HDNLS: Hybrid Deep-Learning and Non-Linear Least Squares-Based Method for Fast Multi-Component T1ρ Mapping in the Knee Joint
Abstract
1. Introduction
1.1. Research Motivation
1.2. AI-Based Exponential Fitting
1.3. Proposed Solution
1.4. Contributions
- (a)
- Initially, a DL-based multi-component T1ρ mapping method is proposed. This approach utilizes synthetic data for training, thereby eliminating the need for reference MRI data.
- (b)
- The HDNLS method is proposed for fast multi-component T1ρ mapping in the knee joint. This method integrates DL and NLS. It effectively addresses key limitations of NLS, including sensitivity to initial guesses, poor convergence speed, and high computational cost.
- (c)
- Since HDNLS’s performance depends on the number of NLS iterations, it becomes a Pareto optimization problem. To address this, a comprehensive analysis of the impact of NLS iterations on HDNLS performance is conducted.
- (d)
- Four variants of HDNLS are suggested that balance accuracy and computational speed. These variants are Ultrafast-NLS, Superfast-HDNLS, HDNLS, and Relaxed-HDNLS. These methods enable users to select a suitable configuration based on their specific speed and performance requirements.
2. Proposed Methodology
2.1. Multi-Component Relaxation Models
2.2. Deep Learning-Based Multi-Component Relaxation Models
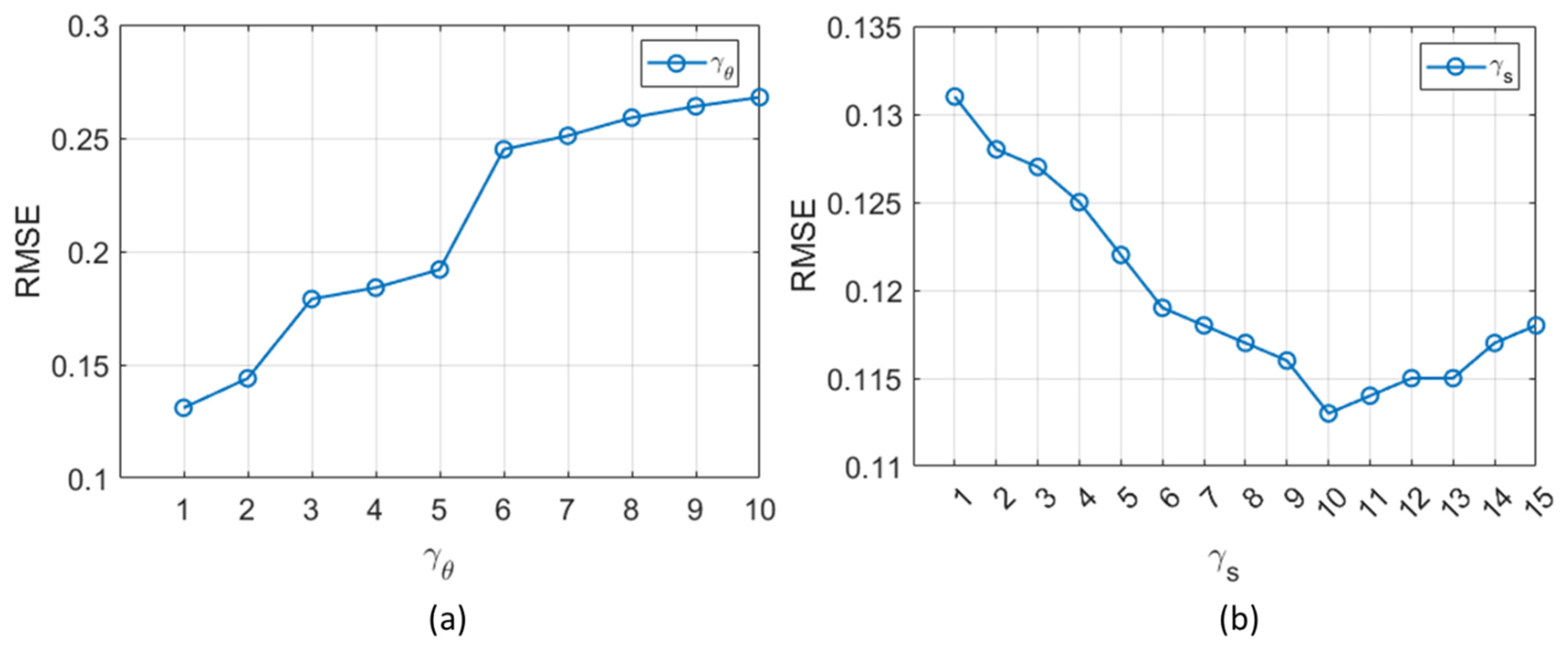
2.3. Sensitivity Analysis of DL Model
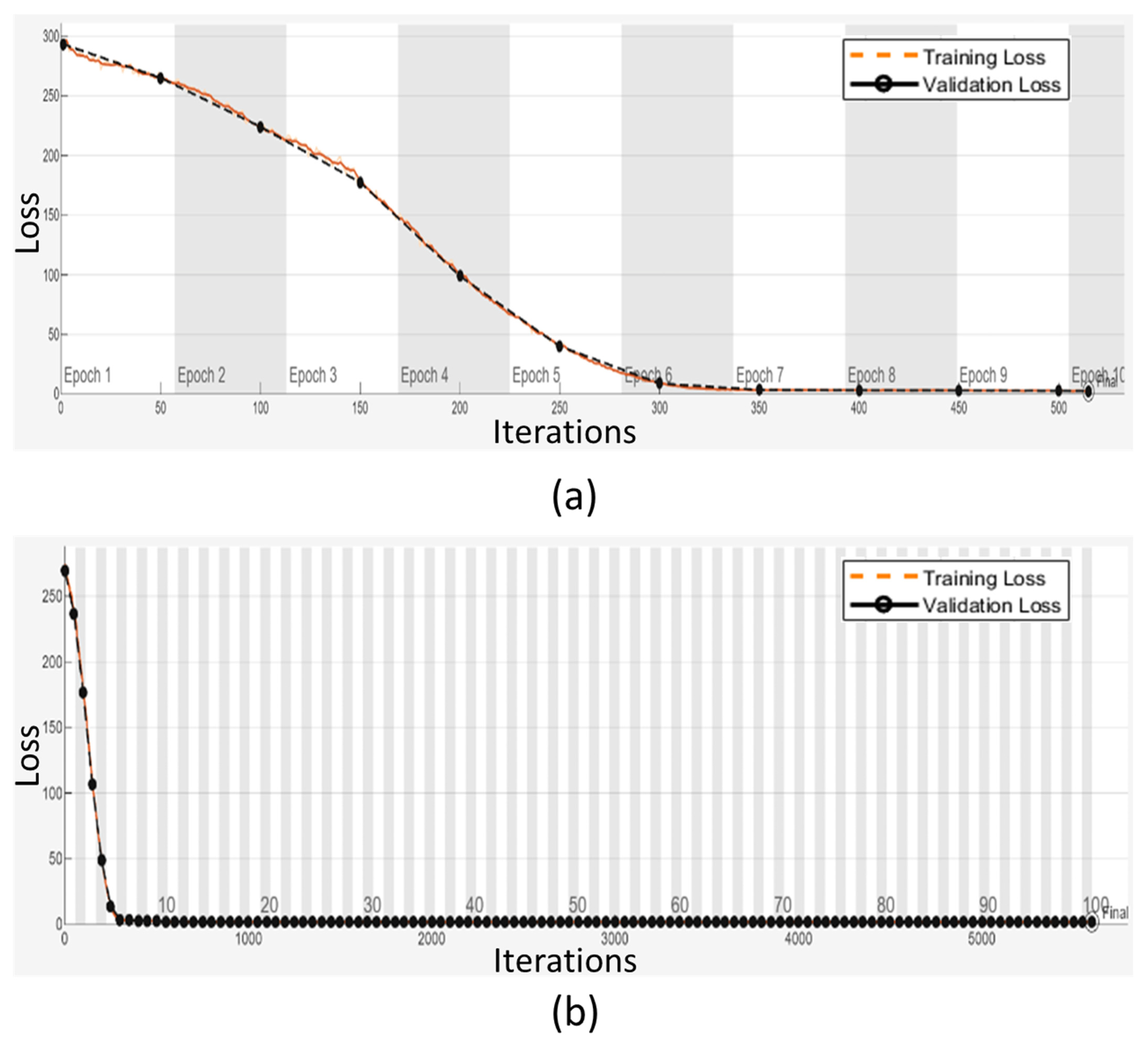
