09/ensemble-learning-improve-machine-learning-results.html (accessed on 2 March 2022).
- Narayanan, B.N.; De Silva, M.S.; Hardie, R.C.; Ali, R. Ensemble Method of Lung Segmentation in Chest Radiographs. In Proceedings of the NAECON 2021—IEEE National Aerospace and Electronics Conference, Dayton, OH, USA, 16–19 August 2021; pp. 382–385. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), Munich, Germany, 5–9 October 2015. [Google Scholar]
- Candemir, S.; Jaeger, S.; Antani, S.; Bagci, U.; Folio, L.R.; Xu, Z.; Thoma, G. Atlas-Based Rib-Bone Detection in Chest X-rays. Comput. Med. Imaging Graph. 2016, 51, 32–39. [Google Scholar] [CrossRef] [Green Version]
- Jaeger, S.; Candemir, S.; Antani, S.; Wang, Y.-X.J.; Lu, P.-X.; Thoma, G. Two Public Chest X-ray Datasets for Computer-Aided Screening of Pulmonary Diseases. Quant. Imaging Med. Surg. 2014, 4, 475–477. [Google Scholar] [CrossRef]
- Ali, R.; Hardie, R.C.; Ragb, H.K. Ensemble Lung Segmentation System Using Deep Neural Networks. In Proceedings of the 2020 IEEE Applied Imagery Pattern Recognition Workshop (AIPR), Washington, DC, USA, 13–15 October 2020; pp. 1–5. [Google Scholar] [CrossRef]
- Abedalla, A.; Abdullah, M.; Al-Ayyoub, M.; Benkhelifa, E. Chest X-ray Pneumothorax Segmentation Using U-Net with EfficientNet and ResNet Architectures. PeerJ Comput. Sci. 2021, 7, 1–36. [Google Scholar] [CrossRef]
- Rajaraman, S.; Cohen, G.; Spear, L.; Folio, L.; Antani, S. DeBoNet: A Deep Bone Suppression Model Ensemble to Improve Disease Detection in Chest Radiographs. PLoS ONE 2022, 17, 1–22. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; Volume 7, pp. 770–778. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the Proceedings —30th IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Tan, M.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the 36th International Conference on Machine Learning (ICML), Long Beach, CA, USA, 10–15 June 2019. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-Excitation Networks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar] [CrossRef] [Green Version]
- Bloice, M.D.; Roth, P.M.; Holzinger, A. Biomedical Image Augmentation Using Augmentor. Bioinformatics 2019, 35, 4522–4524. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Liao, Q.; Ma, L.; Jin, X. Gated Feature Pyramid Network for Object Detection; Springer: Berlin/Heidelberg, Germany, 2018; Volume 11259, pp. 199–208. [Google Scholar] [CrossRef]
- Yeung, M.; Yang, G.; Sala, E.; Schönlieb, C.-B.; Rundo, L. Incorporating Boundary Uncertainty into Loss Functions for Biomedical Image Segmentation. arXiv 2021, arXiv:2111.00533v1. [Google Scholar]
- Abraham, N.; Khan, N.M. A Novel Focal Tversky Loss Function with Improved Attention U-Net for Lesion Segmentation. In Proceedings of the International Symposium on Biomedical Imaging, Venezia, Italy, 8–11 April 2019. [Google Scholar]
- Altman, D.G.; Bland, J.M. Statistics Notes: How to Obtain the P Value from a Confidence Interval. BMJ 2011, 343, d2304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bae, K.; Oh, D.Y.; Yun, I.D.; Jeon, K.N. Bone Suppression on Chest Radiographs for Pulmonary Nodule Detection: Comparison between a Generative Adversarial Network and Dual-Energy Subtraction. Korean J. Radiol. 2022, 23, 139–149. [Google Scholar] [CrossRef]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Zamzmi, G.; Rajaraman, S.; Sachdev, V.; Antani, S. Trilateral Attention Network for Real-Time Cardiac Region Segmentation. IEEE Access 2021, 9, 118205–118214. [Google Scholar] [CrossRef]
