Progress in Integrative Biomaterial Systems to Approach Three-Dimensional Cell Mechanotransduction
Abstract
1. Introduction
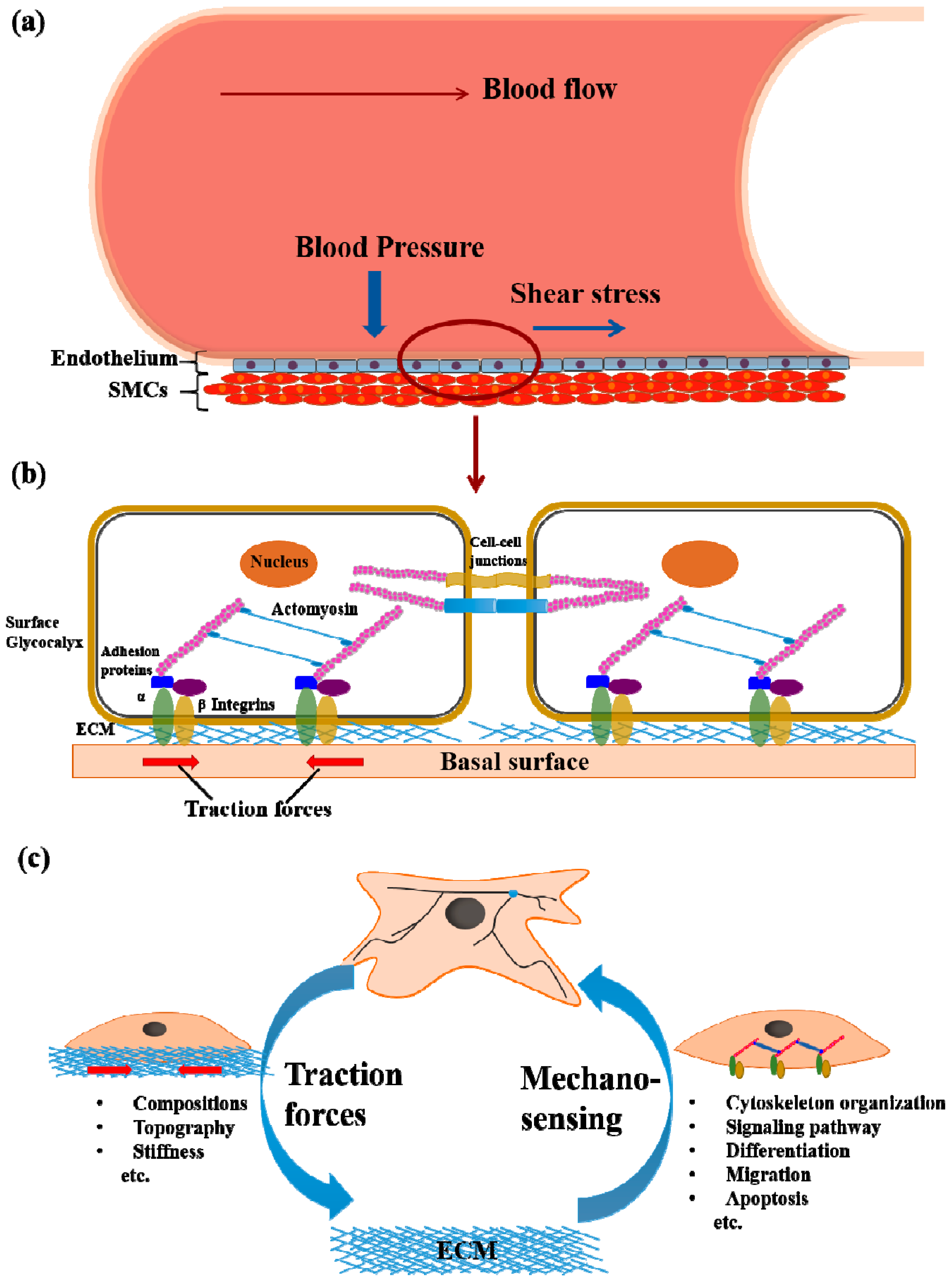
2. Cell Mechanotransduction
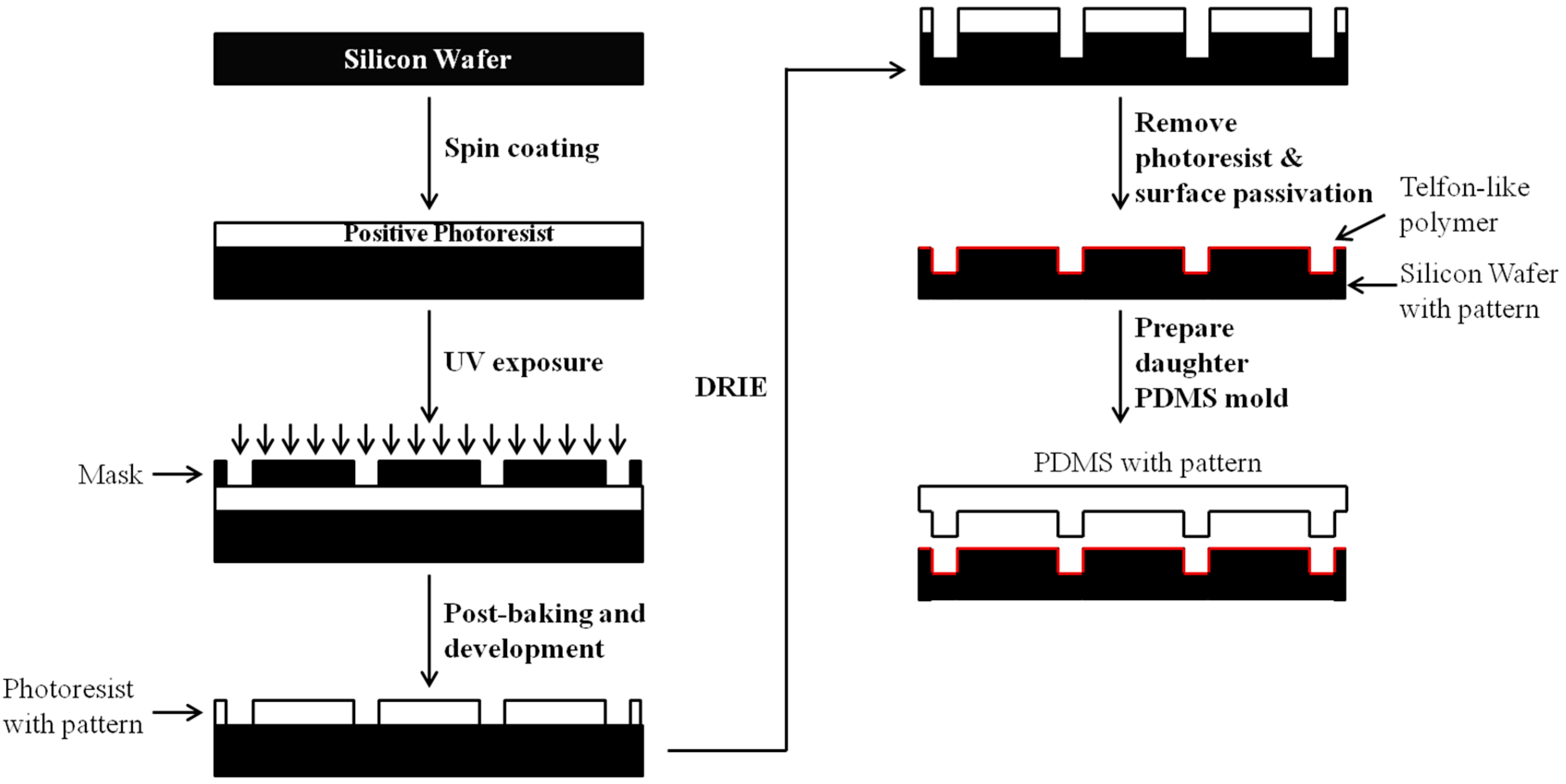
3. 3D Fabrications of Polymeric Biomaterials
4. Recent Progress in Cellular Biomechanics
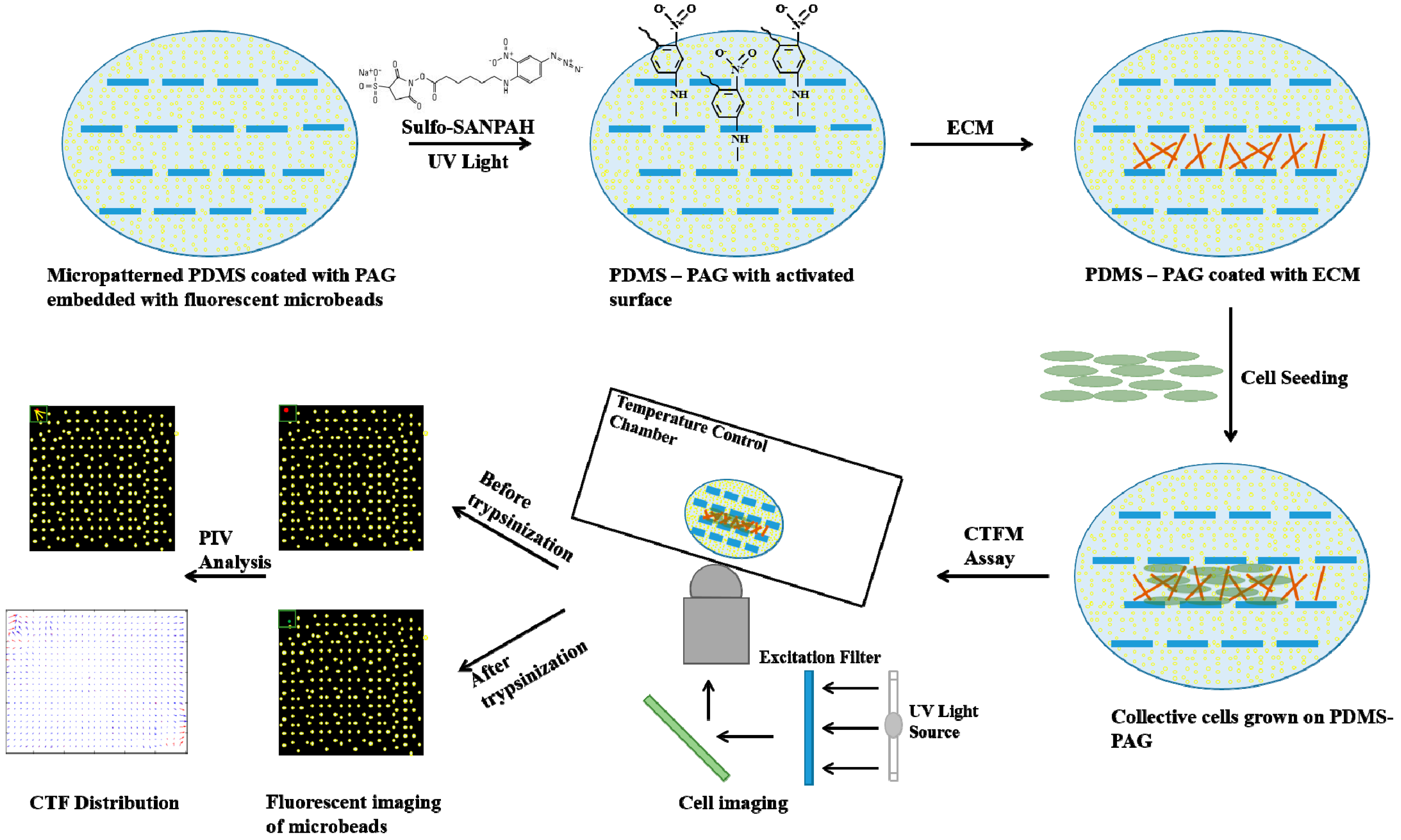
5. Cell Traction Force Measurement
6. Conclusions
Acknowledgments
Conflicts of Interest
References
- Chen, C.S.; Mrksich, M.; Huang, S.; Whitesides, G.M.; Ingber, D.E. Geometric control of cell life and death. Science 1997, 276, 1425–1428. [Google Scholar] [CrossRef] [PubMed]
- Dike, L.E.; Chen, C.S.; Mrksich, M.; Tien, J.; Whitesides, G.M.; Ingber, D.E. Geometric control of switching between growth, apoptosis, and differentiation during angiogenesis using micropatterned substrates. Vitro Cell Dev. Biol. 1999, 35, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Hogrebe, N.J.; Reinhardt, J.W.; Gooch, K.J. Biomaterial microarchitecture: A potent regulator of individual cell behavior and multicellular organization. J. Biomed. Mater. Res. A 2016, 105, 640–661. [Google Scholar] [CrossRef] [PubMed]
- Singhvi, R.; Kumar, A.; Lopez, G.P.; Stephanopoulos, G.N.; Wang, D.I.C.; Whitesides, G.M.; Ingber, D.E. Engineering Cell-Shape and Function. Science 1994, 264, 696–698. [Google Scholar] [CrossRef] [PubMed]
- Delamarche, E.; Bernard, A.; Schmid, H.; Bietsch, A.; Michel, B.; Biebuyck, H. Microfluidic networks for chemical patterning of substrate: Design and application to bioassays. J. Am. Chem. Soc. 1998, 120, 500–508. [Google Scholar] [CrossRef]
- Michel, R.; Lussi, J.W.; Csucs, G.; Reviakine, I.; Danuser, G.; Ketterer, B.; Hubbell, J.A.; Textor, M.; Spencer, N.D. Selective molecular assembly patterning: A new approach to micro- and nanochemical patterning of surfaces for biological applications. Langmuir 2002, 18, 3281–3287. [Google Scholar] [CrossRef]
- Falconnet, D.; Koenig, A.; Assi, T.; Textor, M. A combined photolithographic and molecular-assembly approach to produce functional micropatterns for applications in the biosciences. Adv. Funct. Mater. 2004, 14, 749–756. [Google Scholar] [CrossRef]
- Goessl, A.; Bowen-Pope, D.F.; Hoffman, A.S. Control of shape and size of vascular smooth muscle cells in vitro by plasma lithography. J. Biomed. Mater. Res. 2001, 57, 15–24. [Google Scholar] [CrossRef]
- Falconnet, D.; Csucs, G.; Grandin, H.M.; Textor, M. Surface engineering approaches to micropattern surfaces for cell-based assays. Biomaterials 2006, 27, 3044–3063. [Google Scholar] [CrossRef] [PubMed]
- Desai, T.A. Micro- and nanoscale structures for tissue engineering constructs. Med. Eng. Phys. 2000, 22, 595–606. [Google Scholar] [CrossRef]
- Sachlos, E.; Czernuszka, J.T. Making tissue engineering scaffolds work. Review: The application of solid freeform fabrication technology to the production of tissue engineering scaffolds. Eur. Cells Mater. 2003, 5, 29–39. [Google Scholar] [CrossRef]
- Shin, H. Fabrication methods of an engineered microenvironment for analysis for cell-biomaterial interactions. Biomaterials 2007, 28, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Lammerding, J.; Hsiao, J.; Schulze, P.C.; Kozlov, S.; Stewart, C.L.; Lee, R.T. Abnormal nuclear shape and impaired mechanotransduction in emerin-deficient cells. J. Cell Biol. 2005, 170, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.H.; Laschinger, C.; Arora, P.; Szaszi, K.; Kapus, A.; McCulloch, C.A. Force activates smooth muscle alpha-actin promoter activity through the Rho signaling pathway. J. Cell Sci. 2007, 120, 1801–1809. [Google Scholar] [CrossRef] [PubMed]
- Choquet, D.; Felsenfeld, D.P.; Sheetz, M.P. Extracellular matrix rigidity causes strengthening of integrin-cytoskeleton linkages. Cell 1997, 88, 39–48. [Google Scholar] [CrossRef]
- Hahn, C.; Schwartz, M.A. Mechanotransduction in vascular physiology and atherogenesis. Nat. Rev. Mol. Cell Biol. 2009, 10, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Plopper, G.; Ingber, D.E. Rapid induction and isolation of focal adhesion complexes. Biochem. Biophys. Res. Commun. 1993, 193, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, S.; Akiyama, S.K.; Yamada, K.M. Synergistic roles for receptor occupancy and aggregation in integrin transmembrane function. Science 1995, 267, 883–885. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.P.; Tolic-Norrelykke, I.M.; Fabry, B.; Fredberg, J.J. Traction fields, moments, and strain energy that cells exert on their surroundings. Am. J. Phys. 2002, 282, C595–C605. [Google Scholar] [CrossRef] [PubMed]