2.4. Training and Validation Analysis
2.5. HDNLS
2.6. T1ρ-MRI Data Acquisition and Fitting Methods
2.7. Performance Metrics
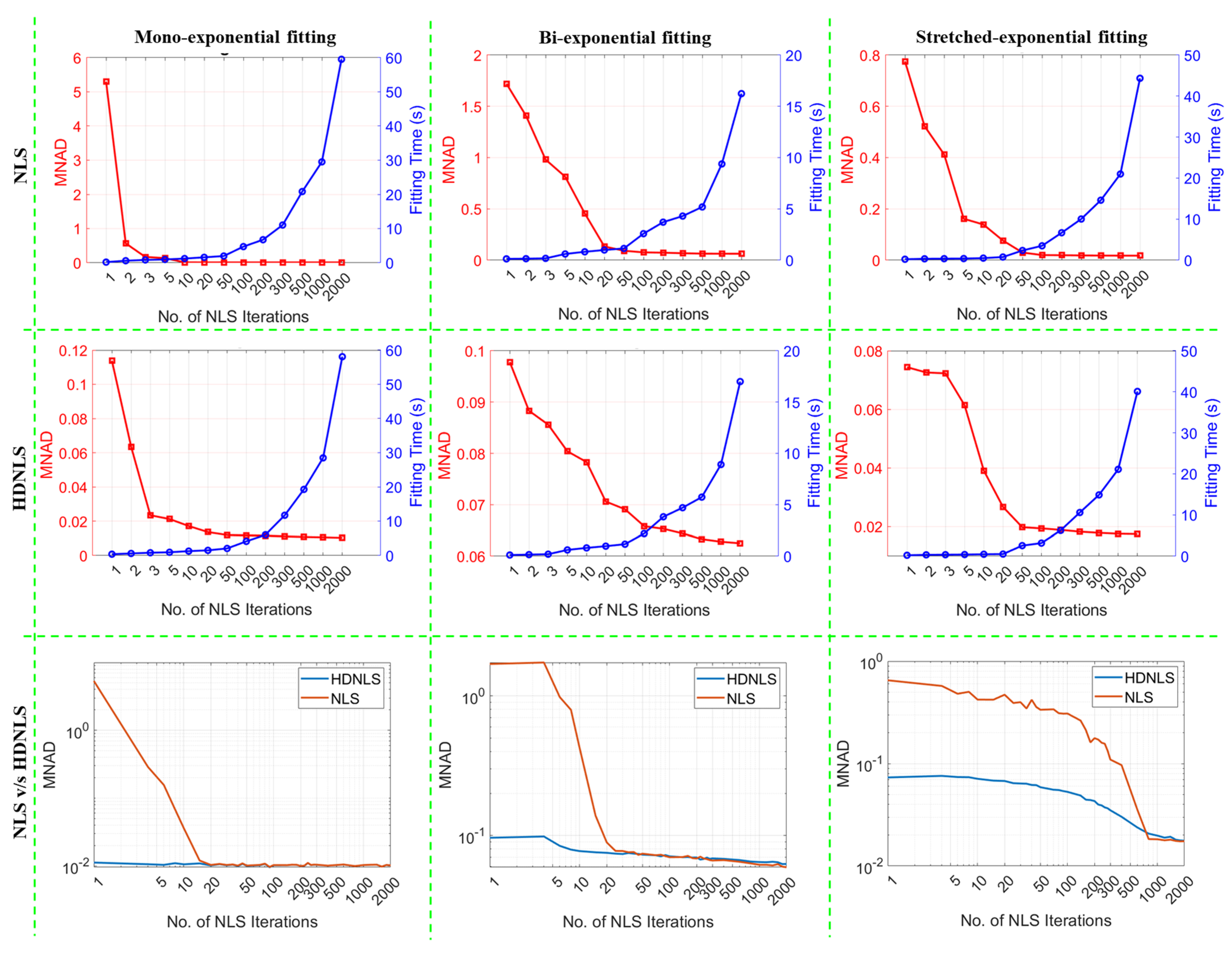
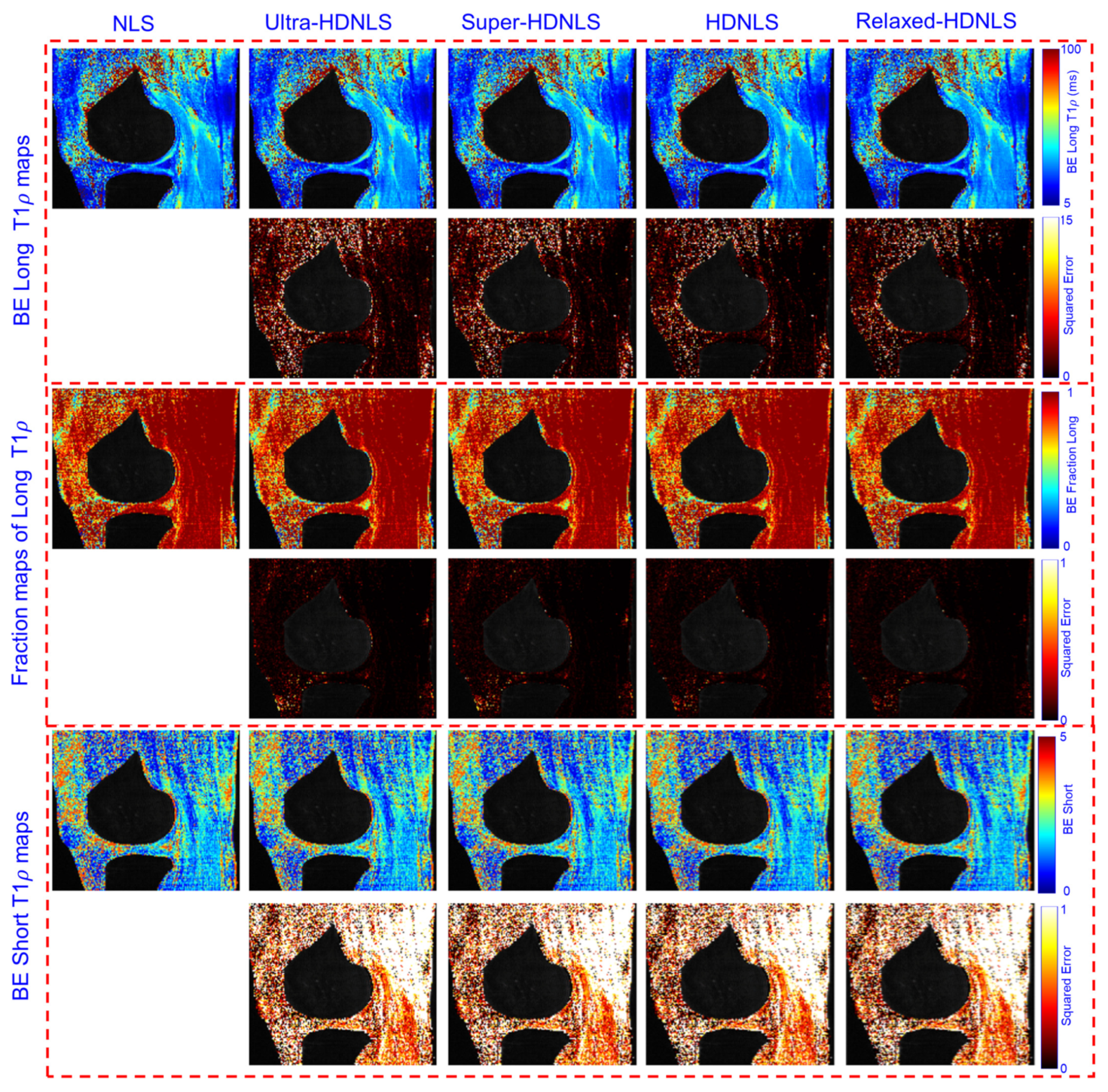
3. Optimal Selection of NLS Iterations for HDNLS
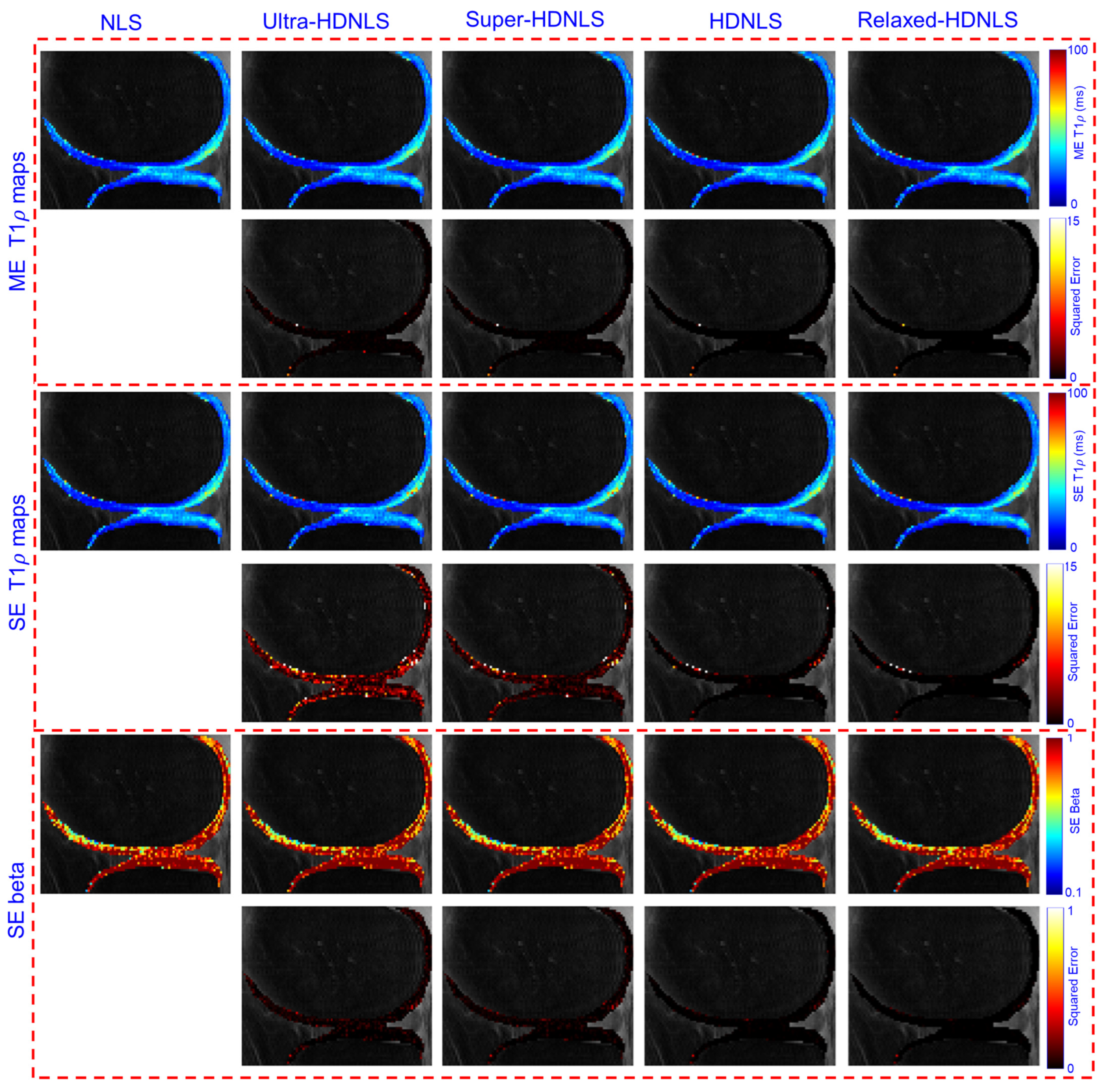
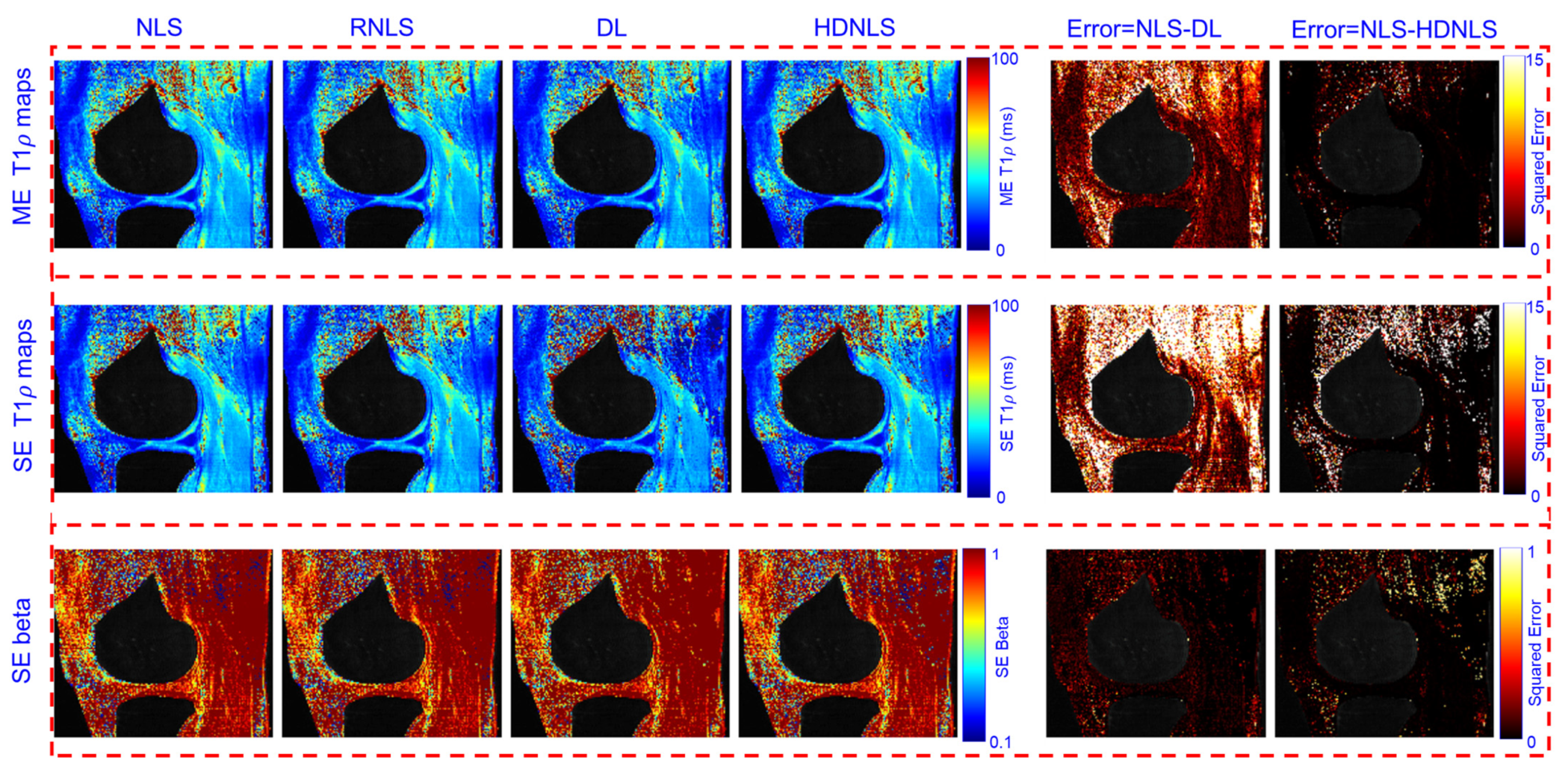
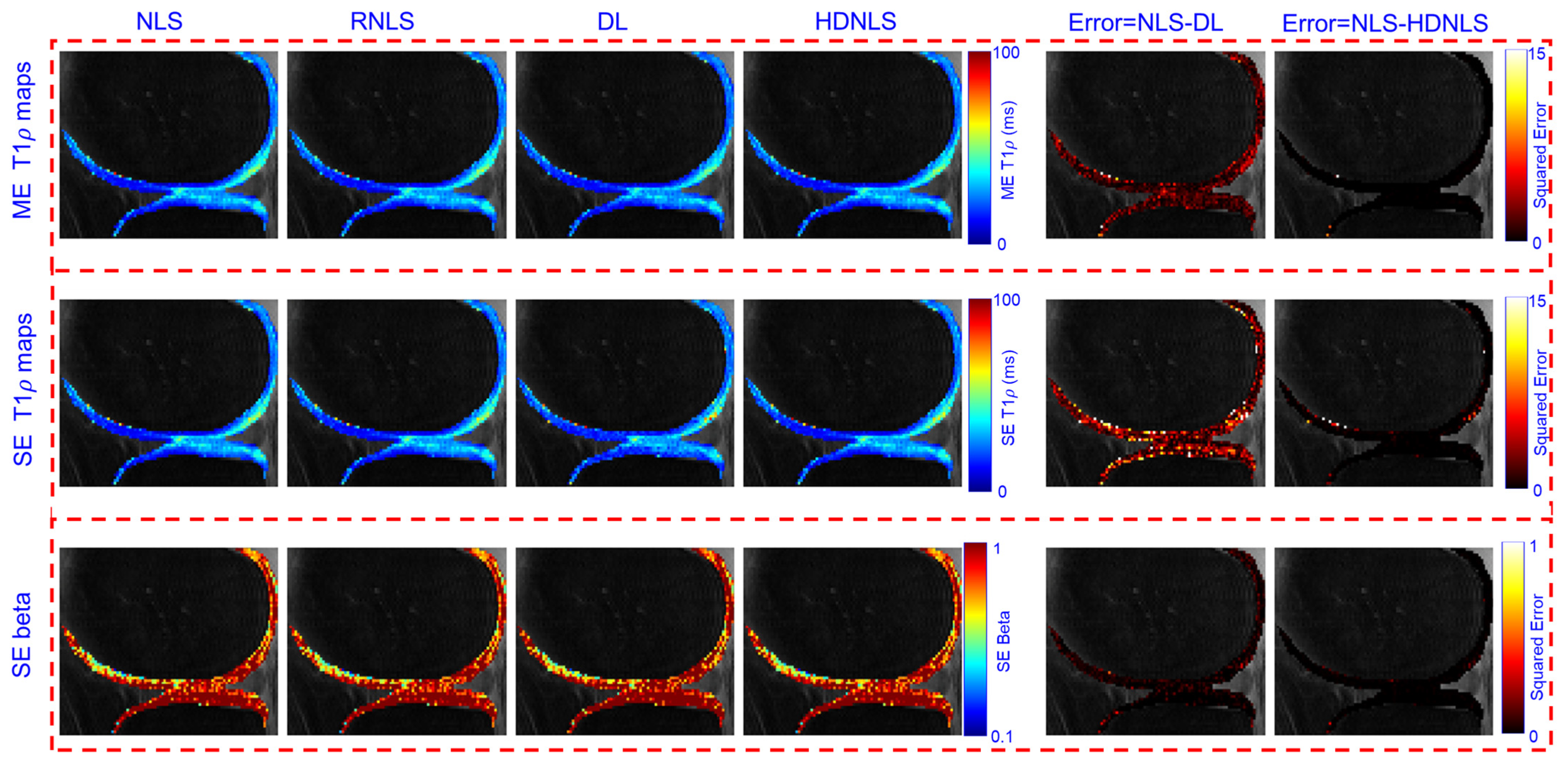
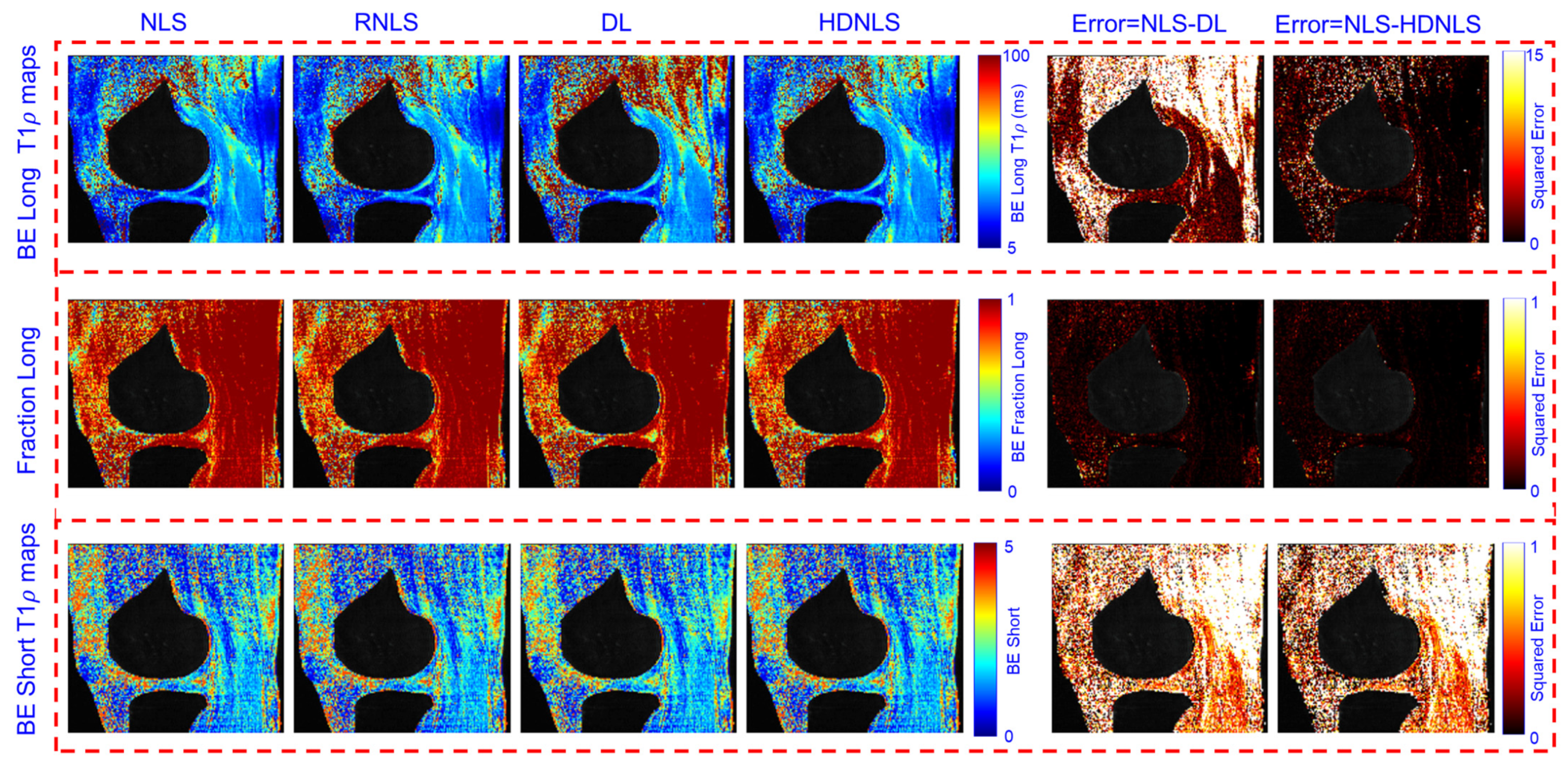
4. Performance Analysis
5. Discussion, Limitations, and Future Directions
5.1. Discussion
5.2. Limitations of HDNLS
- (a)
- The analysis of the BE short component reveals significant challenges due to its susceptibility to noise and rapid decay dynamics. These factors contribute to increased errors in fitting processes, making accurate estimation of BE short relaxation times particularly difficult. The sensitivity of these components to minor parameter variations further complicates the issue.
- (b)
- Despite its computational efficiency compared to traditional NLS and RNLS methods, the iterative nature of HDNLS makes it more computationally demanding than alternative DL approaches.
- (c)
- Although HDNLS demonstrates comparable performance in T1ρ mapping, its applicability to other quantitative MRI parameters, such as T2 mapping or diffusion tensor imaging (DTI), remains unexplored.
- (d)
- The current implementation of HDNLS does not incorporate self-supervised learning techniques. As a result, it may not fully utilize undersampled multi-contrast MRI data.