- Saqib, M.; Anwar, A.; Anwar, S.; Petersson, L.; Sharma, N.; Blumenstein, M. COVID-19 Detection from Radiographs: Is Deep Learning Able to Handle the Crisis? Signals 2022, 3, 296–312. [Google Scholar] [CrossRef]
- Naushad, R.; Kaur, T.; Ghaderpour, E. Deep Transfer Learning for Land Use and Land Cover Classification: A Comparative Study. Sensors 2021, 21, 8083. [Google Scholar] [CrossRef]
- Gao, Q.; Gadekallu, T.R. Design of telemedicine information query system based on wireless sensor network. EAI Endorsed Trans. Perv. Health. Tech. 2022, 8, e1. Available online: https://publications.eai.eu/index.php/phat/article/view/674 (accessed on 15 August 2022). [CrossRef]
- Vyas, A.H.; Mehta, M.A.; Kotecha, K.; Pandya, S.; Alazab, M.; Gadekallu, T.R. Tear film breakup time-based dry eye disease detection using convolutional neural network. Neural Comput. Applic 2022, 1–19. [Google Scholar] [CrossRef]
| Dataset | Train | Validation | Test |
|---|---|---|---|
| Shenzhen TB CXR | 2231 | 66 | 33 |
| Models | IOU | Dice |
|---|---|---|
| ResNet-34 (O) | 0.3599 | 0.5293 (0.3589, 0.6997) |
| ResNet-34 (BS) | 0.3280 | 0.4640 (0.2938, 0.6342) |
| Inception-V3 (O) | 0.3896 | 0.5608 (0.3914, 0.7302) |
| Inception-V3 (BS) | 0.2525 | 0.4032 (0.2358, 0.5706) |
| DenseNet-121 (O) | 0.2996 | 0.4611 (0.2910, 0.6312) |
| DenseNet-121 (BS) | 0.2892 | 0.4486 (0.2789, 0.6183) |
| EfficientNet-B0 (O) | 0.3453 | 0.5134 (0.3428, 0.6840) |
| EfficientNet-B0 (BS) | 0.3381 | 0.5053 (0.3347, 0.6759) |
| SE-ResNext-50 (O) | 0.3201 | 0.4850 (0.3144, 0.6556) |
| SE-ResNext-50 (BS) | 0.2962 | 0.4570 (0.2870, 0.6270) |
| Models | IOU | Dice |
|---|---|---|
| Inception-V3 (O) | 0.3896 | 0.5608 (0.3914, 0.7302) |
| Top-3 ensemble | ||
| Stacking | 0.4028 | 0.5743 (0.4055, 0.7431) |
| Bitwise-AND | 0.3829 | 0.5538 (0.3841, 0.7235) |
| Bitwise-OR | 0.3558 | 0.5249 (0.3545, 0.6953) |
| Bitwise-MAX | 0.3343 | 0.5011 (0.3305, 0.6717) |
| Top-4 ensemble | ||
| Stacking | 0.3962 | 0.5675 (0.3984, 0.7366) |
| Bitwise-AND | 0.3534 | 0.5222 (0.3517, 0.6927) |
| Bitwise-OR | 0.3088 | 0.4718 (0.3014, 0.6422) |
| Bitwise-MAX | 0.2971 | 0.4581 (0.2881, 0.6281) |
| Top-5 ensemble | ||
| Stacking | 0.3974 | 0.5687 (0.3997, 0.7377) |
| Bitwise-AND | 0.3534 | 0.5222 (0.3517, 0.6927) |
| Bitwise-OR | 0.3088 | 0.4718 (0.3014, 0.6422) |
| Bitwise-MAX | 0.2744 | 0.4306 (0.2616, 0.5996) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajaraman, S.; Yang, F.; Zamzmi, G.; Xue, Z.; Antani, S.K. A Systematic Evaluation of Ensemble Learning Methods for Fine-Grained Semantic Segmentation of Tuberculosis-Consistent Lesions in Chest Radiographs. Bioengineering 2022, 9, 413. https://doi.org/10.3390/bioengineering9090413
Rajaraman S, Yang F, Zamzmi G, Xue Z, Antani SK. A Systematic Evaluation of Ensemble Learning Methods for Fine-Grained Semantic Segmentation of Tuberculosis-Consistent Lesions in Chest Radiographs. Bioengineering. 2022; 9(9):413. https://doi.org/10.3390/bioengineering9090413
Chicago/Turabian StyleRajaraman, Sivaramakrishnan, Feng Yang, Ghada Zamzmi, Zhiyun Xue, and Sameer K. Antani. 2022. "A Systematic Evaluation of Ensemble Learning Methods for Fine-Grained Semantic Segmentation of Tuberculosis-Consistent Lesions in Chest Radiographs" Bioengineering 9, no. 9: 413. https://doi.org/10.3390/bioengineering9090413
APA StyleRajaraman, S., Yang, F., Zamzmi, G., Xue, Z., & Antani, S. K. (2022). A Systematic Evaluation of Ensemble Learning Methods for Fine-Grained Semantic Segmentation of Tuberculosis-Consistent Lesions in Chest Radiographs. Bioengineering, 9(9), 413. https://doi.org/10.3390/bioengineering9090413