- Chicurel, M.E.; Chen, C.S.; Ingber, D.E. Cellular control lies in the balance of forces. Curr. Opin. Cell Biol. 1998, 10, 232–239. [Google Scholar] [CrossRef]
- Fouchard, J.; Bimbard, C.; Bufi, N.; Durand-Smet, P.; Proag, A.; Richert, A.; Cardoso, O.; Asnacios, A. Three-dimensional cell body shape dictates the onset of traction force generation and growth of focal adhesions. Proc. Natl. Acad. Sci. USA 2014, 111, 13075–13080. [Google Scholar] [CrossRef] [PubMed]
- Pirone, D.M.; Liu, W.F.; Ruiz, S.A.; Gao, L.; Raghavan, S.; Lemmon, C.A.; Romer, L.H.; Chen, C.S. An inhibitory role for FAK in regulating proliferation: A link between limited adhesion and RhoA-ROCK signaling. J. Cell Biol. 2006, 174, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, N.; Kato, T.; Fujita, A.; Ishizaki, T.; Narumiya, S. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat. Cell Biol. 1999, 1, 136–143. [Google Scholar] [PubMed]
- Sun, Y.; Chen, C.S.; Fu, J. Forcing Stem cells to behave: A biophysical perspective of the cellular microenvironment. Ann. Rev. Biophys. 2012, 41, 519–542. [Google Scholar] [CrossRef] [PubMed]
- Elosegui-Artola, A.; Oria, R.; Chen, Y.; Kosmalska, A.; Pérez-González, C.; Castro, N.; Zhu, C.; Trepat, X.; Roca-Cusachs, P. Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat. Cell Biol. 2016, 18, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Schulte, C.; Rodighiero, S.; Cappelluti, M.A.; Puricelli, L.; Maffioli, E.; Borghi, F.; Negri, A.; Sogne, E.; Galluzzi, M.; Piazzoni, C.; et al. Conversion of nanoscale topographical information of cluster-assembled zirconia surfaces into mechanotransductive events promotes neuronal differentiation. J. Nanobiotechnol. 2016, 14, 18. [Google Scholar] [CrossRef] [PubMed]
- Cukierman, E.; Pankov, R.; Yamada, K.M. Cell interactions with three-dimensional matrices. Curr. Opin. Cell Biol. 2002, 14, 633–639. [Google Scholar] [CrossRef]
- Li, S.; Lao, J.; Chen, B.P.; Li, Y.S.; Zhao, Y.; Chu, J.; Chen, K.D.; Tsou, T.C.; Peck, K.; Chien, S. Genomic analysis of smooth muscle cells in 3-dimensional collagen matrix. Faseb. J. 2003, 17, 97–99. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.; McCullough, C.M.; Stegemann, J.P. The role of ERK signaling in protein hydrogel remodeling by vascular smooth muscle cells. Biomaterials 2007, 28, 3824–3833. [Google Scholar] [CrossRef] [PubMed]
- Provenzano, P.P.; Inman, D.R.; Eliceiri, K.W.; Keely, P.J. Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK-ERK linkage. Oncogene 2009, 28, 4326–4343. [Google Scholar] [CrossRef] [PubMed]
- Petrie, R.J.; Gavara, N.; Chadwick, R.S.; Yamada, K.M. Nonpolarized signaling reveals two distinct modes of 3D cell migration. J. Cell Biol. 2012, 197, 439–455. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Y.; Stevens, K.R.; Schwartz, R.E.; Alejandro, B.S.; Huang, J.H.; Bhatia, S.N. Micropatterned Cell-Cell Interactions Enable Functional Encapsulation of Primary Hepatocytes in Hydrogel Microtissues. Tissue Eng. Part A 2014, 20, 2200–2212. [Google Scholar] [CrossRef] [PubMed]
- Mertz, A.F.; Banerjee, S.; Che, Y.L.; German, G.K.; Xu, Y.; Hyland, C.; Marchetti, M.C.; Horsley, V.; Dufresne, E.R. Scaling of Traction Forces with the Size of Cohesive Cell Colonies. Phys. Rev. Lett. 2012, 108, 198101. [Google Scholar] [CrossRef] [PubMed]
- Trepat, X.; Wasserman, M.R.; Angelini, T.E.; Millet, E.; Weitz, D.A.; Butler, J.P.; Fredberg, J.J. Physical forces during collective cell migration. Nat. Phys. 2009, 5, 426–430. [Google Scholar] [CrossRef]
- Serra-Picamal, X.; Conte, V.; Sunyer, R.; Muñoz, J.J.; Trepat, X. Mapping forces and kinematics during collective cell migration. Methods Cell Biol. 2015, 125, 309–330. [Google Scholar] [PubMed]
- Sunyer, R.; Conte, V.; Escribano, J.; Elosegui-Artola, A.; Labernadie, A.; Valon, L.; Navajas, D.; García-Aznar, J.M.; Muñoz, J.J.; Roca-Cusachs, P.; Trepat, X. Collective cell durotaxis emerges from long-range intercellular force transmission. Science 2016, 353, 1157–1161. [Google Scholar] [CrossRef] [PubMed]
- Malinverno, C.; Corallino, S.; Giavazzi, F.; Bergert, M.; Li, Q.; Leoni, M.; Disanza, A.; Frittoli, E.; Oldani, A.; Martini, E.; et al. Endocytic reawakening of motility in jammed epithelia. Nat. Mater. 2017, 16, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Guilak, F.; Butler, D.L.; Goldstein, S.A.; Baaijens, F.P.T. Biomechanics and mechanobiology in functional tissue engineering. J. Biomech. 2014, 47, 1933–1940. [Google Scholar] [CrossRef] [PubMed]