5.3. Future Directions
- (a)
- Further studies are essential to evaluate the performance of HDNLS across diverse patient populations and imaging protocols. Testing the model with data from various MRI scanners is crucial. This will help identify potential limitations and improve generalizability in clinical settings.
- (b)
- While HDNLS primarily targets T1ρ mapping, future research could adapt its methodology for other quantitative MRI parameters, such as T2 mapping and diffusion tensor imaging (DTI). This broader application could provide valuable insights into tissue characterization and significantly enhance diagnostic capabilities.
- (c)
- Self-supervised learning techniques could minimize reliance on labeled and synthetic data. This would enhance the training efficiency of HDNLS. Additionally, this approach may improve HDNLS’s ability to learn from raw or undersampled multi-contrast MRI data. Consequently, we can develop an end-to-end HDNLS model for parameter fitting.
- (d)
- Optimizing the DL architecture or incorporating advanced techniques like attention mechanisms or transformer networks could enhance HDNLS’s ability to capture complex data patterns. This improvement would reduce the number of NLS iterations required. As a result, the overall process would become faster. Additionally, using advanced regularization techniques could make HDNLS more robust to noise and outliers. This would ensure more accurate parameter estimation in challenging scenarios.
- (e)
- The BE short component is inherently challenging to estimate due to its rapid decay dynamics and low signal intensity, making it more susceptible to noise. We propose the following steps to address this issue in the near future: (i) Implement regularization techniques, such as spatial smoothness constraints and multi-voxel regularization, to stabilize parameter estimation across neighboring voxels. (ii) Integrate advanced noise-aware DL architectures, incorporating uncertainty quantification, into the DL component of HDNLS. These architectures will explicitly account for input data variability, improving robustness and reliability under noisy conditions. (iii) Explore adaptive weighting schemes in the loss function during training for BE fitting to assign appropriate emphasis to the short component. This approach aims to reduce the sensitivity of the short component to noise while maintaining overall model performance.
- (f)
- HDNLS can be extended using a pre-trained model on similar domains, such as quantitative MRI or exponential model fitting tasks. This may accelerate convergence, improve generalization, and potentially expand the range of applicability for voxel-wise multi-component T1ρ fitting.
- (g)
- As part of future studies, we will evaluate the HDNLS framework in clinical workflows to assess its utility and impact on patient diagnosis and treatment planning.
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vergeldt, F.J.; Prusova, A.; Fereidouni, F.; van Amerongen, H.; Van As, H.; Scheenen, T.W.J.; Bader, A.N. Multi-component quantitative magnetic resonance imaging by phasor representation. Sci. Rep. 2017, 7, 861. [Google Scholar] [CrossRef] [PubMed]
- Rosenkrantz, A.B.; Mendiratta-Lala, M.; Bartholmai, B.J.; Ganeshan, D.; Abramson, R.G.; Burton, K.R.; John-Paul, J.Y.; Scalzetti, E.M.; Yankeelov, T.E.; Subramaniam, R.M.; et al. Clinical utility of quantitative imaging. Acad. Radiol. 2015, 22, 33–49. [Google Scholar] [CrossRef] [PubMed]
- Keenan, K.E.; Biller, J.R.; Delfino, J.G.; Boss, M.A.; Does, M.D.; Evelhoch, J.L.; Griswold, M.A.; Gunter, J.L.; Hinks, R.S.; Hoffman, S.W.; et al. Recommendations towards standards for quantitative MRI (qMRI) and outstanding needs. J. Magn. Reson. Imaging JMRI 2019, 49, e26. [Google Scholar] [CrossRef] [PubMed]
- Richter, R.H.; Byerly, D.; Schultz, D.; Mansfield, L.T. Challenges in the interpretation of MRI examinations without radiographic correlation: Pearls and pitfalls to avoid. Cureus 2021, 13, e16419. [Google Scholar] [CrossRef]
- Cercignani, M.; Dowell, N.G.; Tofts, P.S. Quantitative MRI of the Brain: Principles of Physical Measurement; CRC Press: Boca Raton, FL, USA, 2018. [Google Scholar]
- Brown, R.W.; Cheng, Y.-C.N.; Haacke, E.M.; Thompson, M.R.; Venkatesan, R. Magnetic Resonance Imaging: Physical Principles and Sequence Design; John Wiley & Sons: Hoboken, NJ, USA, 2014. [Google Scholar]
- Kuperman, V. Magnetic Resonance Imaging: Physical Principles and Applications; Elsevier: Amsterdam, The Netherlands, 2000. [Google Scholar]
- Jelescu, I.O.; Veraart, J.; Fieremans, E.; Novikov, D.S. Degeneracy in model parameter estimation for multi-compartmental diffusion in neuronal tissue. NMR Biomed. 2016, 29, 33–47. [Google Scholar] [CrossRef]
- Madsen, K.; Nielsen, H.B.; Tingleff, O. Methods for Non-Linear Least Squares Problems; Technical University of Denmark: Kongens Lyngby, Denmark, 2004. [Google Scholar]
- Dong, G.; Flaschel, M.; Hintermüller, M.; Papafitsoros, K.; Sirotenko, C.; Tabelow, K. Data-driven methods for quantitative imaging. arXiv 2024, arXiv:2404.07886. [Google Scholar] [CrossRef]
- Levenberg, K. A method for the solution of certain non-linear problems in least squares. Q. Appl. Math. 1944, 2, 164–168. [Google Scholar] [CrossRef]
- Gavin, H.P. The Levenberg-Marquardt Algorithm for Nonlinear Least Squares Curve-Fitting Problems; Department of Civil and Environmental Engineering Duke University: Durham, NC, USA, 3 August 2019. [Google Scholar]
- Shterenlikht, A.; Alexander, N.A. Levenberg–Marquardt vs Powell’s dogleg method for Gurson–Tvergaard–Needleman plasticity model. Comput. Methods Appl. Mech. Eng. 2012, 237, 1–9. [Google Scholar] [CrossRef]
- Marquardt, D.W. An algorithm for least-squares estimation of nonlinear parameters. J. Soc. Ind. Appl. Math. 1963, 11, 431–441. [Google Scholar] [CrossRef]
- Nelder, J.A.; Mead, R. A simplex method for function minimization. Comput. J. 1965, 7, 308–313. [Google Scholar] [CrossRef]
- Hwang, D.; Du, Y.P. Improved myelin water quantification using spatially regularized non-negative least squares algorithm. J. Magn. Reson. Imaging 2009, 30, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Fenrich, F.R.E.; Beaulieu, C.; Allen, P.S. Relaxation times and microstructures. NMR Biomed. 2001, 14, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Zibetti, M.V.W.; Helou, E.S.; Sharafi, A.; Regatte, R.R. Fast multicomponent 3D-T1ρ relaxometry. NMR Biomed. 2020, 33, e4318. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, J. Optimization and Regularization of Nonlinear Least Squares Problems; Verlag nicht ermittelbar: Jerusalem, Israel, 1996. [Google Scholar]
- Singh, D.; Regatte, R.R.; Zibetti, M.V.W. Self-Supervised Deep-Learning Networks for Mono and Bi-exponential T1ρ Fitting in the Knee Joint. In Proceedings of the ISMRM, Singapore, 4–9 May 2024; 2024. Available online: https://submissions.mirasmart.com/ISMRM2024/Itinerary/?Refresh=1&ses=D-162 (accessed on 3 October 2024).