- Chin, L.; Xia, Y.; Discher, D.E.; Janmey, P.A. Mechanotransduction in cancer. Curr. Opin. Chem. Eng. 2016, 11, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Vasioukhin, V.; Fuchs, E. Actin dynamics and cell-cell adhesion in epithelia. Curr. Opin. Cell Biol. 2001, 13, 76–84. [Google Scholar] [CrossRef]
- Dejana, E. Endothelial cell-cell junctions: Happy together. Nat. Rev. Mol. Cell Biol. 2004, 5, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Leckband, D.E.; le Duc, Q.; Wang, N.; de Rooij, J. Mechanotransduction at cadherin-mediated adhesions. Curr. Opin. Cell Biol. 2011, 23, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Abercrombie, M. Contact inhibition: The phenomenon and its biological implications. Natl. Cancer Inst. Monogr. 1967, 26, 249–277. [Google Scholar]
- Castor, L.N. Contact regulation of cell division in an epithelial-like cell line. J. Cell Phys. 1968, 72, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Abercrombie, M. Contact inhibition in tissue culture. Vitro 1970, 6, 128–142. [Google Scholar] [CrossRef]
- Martz, E.; Steinberg, M.S. The role of cell-cell contact in “contact” inhibition of cell division: A review and new evidence. J. Cell Phys. 1972, 79, 189–210. [Google Scholar] [CrossRef] [PubMed]
- Puliafito, A.; Hufnagel, L.; Neveu, P.; Streichan, S.; Sigal, A.; Fygenson, D.K.; Shraiman, B.I. Collective and single cell behavior in epithelial contact inhibition. Proc. Natl. Acad. Sci. USA 2012, 109, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Shraiman, B.I. Mechanical feedback as a possible regulator of tissue growth. Proc. Natl. Acad. Sci. USA 2005, 102, 3318–3323. [Google Scholar] [CrossRef] [PubMed]
- Tambe, D.T.; Hardin, C.C.; Angelini, T.E.; Rajendran, K.; Park, C.Y.; Serra-Picamal, X.; Zhou, E.H.H.; Zaman, M.H.; Butler, J.P.; Weitz, D.A.; et al. Collective cell guidance by cooperative intercellular forces. Nat. Mater. 2011, 10, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Theveneau, E.; Mayor, R. Collective cell migration of epithelial and mesenchymal cells. Cell. Mol. Life Sci. 2013, 70, 3481–3492. [Google Scholar] [CrossRef] [PubMed]
- Theveneau, E.; Steventon, B.; Scarpa, E.; Garcia, S.; Trepat, X.; Streit, A.; Mayor, R. Chase-and-run between adjacent cell populations promotes directional collective migration. Nat. Cell Biol. 2013, 15, 763–772. [Google Scholar] [CrossRef] [PubMed]
- Szabo, B.; Szollosi, G.J.; Gonci, B.; Juranyi, Z.; Selmeczi, D.; Vicsek, T. Phase transition in the collective migration of tissue cells: Experiment and model. Phys. Rev. E 2006, 74, 061908. [Google Scholar] [CrossRef] [PubMed]
- Szabo, A.; Unnep, R.; Mehes, E.; Twal, W.O.; Argraves, W.S.; Cao, Y.; Czirok, A. Collective cell motion in endothelial monolayers. Phys. Biol. 2010, 7, 046007. [Google Scholar] [CrossRef] [PubMed]
- Kabla, A.J. Collective cell migration: Leadership, invasion and segregation. J. R. Soc. Interf. 2012, 9, 3268–3278. [Google Scholar] [CrossRef] [PubMed]
- Maruthamuthu, V.; Sabass, B.; Schwarz, U.S.; Gardel, M.L. Cell-ECM traction force modulates endogenous tension at cell-cell contacts. Proc. Natl. Acad. Sci. USA 2011, 108, 4708–4713. [Google Scholar] [CrossRef] [PubMed]
- Jasaitis, A.; Estevez, M.; Heysch, J.; Ladoux, B.; Dufour, S. E-Cadherin-Dependent Stimulation of Traction Force at Focal Adhesions via the Src and PI3K Signaling Pathways. Biophys. J. 2012, 103, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Mertz, A.F.; Che, Y.; Banerjee, S.; Goldstein, J.M.; Rosowski, K.A.; Revilla, S.F.; Niessen, C.M.; Marchetti, M.C.; Dufresne, E.R.; Horsley, V. Cadherin-based intercellular adhesions organize epithelial cell-matrix traction forces. Proc. Natl. Acad. Sci. USA 2013, 110, 842–847. [Google Scholar] [CrossRef] [PubMed]
- Giordano, R.A.; Wu, B.M.; Borland, S.W.; Cima, L.G.; Sachs, E.M.; Cima, M.J. Mechanical properties of dense polylactic acid structures fabricated by three dimensional printing. J. Biomater. Sci. Polym. E 1996, 8, 63–75. [Google Scholar] [CrossRef]
- Sarkar, S.; Lee, G.Y.; Wong, J.Y.; Desai, T.A. Development and characterization of a porous micro-patterned scaffold for vascular tissue engineering applications. Biomaterials 2006, 27, 4775–4782. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, J.; Motiagh, D.; Russell, B.; Desai, T.A. Fabrication of microtextured membranes for cardiac myocyte attachment and orientation. J. Biomed. Mater. Res. 2000, 53, 267–275. [Google Scholar] [CrossRef]