- Nedjati-Gilani, G.L.; Schneider, T.; Hall, M.G.; Cawley, N.; Hill, I.; Ciccarelli, O.; Drobnjak, I.; Gandini Wheeler-Kingshott, C.A.M.; Alexander, D.C. Machine learning based compartment models with permeability for white matter microstructure imaging. NeuroImage 2017, 150, 119–135. [Google Scholar] [CrossRef]
- Barbieri, S.; Gurney-Champion, O.J.; Klaassen, R.; Thoeny, H.C. Deep learning how to fit an intravoxel incoherent motion model to diffusion-weighted MRI. Magn. Reson. Med. 2020, 83, 312–321. [Google Scholar] [CrossRef]
- Sjölund, J.; Eklund, A.; Özarslan, E.; Herberthson, M.; Bänkestad, M.; Knutsson, H. Bayesian uncertainty quantification in linear models for diffusion MRI. NeuroImage 2018, 175, 272–285. [Google Scholar] [CrossRef]
- Liu, H.; Li, D.K.B. Myelin water imaging data analysis in less than one minute. NeuroImage 2020, 210, 116551. [Google Scholar] [CrossRef]
- Rafati, J.; Marcia, R.F. Improving L-BFGS initialization for trust-region methods in deep learning. In Proceedings of the 2018 17th IEEE International Conference on Machine Learning and Applications (ICMLA), Orlando, FL, USA, 17–20 December 2018; IEEE: New York, NY, USA, 2018; pp. 501–508. [Google Scholar]
- Bishop, C.M.; Roach, C.M. Fast curve fitting using neural networks. Rev. Sci. Instrum. 1992, 63, 4450–4456. [Google Scholar] [CrossRef]
- Gözcü, B.; Mahabadi, R.K.; Li, Y.-H.; Ilıcak, E.; Cukur, T.; Scarlett, J.; Cevher, V. Learning-based compressive MRI. IEEE Trans. Med. Imaging 2018, 37, 1394–1406. [Google Scholar] [CrossRef]
- Jung, S.; Lee, H.; Ryu, K.; Song, J.E.; Park, M.; Moon, W.-J.; Kim, D.-H. Artificial neural network for multi-echo gradient echo–based myelin water fraction estimation. Magn. Reson. Med. 2021, 85, 380–389. [Google Scholar] [CrossRef]
- Zibetti, M.V.W.; Herman, G.T.; Regatte, R.R. Fast data-driven learning of parallel MRI sampling patterns for large scale problems. Sci. Rep. 2021, 11, 19312. [Google Scholar] [CrossRef] [PubMed]
- Bertleff, M.; Domsch, S.; Weingärtner, S.; Zapp, J.; O’Brien, K.; Barth, M.; Schad, L.R. Diffusion parameter mapping with the combined intravoxel incoherent motion and kurtosis model using artificial neural networks at 3 T. NMR Biomed. 2017, 30, e3833. [Google Scholar] [CrossRef] [PubMed]
- Domsch, S.; Mürle, B.; Weingärtner, S.; Zapp, J.; Wenz, F.; Schad, L.R. Oxygen extraction fraction mapping at 3 Tesla using an artificial neural network: A feasibility study. Magn. Reson. Med. 2018, 79, 890–899. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanyam, C.; Ajay, M.S.; Spandana, K.R.; Shetty, A.B.; Seetharamu, K.N. Curve fitting for coarse data using artificial neural network. WSEAS Trans. Math. 2014, 13, 406–415. [Google Scholar]
- Liu, F.; Feng, L.; Kijowski, R. MANTIS: Model-augmented neural network with incoherent k-space sampling for efficient MR parameter mapping. Magn. Reson. Med. 2019, 82, 174–188. [Google Scholar] [CrossRef]
- Fu, Z.; Mandava, S.; Keerthivasan, M.B.; Li, Z.; Johnson, K.; Martin, D.R.; Altbach, M.I.; Bilgin, A. A multi-scale residual network for accelerated radial MR parameter mapping. Magn. Reson. Imaging 2020, 73, 152–162. [Google Scholar] [CrossRef]
- Li, H.; Yang, M.; Kim, J.H.; Zhang, C.; Liu, R.; Huang, P.; Liang, D.; Zhang, X.; Li, X.; Ying, L. SuperMAP: Deep ultra-fast MR relaxometry with joint spatiotemporal undersampling. Magn. Reson. Med. 2023, 89, 64–76. [Google Scholar] [CrossRef]
- Liu, F.; Samsonov, A.; Chen, L.; Kijowski, R.; Feng, L. SANTIS: Sampling-augmented neural network with incoherent structure for MR image reconstruction. Magn. Reson. Med. 2019, 82, 1890–1904. [Google Scholar] [CrossRef]
- Liu, F.; Kijowski, R.; El Fakhri, G.; Feng, L. Magnetic resonance parameter mapping using model-guided self-supervised deep learning. Magn. Reson. Med. 2021, 85, 3211–3226. [Google Scholar] [CrossRef]
- Liu, F.; Kijowski, R.; Feng, L.; El Fakhri, G. High-performance rapid MR parameter mapping using model-based deep adversarial learning. Magn. Reson. Imaging 2020, 74, 152–160. [Google Scholar] [CrossRef]
- Feng, R.; Zhao, J.; Wang, H.; Yang, B.; Feng, J.; Shi, Y.; Zhang, M.; Liu, C.; Zhang, Y.; Zhuang, J.; et al. MoDL-QSM: Model-based deep learning for quantitative susceptibility mapping. NeuroImage 2021, 240, 118376. [Google Scholar] [CrossRef] [PubMed]
- Jun, Y.; Shin, H.; Eo, T.; Kim, T.; Hwang, D. Deep model-based magnetic resonance parameter mapping network (DOPAMINE) for fast T1 mapping using variable flip angle method. Med. Image Anal. 2021, 70, 102017. [Google Scholar] [CrossRef] [PubMed]
- Bian, W.; Jang, A.; Liu, F. Improving quantitative MRI using self-supervised deep learning with model reinforcement: Demonstration for rapid T1 mapping. Magn. Reson. Med. 2024, 92, 98–111. [Google Scholar] [CrossRef] [PubMed]
- Gabrielle Blumenkrantz, B.; Lozano, B.J.; Julio, C.-G.; Ries, M.; Majumdar, S. In vivo T1Rho and T2 mapping of articular cartilage in osteoarthritis of the knee using 3 tesla MRI. Osteoarthr. Cartil. 2007, 15, 789–797. [Google Scholar]
- Milford, D.; Rosbach, N.; Bendszus, M.; Heiland, S. Mono-exponential fitting in T2-relaxometry: Relevance of offset and first echo. PLoS ONE 2015, 10, e0145255. [Google Scholar] [CrossRef]
- Souza, R.B.; Feeley, B.T.; Zarins, Z.A.; Link, T.M.; Li, X.; Majumdar, S. T1rho MRI relaxation in knee OA subjects with varying sizes of cartilage lesions. Knee 2013, 20, 113–119. [Google Scholar] [CrossRef][Green Version]
- Shao, H.; Chang, E.; Pauli, C.; Zanganeh, S.; Bae, W.; Chung, C.; Tang, G.; Du, J. UTE bi-component analysis of T2* relaxation in articular cartilage. Osteoarthr. Cartil. 2016, 24, 364–373. [Google Scholar] [CrossRef]
- Sharafi, A.; Xia, D.; Chang, G.; Regatte, R.R. Biexponential T1ρ relaxation mapping of human knee cartilage in vivo at 3 T. NMR Biomed. 2017, 30, e3760. [Google Scholar] [CrossRef]
- Bakker, C.J.G.; Vriend, J. Multi-exponential water proton spin-lattice relaxation in biological tissues and its implications for quantitative NMR imaging. Phys. Med. Biol. 1984, 29, 509–518. [Google Scholar] [CrossRef]
- Reiter, D.A.; Magin, R.L.; Li, W.; Trujillo, J.J.; Velasco, M.P.; Spencer, R.G. Anomalous T2 relaxation in normal and degraded cartilage. Magn. Reson. Med. 2016, 76, 953–962. [Google Scholar] [CrossRef]
- Wilson, R.; Bowen, L.; Kim, W.; Reiter, D.; Neu, C. Stretched-Exponential Modeling of Anomalous T1ρ and T2 Relaxation in the Intervertebral Disc In Vivo. bioRxiv 2020, preprint. [Google Scholar] [CrossRef]
- Johnston, D.C. Stretched exponential relaxation arising from a continuous sum of exponential decays. Phys. Rev. B 2006, 74, 184430. [Google Scholar] [CrossRef]
- Steihaug, T. The conjugate gradient method and trust regions in large scale optimization. SIAM J. Numer. Anal. 1983, 20, 626–637. [Google Scholar] [CrossRef]
- Li, X.; Han, E.T.; Busse, R.F.; Majumdar, S. In vivo T1ρ mapping in cartilage using 3D magnetization-prepared angle-modulated partitioned k-space spoiled gradient echo snapshots (3D MAPSS). Magn. Reson. Med. 2008, 59, 298–307. [Google Scholar] [CrossRef]
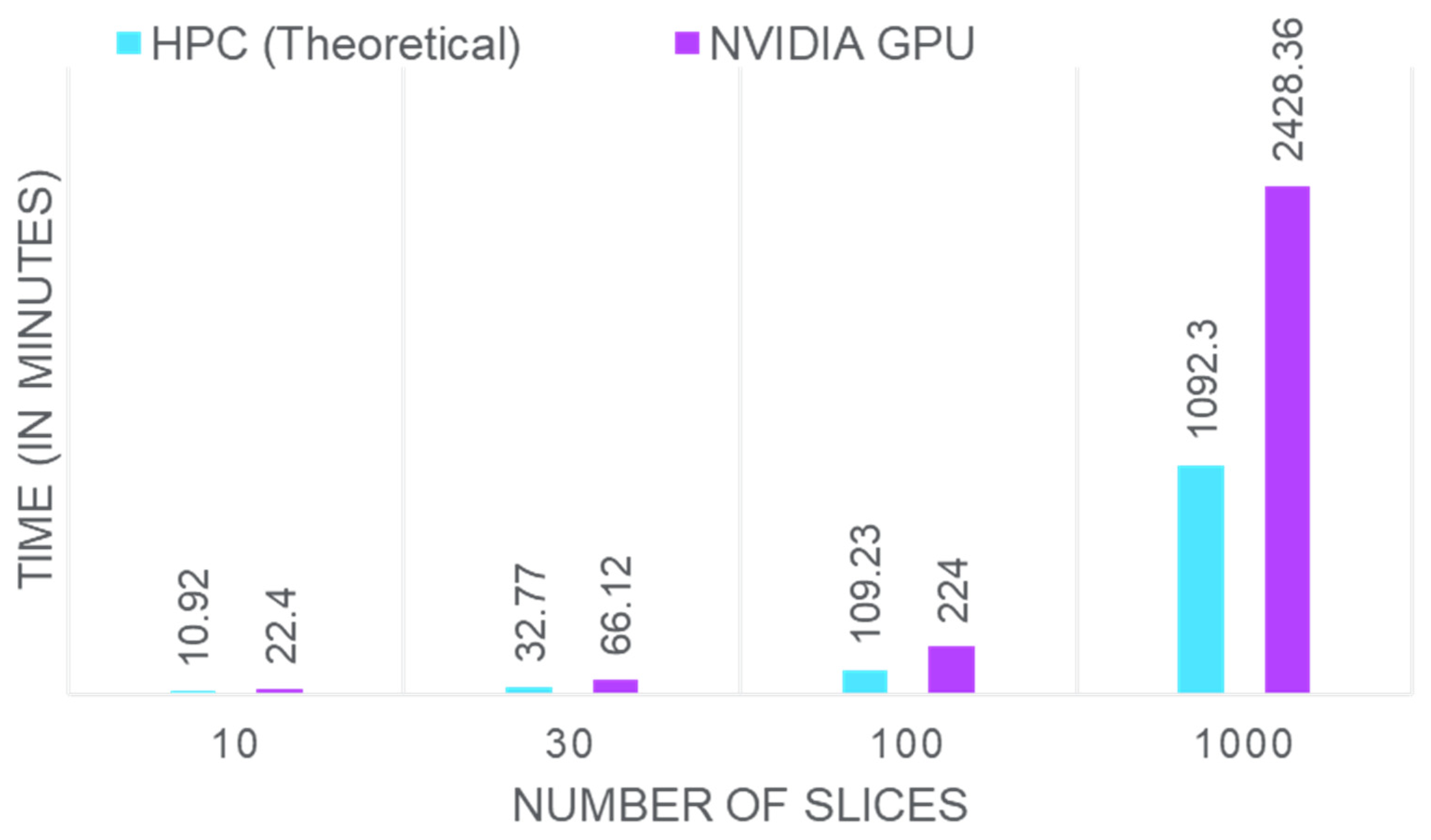
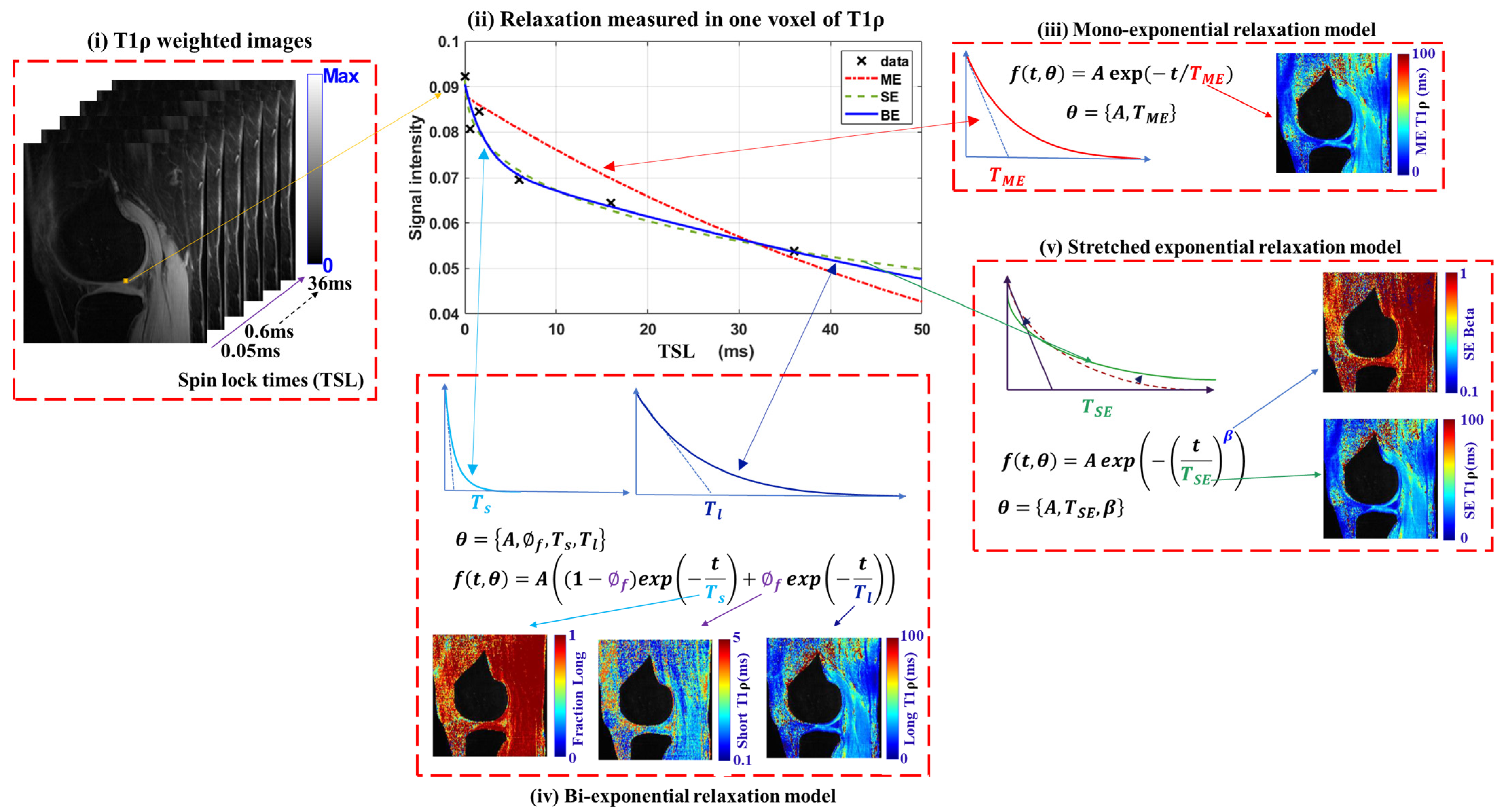
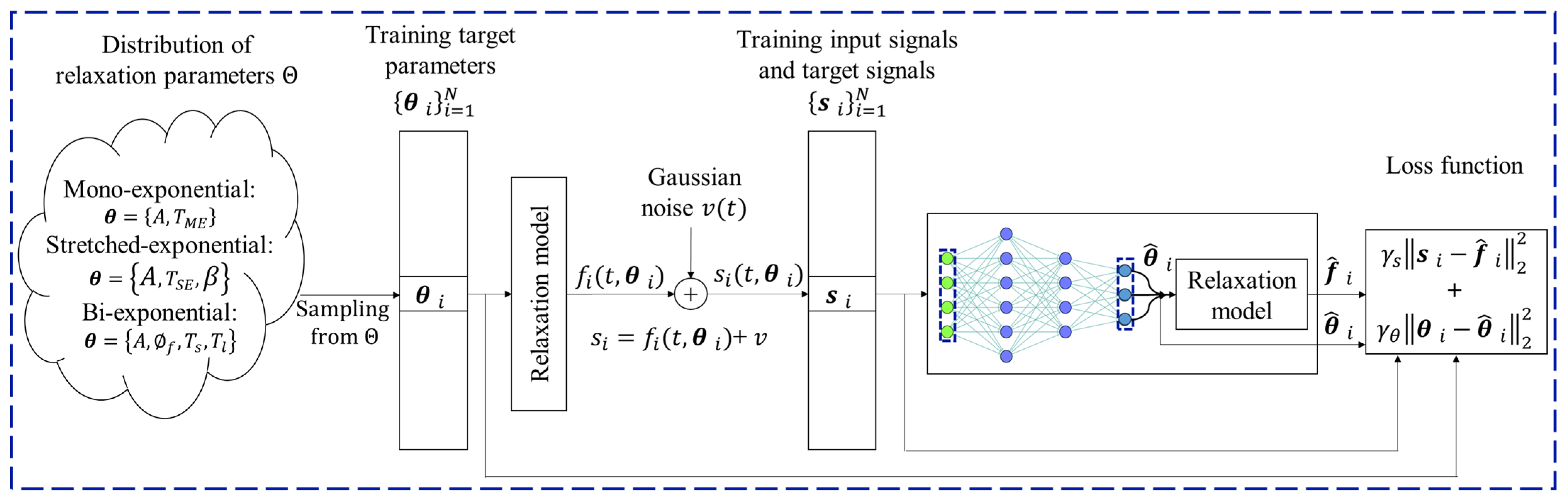

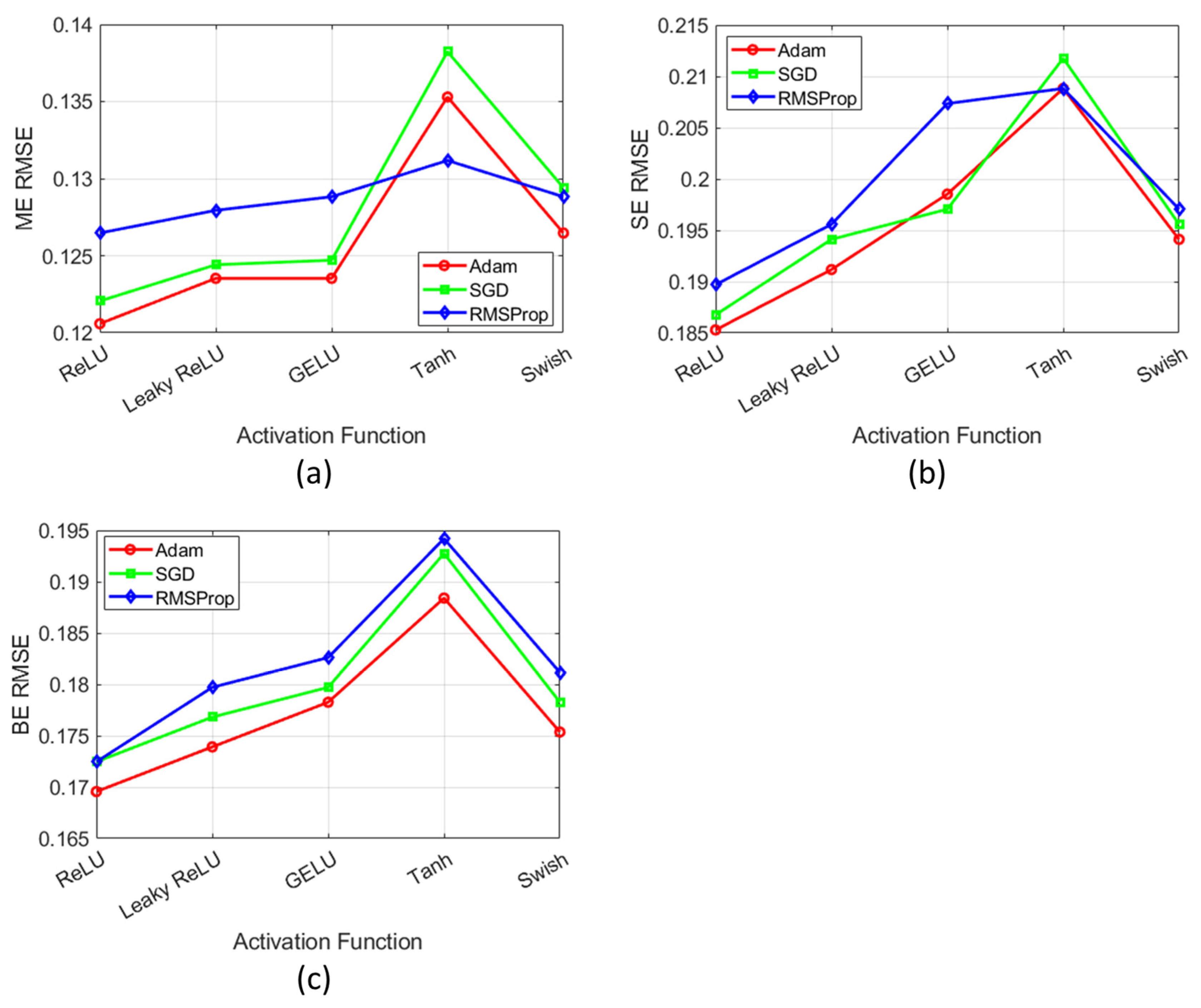





| Ref. | Year | Method | Features | Open Challenges |
|---|---|---|---|---|
| [21] | 2017 | Monte Carlo simulations and a random forest regressor |
|
|
| [22] | 2019 | Deep neural network (DNN) |
|
|
| [24] | 2020 | DNN |
|
|
| [33] | 2019 | MANTIS |
|
|
| [38] | 2020 | Model-based deep adversarial learning |
|
|
| [39] | 2021 | MoDL-QSM |
|
|
| [40] | 2021 | DOPAMINE |
|
|
| [37] | 2021 | RELAX |
|
|
| [35] | 2023 | SuperMAP |
|
|
| [41] | 2024 | RELAX-MORE |
|
|
| Model | Equation | Description | Range | |
|---|---|---|---|---|
| ME [20,42,43,44] | Assumes tissue is homogeneous with a single relaxation time. is the complex-valued amplitude and is the real-valued relaxation time. | (for knee cartilage ≥ 5 ms) | ||
| BE [45,46,47] | Accounts for two compartments with distinct relaxation times, (short) and (long). is the fraction of the long compartment. | |||
| SE [48,49,50] | Models heterogeneous environments with distributed relaxation rates. is the relaxation time, and is the stretching exponent. |
| Model | Features | Advantages | Disadvantages |
|---|---|---|---|
| ME |
|
|
|
| BE |
|
|
|
| SE |
|
|
|
| Metric | Description | Equation | Symbols |
|---|---|---|---|
| MNAD | Measures the median of the normalized absolute difference between estimated and reference T1ρ parameters across voxels. | and are the estimated and reference parameters p of the voxel indexed by i that belongs to the region of interest or set of voxels I, and . | |