- Hahn, M.S.; Taite, L.J.; Moon, J.J.; Rowland, M.C.; Ruffino, K.A.; West, J.L. Photolithographic patterning of polyethylene glycol hydrogels. Biomaterials 2006, 27, 2519–2524. [Google Scholar] [CrossRef] [PubMed]
- Athanasiou, K.A.; Niederauer, G.G.; Agrawal, C.M. Sterilization, toxicity, biocompatibility and clinical applications of polylactic acid polyglycolic acid copolymers. Biomaterials 1996, 17, 93–102. [Google Scholar] [CrossRef]
- Cao, H.; Kuboyama, N. A biodegradable porous composite scaffold of PGA/beta-TCP for bone tissue engineering. Bone 2010, 46, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Peters, M.C.; Polverini, P.J.; Mooney, D.J. Engineering vascular networks in porous polymer matrices. J. Biomed. Mater. Res. 2002, 60, 668–678. [Google Scholar] [CrossRef] [PubMed]
- Kasuya, J.; Sudo, R.; Tamogami, R.; Masuda, G.; Mitaka, T.; Ikeda, M.; Tanishita, K. Reconstruction of 3D stacked hepatocyte tissues using degradable, microporous poly(D,L-lactide-co-glycolide) membranes. Biomaterials 2012, 33, 2693–2700. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.Y.; Chan-Park, M.B.; Zhu, A.P.; Zhu, X.; Beuerman, R.W.; Yang, E.B.; Chen, W.; Chan, V. Three-dimensional microchannels in biodegradable polymeric films for control orientation and phenotype of vascular smooth muscle cells. Tissue Eng. 2006, 12, 2229–2240. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.Y.; Chan-Park, M.B.E.; Feng, Z.O.; Chan, V.; Feng, Z.W. UV-embossed microchannel in biocompatible polymeric film: Application to control of cell shape and orientation of muscle cells. J. Biomed. Mater. Res. B 2006, 77B, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Chan-Park, M.B.; Yan, Y.H.; Neo, W.K.; Zhou, W.X.; Zhang, J.; Yue, C.Y. Fabrication of high aspect ratio poly(ethylene glycol)-containing microstructures by UV embossing. Langmuir 2003, 19, 4371–4380. [Google Scholar] [CrossRef]
- Feng, J.; Chan-Park, M.B.; Shen, J.Y.; Chan, V. Quick layer-by-layer assembly of aligned multilayers of vascular smooth muscle cells in deep microchannels. Tissue Eng. 2007, 13, 1003–1012. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Isenberg, B.C.; Hodis, E.; Leach, J.B.; Desai, T.A.; Wong, J.Y. Fabrication of a layered microstructured polycaprolactone construct for 3-D tissue engineering. J. Biomater. Sci. Polym. E 2008, 19, 1347–1362. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Dadhania, M.; Rourke, P.; Desai, T.A.; Wong, J.Y. Vascular tissue engineering: Microtextured scaffold templates to control organization of vascular smooth muscle cells and extracellular matrix. Acta Biomater. 2005, 1, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Revzin, A.; Russell, R.J.; Yadavalli, V.K.; Koh, W.G.; Deister, C.; Hile, D.D.; Mellott, M.B.; Pishko, M.V. Fabrication of poly(ethylene glycol) hydrogel microstructures using photolithography. Langmuir 2001, 17, 5440–5447. [Google Scholar] [CrossRef] [PubMed]
- Koh, W.G.; Revzin, A.; Pishko, M.V. Poly(ethylene glycol) hydrogel microstructures encapsulating living cells. Langmuir 2002, 18, 2459–2462. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Xia, B.; McBride, R.; Oakey, J. A microfluidic-based cell encapsulation platform to achieve high long-term cell viability in photopolymerized PEGNB hydrogel microspheres. J. Mater. Chem. B 2017, 5, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.M.; Sarvestani, S.K.; Moeinzadeh, S.; He, X.Z.; Jabbari, E. Three-Dimensional-Engineered Matrix to Study Cancer Stem Cells and Tumorsphere Formation: Effect of Matrix Modulus. Tissue Eng. Part A 2013, 19, 669–684. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M.S.; Miller, J.S.; West, J.L. Laser scanning lithography for surface micropatterning on hydrogels. Adv. Mater. 2005, 17. [Google Scholar] [CrossRef]
- Cuchiara, M.P.; Allen, A.C.B.; Chen, T.M.; Miller, J.S.; West, J.L. Multilayer microfluidic PEGDA hydrogels. Biomaterials 2010, 31, 5491–5497. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.M.; Tang, C.; Kottke-Marchant, K.; Marchant, R.E. Design and Synthesis of Biomimetic Hydrogel Scaffolds with Controlled Organization of Cyclic RGD Peptides. Bioconj. Chem. 2009, 20, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Oliviero, O.; Ventre, M.; Netti, P.A. Functional porous hydrogels to study angiogenesis under the effect of controlled release of vascular endothelial growth factor. Acta Biomater. 2012, 8, 3294–3301. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Shao, Y.; Li, X.; Zhao, G.; Fu, J. Nanotopographical surfaces for stem cell fate control: Engineering mechanobiology from the bottom. Nano Today 2014, 9, 759–784. [Google Scholar] [CrossRef] [PubMed]