| NRMSR | Assesses the size of the residual between MR measurements and predicted MR signals. | is the measured MR signal at the voxel indexed by i, and is the predicted MR signal at the same voxel with . | |
| NRMSE | Assesses the normalized value of the root-mean-squared error (RMSE) across the reference parameters. | and are the estimated and reference parameters p of the voxel indexed by i that belongs to the region of interest or set of voxels I, and . | |
| Fitting Time | Represents the time required by models (e.g., NLS, RNLS, HDNLS, and DL) to compute the T1ρ map for one MRI slice, measured on the same hardware. | Start time of the fitting process. End time of the fitting process. |
| Method | Metric | Ultra-HDNLS | Super-HDNLS | HDNLS | Relaxed-HDNLS |
|---|---|---|---|---|---|
| ME | MNAD (%) | 4.68 | 4.22 | 4.03 | 3.82 |
| Fitting time (s) | 0.99 | 1.76 | 5.13 | 10.96 | |
| NRMSR | 0.03 | 0.03 | 0.02 | 0.02 | |
| SE | MNAD (%) | 13.49 | 8.92 | 7.18 | 6.75 |
| Fitting time (s) | 1.12 | 1.85 | 3.42 | 7.12 | |
| NRMSR | 1.09 | 0.70 | 0.18 | 0.15 | |
| BE | MNAD (%) | 21.38 | 20.26 | 19.87 | 19.24 |
| Fitting time (s) | 1.22 | 2.14 | 3.96 | 7.73 | |
| NRMSR | 0.03 | 0.03 | 0.02 | 0.02 |
| Method | Metric | Ultra-HDNLS | Super-HDNLS | HDNLS | Relaxed-HDNLS |
|---|---|---|---|---|---|
| ME | MNAD (%) | 4.36 | 4.16 | 3.86 | 3.74 |
| Fitting time (s) | 0.52 | 0.95 | 2.38 | 5.73 | |
| NRMSR | 0.03 | 0.02 | 0.02 | 0.02 | |
| SE | MNAD (%) | 13.24 | 8.73 | 7.12 | 6.67 |
| Fitting time (s) | 0.51 | 0.87 | 2.28 | 3.71 | |
| NRMSR | 1.09 | 0.70 | 0.18 | 0.14 | |
| BE | MNAD (%) | 20.34 | 19.77 | 19.28 | 18.94 |
| Fitting time (s) | 0.46 | 0.87 | 1.85 | 3.26 | |
| NRMSR | 0.03 | 0.02 | 0.02 | 0.02 |
| Method | Metric | NLS | RNLS | DL | HDNLS |
|---|---|---|---|---|---|
| ME | MNAD (%) | 3.65 | 3.69 | 4.29 | 3.70 |
| Fitting time (s) | 37.48 | 32.83 | 0.79 | 4.94 | |
| NRMSR | 0.02 | 0.02 | 0.03 | 0.02 | |
| SE | MNAD (%) | 6.18 | 6.27 | 7.61 | 6.32 |
| Fitting time (s) | 41.9 | 37.75 | 0.61 | 4.17 | |
| NRMSR | 0.15 | 0.15 | 0.18 | 0.16 | |
| BE | MNAD (%) | 16.19 | 16.36 | 18.37 | 16.47 |
| Fitting time (s) | 11.73 | 11.16 | 0.52 | 2.48 | |
| NRMSR | 0.02 | 0.02 | 0.02 | 0.02 |
| Method | Metric | Full Knee | Knee Cartilage | ||
|---|---|---|---|---|---|
| DL | HDNLS | DL | HDNLS | ||
| ME | MNAD (%) | 22.36 | 18.93 | 19.32 | 17.20 |
| NRMSE (%) | 24.05 | 21.25 | 21.20 | 19.31 | |
| SE | MNAD (%) | 26.38 | 23.94 | 23.38 | 21.64 |
| NRMSE (%) | 25.81 | 22.94 | 22.46 | 19.85 | |
| BE | MNAD (%) | 17.70 | 13.02 | 14.09 | 12.24 |
| NRMSE (%) | 20.16 | 16.36 | 18.37 | 14.85 | |
| Method | Metric | NLS | RNLS | DL | HDNLS |
|---|---|---|---|---|---|
| ME | Full Knee | 274.68 | 234.22 | 1.35 | 19.32 |
| Knee Cartilage | 76.37 | 65.82 | 0.92 | 6.78 | |
| SE | Full Knee | 293.49 | 262.92 | 1.78 | 22.50 |
| Knee Cartilage | 91.12 | 88.39 | 1.67 | 7.51 | |
| BE | Full Knee | 257.38 | 229.13 | 1.21 | 18.14 |
| Knee Cartilage | 70.26 | 67.14 | 0.84 | 6.13 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, D.; Regatte, R.R.; Zibetti, M.V.W. HDNLS: Hybrid Deep-Learning and Non-Linear Least Squares-Based Method for Fast Multi-Component T1ρ Mapping in the Knee Joint. Bioengineering 2025, 12, 8. https://doi.org/10.3390/bioengineering12010008
Singh D, Regatte RR, Zibetti MVW. HDNLS: Hybrid Deep-Learning and Non-Linear Least Squares-Based Method for Fast Multi-Component T1ρ Mapping in the Knee Joint. Bioengineering. 2025; 12(1):8. https://doi.org/10.3390/bioengineering12010008
Chicago/Turabian StyleSingh, Dilbag, Ravinder R. Regatte, and Marcelo V. W. Zibetti. 2025. "HDNLS: Hybrid Deep-Learning and Non-Linear Least Squares-Based Method for Fast Multi-Component T1ρ Mapping in the Knee Joint" Bioengineering 12, no. 1: 8. https://doi.org/10.3390/bioengineering12010008
APA StyleSingh, D., Regatte, R. R., & Zibetti, M. V. W. (2025). HDNLS: Hybrid Deep-Learning and Non-Linear Least Squares-Based Method for Fast Multi-Component T1ρ Mapping in the Knee Joint. Bioengineering, 12(1), 8. https://doi.org/10.3390/bioengineering12010008