- Chevallay, B.; Herbage, D. Collagen-based biomaterials as 3D scaffold for cell cultures: Applications for tissue engineering and gene therapy. Med. Biol. Eng. Comput. 2000, 38, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.Z.; Xia, Z.D.; Han, Z.W.; Hulley, P.A.; Triffitt, J.T.; Czernuszka, J.T. Novel 3D collagen scaffolds fabricated by indirect printing technique for tissue engineering. J. Biomed. Mater. Res. B 2008, 85B, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Sachlos, E.; Reis, N.; Ainsley, C.; Derby, B.; Czernuszka, J.T. Novel collagen scaffolds with predefined internal morphology made by solid freeform fabrication. Biomaterials 2003, 24, 1487–1497. [Google Scholar] [CrossRef]
- Hutmacher, D.W.; Sittinger, M.; Risbud, M.V. Scaffold-based tissue engineering: Rationale for computer-aided design and solid free-form fabrication systems. Trends Biotechnol. 2004, 22, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Taboas, J.M.; Maddox, R.D.; Krebsbach, P.H.; Hollister, S.J. Indirect solid free form fabrication of local and global porous, biomimetic and composite 3D polymer-ceramic scaffolds. Biomaterials 2003, 24, 181–194. [Google Scholar] [CrossRef]
- O’Brien, F.J.; Harley, B.A.; Yannas, I.V.; Gibson, L. Influence of freezing rate on pore structure in freeze-dried collagen-GAG scaffolds. Biomaterials 2004, 25, 1077–1086. [Google Scholar] [CrossRef]
- O’rien, F.J.; Harley, B.A.; Yannas, I.V.; Gibson, L.J. The effect of pore size on cell adhesion in collagen-GAG scaffolds. Biomaterials 2005, 26, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Chung, T.W.; Yang, J.; Akaike, T.; Cho, K.Y.; Nah, J.W.; Kim, S.I.; Cho, C.S. Preparation of alginate/galactosylated chitosan scaffold for hepatocyte attachment. Biomaterials 2002, 23, 2827–2834. [Google Scholar] [CrossRef]
- Zhao, F.; Yin, Y.J.; Lu, W.W.; Leong, J.C.; Zhang, W.J.; Zhang, J.Y.; Zhang, M.F.; Yao, K.D. Preparation and histological evaluation of biomimetic three-dimensional hydroxyapatite/chitosan-gelatin network composite scaffolds. Biomaterials 2002, 23, 3227–3234. [Google Scholar] [CrossRef]
- Ang, T.H.; Sultana, F.S.A.; Hutmacher, D.W.; Wong, Y.S.; Fuh, J.Y.H.; Mo, X.M.; Loh, H.T.; Burdet, E.; Teoh, S.H. Fabrication of 3D chitosan-hydroxyapatite scaffolds using a robotic dispensing system. Mater. Sci. Eng. C 2002, 20, 35–42. [Google Scholar] [CrossRef]
- Zhang, S.; Holmes, T.C.; DiPersio, C.M.; Hynes, R.O.; Su, X.; Rich, A. Self-complementary oligopeptide matrices support mammalian cell attachment. Biomaterials 1995, 16, 1385–1393. [Google Scholar] [CrossRef]
- Gelain, F.; Bottai, D.; Vescovi, A.; Zhang, S. Designer self-assembling peptide nanofiber scaffolds for adult mouse neural stem cell 3-dimensional cultures. PLoS ONE 2006, 1, e119. [Google Scholar] [CrossRef] [PubMed]
- Tsutsumi, H.; Mihara, H. Soft materials based on designed self-assembling peptides: From design to application. Mol. Biol. Syst. 2013, 9, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.E.; Motion, J.P.; Narmoneva, D.A.; Takahashi, T.; Hakuno, D.; Kamm, R.D.; Zhang, S.; Lee, R.T. Injectable self-assembling peptide nanofibers create intramyocardial microenvironments for endothelial cells. Circulation 2005, 111, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Discher, D.E.; Janmey, P.; Wang, Y.L. Tissue cells feel and respond to the stiffness of their substrate. Science 2005, 310, 1139–1143. [Google Scholar] [CrossRef] [PubMed]
- Pelham, R.J.; Wang, Y.L. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl. Acad. Sci. USA 1997, 94, 13661–13665. [Google Scholar] [CrossRef] [PubMed]
- Lo, C.M.; Wang, H.B.; Dembo, M.; Wang, Y.L. Cell movement is guided by the rigidity of the substrate. Biophys. J. 2000, 79, 144–152. [Google Scholar] [CrossRef]
- Bazellières, E.; Conte, V.; Elosegui-Artola, A.; Serra-Picamal, X.; Bintanel-Morcillo, M.; Roca-Cusachs, P.; Muñoz, J.J.; Sales-Pardo, M.; Guimerà, R.; Trepat, X. Control of cell-cell forces and collective cell dynamics by the intercellular adhesome. Nat. Cell Biol. 2015, 17, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Mann, J.M.; Lam, R.H.W.; Weng, S.N.; Sun, Y.B.; Fu, J.P. A silicone-based stretchable micropost array membrane for monitoring live-cell subcellular cytoskeletal response. Lab Chip 2012, 12, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix elasticity directs stem cell lineage specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Bidone, T.C.; Jung, W.; Maruri, D.; Borau, C.; Kamm, R.D.; Kim, T. Morphological Transformation and Force Generation of Active Cytoskeletal Networks. PLoS Comput. Biol. 2017, 13, e1005277. [Google Scholar] [CrossRef] [PubMed]
- Iskratsch, T.; Wolfenson, H.; Sheetz, M.P. Appreciating force and shape: The rise of mechanotransduction in cell biology. Nat. Rev. Mol. Cell Biol. 2014, 15, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Dimilla, P.A.; Barbee, K.; Lauffenburger, D.A. Mathematical model for the effects of adhesion and mechanics on cell migration speed. Biophys. J. 1991, 60, 15–37. [Google Scholar] [CrossRef]
- Peyton, S.R.; Putnam, A.J. Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J. Cell Phys. 2005, 204, 198–209. [Google Scholar] [CrossRef] [PubMed]
- Lauridsen, H.M.; Gonzalez, A.L. Biomimetic, ultrathin and elastic hydrogels regulate human neutrophil extravasation across endothelial-pericyte bilayers. PLoS One 2017, 12, e0171386. [Google Scholar] [CrossRef] [PubMed]
- Paul, R.; Heil, P.; Spatz, J.P.; Schwarz, U.S. Propagation of mechanical stress through the actin cytoskeleton toward focal adhesions: Model and experiment. Biophys. J. 2008, 94, 1470–1482. [Google Scholar] [CrossRef] [PubMed]
- Schulte, C.; Ripamonti, M.; Maffioli, E.; Cappelluti, M.A.; Nonnis, S.; Puricelli, L.; Lamanna, J.; Piazzoni, C.; Podestà, A.; Lenardi, C.; et al. Scale invariant disordered nanotopography promotes hippocampal neuron development and maturation with involvement of mechanotransductive pathways. Front. Cell Neurosci. 2016, 10, 267. [Google Scholar] [CrossRef] [PubMed]
- Harris, A.K.; Wild, P.; Stopak, D. Silicone rubber substrata: A new wrinkle in the study of cell locomotion. Science 1980, 208, 177–179. [Google Scholar] [CrossRef] [PubMed]
- Oliver, T.; Jacobson, K.; Dembo, M. Design and use of substrata to measure traction forces exerted by cultured cells. Method Enzymol. 1998, 298, 497–521. [Google Scholar]
- Oliver, T.; Dembo, M.; Jacobson, K. Traction Forces in Locomoting Cells. Cell Motil. Cytoskel. 1995, 31, 225–240. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.Q.; Schwarz, U.S.; Riveline, D.; Goichberg, P.; Tzur, G.; Sabanay, I.; Lamanna, J.; Piazzoni, C.; Podestà, A.; Lenardi, C.; et al. Force and focal adhesion assembly: A close relationship studied using elastic micropatterned substrates. Nat. Cell Biol. 2001, 3, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.L.; Tien, J.; Pirone, D.M.; Gray, D.S.; Bhadriraju, K.; Chen, C.S. Cells lying on a bed of microneedles: An approach to isolate mechanical force. Proc. Natl. Acad. Sci. USA 2003, 100, 1484–1489. [Google Scholar] [CrossRef] [PubMed]
- Cesa, C.M.; Kirchgessner, N.; Mayer, D.; Schwarz, U.S.; Hoffmann, B.; Merkel, R. Micropatterned silicone elastomer substrates for high resolution analysis of cellular force patterns. Rev. Sci. Instrum. 2007, 78. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.P.; Wang, Y.K.; Yang, M.T.; Desai, R.A.; Yu, X.A.; Liu, Z.J.; Chen, C.S. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat. Methods 2010, 7. [Google Scholar] [CrossRef] [PubMed]
- Van Hoorn, H.; Harkes, R.; Spiesz, E.M.; Storm, C.; van Noort, D.; Ladoux, B.; Schmidt, T. The Nanoscale Architecture of Force-Bearing Focal Adhesions. Nano Lett. 2014, 14, 4257–4262. [Google Scholar] [CrossRef] [PubMed]
- Bendell, A.C.; Williamson, E.K.; Chen, C.S.; Burkhardt, J.K.; Hammer, D.A. The Arp2/3 complex binding protein HS1 is required for efficient dendritic cell random migration and force generation. Integr. Biol. 2017, 9, 695–708. [Google Scholar] [CrossRef] [PubMed]
- Lam, R.H.W.; Weng, S.N.; Lu, W.; Fu, J.P. Live-cell subcellular measurement of cell stiffness using a microengineered stretchable micropost array membrane. Integr. Biol. UK 2012, 4, 1289–1298. [Google Scholar] [CrossRef] [PubMed]
- Chandra, D.; Taylor, J.A.; Yang, S. Replica molding of high-aspect-ratio (sub-)micron hydrogel pillar arrays and their stability in air and solvents. Soft Matter. 2008, 4, 979–984. [Google Scholar] [CrossRef]
- Chandra, D.; Yang, S. Stability of High-Aspect-Ratio Micropillar Arrays against Adhesive and Capillary Forces. Accounts Chem. Res. 2010, 43, 1080–1091. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.L.; Pelham, R.J. Preparation of a flexible, porous polyacrylamide substrate for mechanical studies of cultured cells. Method Enzymol. 1998, 298, 489–496. [Google Scholar]
- Kandow, C.E.; Georges, P.C.; Janmey, P.A.; Beningo, K.A. Polyacrylamidc hydrogels for cell mechanics: Steps toward optimization and alternative uses. Method Cell Biol. 2007, 83. [Google Scholar] [CrossRef]
- Beningo, K.A.; Lo, C.M.; Wang, Y.L. Flexible polyacrylamide substrata for the analysis of mechanical interactions at cell-substratum adhesions. Methods Cell Matrix Adhes. 2002, 69, 325–339. [Google Scholar]
- Lin, G.; Pister, K.S.J.; Roos, K.P. Surface micromachined polysilicon heart cell force transducer. J. Microelectromech. S 2000, 9, 9–17. [Google Scholar] [CrossRef]
- Galbraith, C.G.; Sheetz, M.P. A micromachined device provides a new bend on fibroblast traction forces. Proc. Natl. Acad. Sci. USA 1997, 94, 9114–9118. [Google Scholar] [CrossRef] [PubMed]
- Petronis, S.; Gold, J.; Kasemo, B. Microfabricated force-sensitive elastic substrates for investigation of mechanical cell-substrate interactions. J. Micromech. Microeng. 2003, 13, 900–913. [Google Scholar] [CrossRef]
- Even-Ram, S.; Yamada, K.M. Cell migration in 3D matrix. Curr. Opin. Cell Biol. 2005, 17, 524–532. [Google Scholar] [CrossRef] [PubMed]
- Zaman, M.H.; Trapani, L.M.; Sieminski, A.L.; Mackellar, D.; Gong, H.; Kamm, R.D.; Wells, A.; Lauffenburger, D.A.; Matsudaira, P. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis. Proc. Natl. Acad. Sci. USA 2006, 103, 10889–10894. [Google Scholar] [CrossRef] [PubMed]
- Critser, P.J.; Kreger, S.T.; Voytik-Harbin, S.L.; Yoder, M.C. Collagen matrix physical properties modulate endothelial colony forming cell-derived vessels in vivo. Microvasc. Res. 2010, 80, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Ehrbar, M.; Sala, A.; Lienemann, P.; Ranga, A.; Mosiewicz, K.; Bittermann, A.; Rizzi, S.C.; Weber, F.E.; Lutolf, M.P. Elucidating the role of matrix stiffness in 3D cell migration and remodeling. Biophys. J. 2011, 100, 284–293. [Google Scholar] [CrossRef] [PubMed]
- Mason, B.N.; Starchenko, A.; Williams, R.M.; Bonassar, L.J.; Reinhart-King, C.A. Tuning three-dimensional collagen matrix stiffness independently of collagen concentration modulates endothelial cell behavior. Acta Biomater. 2013, 9, 4635–4644. [Google Scholar] [CrossRef] [PubMed]
- Cukierman, E.; Pankov, R.; Stevens, D.R.; Yamada, K.M. Taking cell-matrix adhesions to the third dimension. Science 2001, 294, 1708–1712. [Google Scholar] [CrossRef] [PubMed]
- Goreczny, G.J.; Ouderkirk-Pecone, J.L.; Olson, E.C.; Krendel, M.; Turner, C.E. Hic-5 remodeling of the stromal matrix promotes breast tumor progression. Oncogene 2017, 36, 2693–2703. [Google Scholar] [CrossRef] [PubMed]
- Fraley, S.I.; Feng, Y.F.; Krishnamurthy, R.; Kim, D.H.; Celedon, A.; Longmore, G.D.; Wirtz, D. A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat. Cell Biol. 2010, 12. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, M.L.; Mcgarry, P. Review on cell mechanics: Experimental and modeling approaches. Appl. Mech. Rev. 2013, 65, 60801. [Google Scholar] [CrossRef]
- Dembo, M.; Wang, Y.L. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys. J. 1999, 76, 2307–2316. [Google Scholar] [CrossRef]
- Yang, Z.C.; Lin, J.S.; Chen, J.X.; Wang, J.H.C. Determining substrate displacement and cell traction fields—a new approach. J. Theor. Biol. 2006, 242, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Franck, C.; Hong, S.; Maskarinec, S.A.; Tirrell, D.A.; Ravichandran, G. Three-dimensional full-field measurements of large deformations in soft materials using confocal microscopy and digital volume correlation. Exp. Mech. 2007, 47, 427–438. [Google Scholar] [CrossRef]
- Franck, C.; Maskarinec, S.A.; Tirrell, D.A.; Ravichandran, G. Three-dimensional traction force microscopy: A new tool for quantifying cell-matrix interactions. PLoS ONE 2011, 6, e17833. [Google Scholar] [CrossRef] [PubMed]
- Maskarinec, S.A.; Franck, C.; Tirrell, D.A.; Ravichandran, G. Quantifying cellular traction forces in three dimensions. Proc. Natl. Acad. Sci. USA 2009, 106, 22108–22113. [Google Scholar] [CrossRef] [PubMed]
- Hur, S.S.; Zhao, Y.; Li, Y.S.; Botvinick, E.; Chien, S. Live Cells Exert 3-Dimensional Traction Forces on Their Substrata. Cell Mol. Bioeng. 2009, 2, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Delanoe-Ayari, H.; Rieu, J.P.; Sano, M. 4D traction force microscopy reveals asymmetric cortical forces in migrating Dictyostelium cells. Phys. Rev. Lett. 2010, 105, 248103. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.S.; Long, R.; Feng, X.Z.; Huang, Y.L.; Hui, C.Y.; Wu, M.M. Toward single cell traction microscopy within 3D collagen matrices. Exp. Cell Res. 2013, 319, 2396–2408. [Google Scholar] [CrossRef] [PubMed]
- Toyjanova, J.; Bar-Kochba, E.; Lopez-Fagundo, C.; Reichner, J.; Hoffman-Kim, D.; Franck, C. High resolution, large deformation 3D traction force microscopy. PLoS ONE 2014, 9, e90976. [Google Scholar] [CrossRef] [PubMed]
- Legant, W.R.; Miller, J.S.; Blakely, B.L.; Cohen, D.M.; Genin, G.M.; Chen, C.S. Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat. Methods 2010, 7, 969–971. [Google Scholar] [CrossRef] [PubMed]
- Gjorevski, N.; Nelson, C.M. Mapping of mechanical strains and stresses around quiescent engineered three-dimensional epithelial tissues. Biophys. J. 2012, 103, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Koch, T.M.; Munster, S.; Bonakdar, N.; Butler, J.P.; Fabry, B. 3D Traction forces in cancer cell invasion. PLoS ONE 2012, 7, e33476. [Google Scholar] [CrossRef] [PubMed]
- Vogel, V.; Sheetz, M. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 2006, 7, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Freikamp, A.; Mehlich, A.; Klingner, C.; Grashoff, C. Investigating piconewton forces in cells by FRET-based molecular force microscopy. J. Struct. Biol. 2017, 197, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ng, S.S.; Wang, Y.L.; Feng, H.X.; Chen, W.N.; Chan-Park, M.B.; Li, C.; Chan, V. Collective cell traction force analysis on aligned smooth muscle cell sheet between three-dimensional microwalls. Interf. Focus 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Hur, S.S.; del Alamo, J.C.; Park, J.S.; Li, Y.S.; Nguyen, H.A.; Teng, D.; Wang, K.C.; Flores, L.; Alonso-Latorre, B.; Lasheras, J.C.; et al. Roles of cell confluency and fluid shear in 3-dimensional intracellular forces in endothelial cells. Proc. Natl. Acad. Sci. USA 2012, 109, 11110–11115. [Google Scholar] [CrossRef] [PubMed]
- Du Roure, O.; Saez, A.; Buguin, A.; Austin, R.H.; Chavrier, P.; Siberzan, P.; Ladoux, B. Force mapping in epithelial cell migration (vol 102, pg 2390, 2005). Proc. Natl. Acad. Sci. USA 2005, 102, 14122. [Google Scholar]
- Tang, X.; Tofangchi, A.; Anand, S.V.; Saif, T.A. A novel cell traction force microscopy to study multi-cellular system. PLoS Comput. Biol. 2014, 10, e1003631. [Google Scholar] [CrossRef] [PubMed]
- Campas, O.; Mammoto, T.; Hasso, S.; Sperling, R.A.; O’Connell, D.; Bischof, A.G.; Maas, R.; Weitz, D.A.; Mahadevan, L.; Ingber, D.E. Quantifying cell-generated mechanical forces within living embryonic tissues. Nat. Methods 2014, 11. [Google Scholar] [CrossRef]
- Kim, D.H.; Provenzano, P.P.; Smith, C.L.; Levchenko, A. Matrix nanotopography as a regulator of cell function. J. Cell Biol. 2012, 197, 351. [Google Scholar] [CrossRef] [PubMed]

| TECHNIQUE | FORCE SENSITIVITY |
|---|---|
| Optical Tweezers | 1–100 pN |
| Atomic Force Microscope | 10–105 pN |
| Magnetic Tweezers | 10–103 pN |
| Gel Wrinkling Method | 10–100 nN |
| Micropost Deformation | 1–100 nN |
| Cell Traction Force Microscope | 10–106 pN |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Liao, K.; Li, C.; Lai, A.C.K.; Foo, J.-J.; Chan, V. Progress in Integrative Biomaterial Systems to Approach Three-Dimensional Cell Mechanotransduction. Bioengineering 2017, 4, 72. https://doi.org/10.3390/bioengineering4030072
Zhang Y, Liao K, Li C, Lai ACK, Foo J-J, Chan V. Progress in Integrative Biomaterial Systems to Approach Three-Dimensional Cell Mechanotransduction. Bioengineering. 2017; 4(3):72. https://doi.org/10.3390/bioengineering4030072
Chicago/Turabian StyleZhang, Ying, Kin Liao, Chuan Li, Alvin C.K. Lai, Ji-Jinn Foo, and Vincent Chan. 2017. "Progress in Integrative Biomaterial Systems to Approach Three-Dimensional Cell Mechanotransduction" Bioengineering 4, no. 3: 72. https://doi.org/10.3390/bioengineering4030072
APA StyleZhang, Y., Liao, K., Li, C., Lai, A. C. K., Foo, J.-J., & Chan, V. (2017). Progress in Integrative Biomaterial Systems to Approach Three-Dimensional Cell Mechanotransduction. Bioengineering, 4(3), 72. https://doi.org/10.3390/bioengineering4030072







