Visualized Lead Selection for Arrhythmia Classification Based on a Lead Activation Heatmap Using Multi-Lead ECGs
Abstract
1. Introduction
- (1)
- The lead selection method can choose valid leads rather than redundant ones in a visual and explainable way.
- (2)
- The proposed LA heatmap enhances the interpretability of deep learning methods in arrhythmias detection.
- (3)
- The ResBiTime network merges Bi-LSTM networks with residual connections to obtain not only the temporal features of ECG signals, but also the complementary information among different leads.
- (4)
- The proposed method achieved satisfactory results in the 9-class classification of arrhythmias within the CPSC 2018 DB.
2. Materials and Methods
2.1. Database
2.2. Method Outline
2.3. Data Preprocessing
2.3.1. Removing Noise
2.3.2. Heartbeats Detection
2.3.3. Data Balancing
2.4. The Visualized Lead Selection Module
2.4.1. The Lead-Wise Network
2.4.2. Determining Valid Leads
2.5. The ResBiTime Network
3. Results
3.1. Evaluation Metrics
3.2. Results
3.3. Comparison with Previous Works for the CPSC 2018 Db
3.4. Comparison with Classical Baselines for the CPSC 2018 DB
4. Discussion
4.1. Ablation Experiment
4.2. Interpretability Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Cardiovascular Diseases (CVDs) Factsheet [Internet]; WHO: Geneva, Switzerland, 2013. [Google Scholar]
- Ansari, Y.; Mourad, O.; Qaraqe, K.; Serpedin, E. Deep learning for ECG Arrhythmia detection and classification: An overview of progress for period 2017–2023. Front. Physiol. 2023, 14, 20. [Google Scholar]
- Kachuee, M.; Fazeli, S.; Sarrafzadeh, M.; Soc, I.C. ECG Heartbeat Classification: A Deep Transferable Representation. In Proceedings of the 6th IEEE International Conference on Healthcare Informatics (ICHI), New York, NY, USA, 4–7 June 2018; IEEE Computer Society: New York, NY, USA, 2018. [Google Scholar]
- Jahmunah, V.; Ng, E.Y.K.; Tan, R.S.; Oh, S.L.; Acharya, U.R. Explainable detection of myocardial infarction using deep learning models with Grad-CAM technique on ECG signals. Comput. Biol. Med. 2022, 146, 19. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.P.; Li, D.; Zhang, Y.T. A deep neural network based on multi-model and multi-scale for arrhythmia classification. Biomed. Signal Process. Control 2023, 85, 8. [Google Scholar] [CrossRef]
- Li, J.H.; Pang, S.P.; Xu, F.Z.; Ji, P.; Zhou, S.W.; Shu, M.L. Two-dimensional ECG-based cardiac arrhythmia classification using DSE-ResNet. Sci. Rep. 2022, 12, 13. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Xie, Y.T.; Liu, X.W.; Yuan, X.C.; Wang, H. An End-to-End 12-Leading Electrocardiogram Diagnosis System Based on Deformable Convolutional Neural Network With Good Antinoise Ability. IEEE Trans. Instrum. Meas. 2021, 70, 13. [Google Scholar] [CrossRef]
- Cai, W.J.; Chen, Y.D.; Guo, J.; Han, B.S.; Shi, Y.J.; Ji, L.; Wang, J.L.; Zhang, G.L.; Luo, J.W. Accurate detection of atrial fibrillation from 12-lead ECG using deep neural network. Comput. Biol. Med. 2020, 116, 9. [Google Scholar] [CrossRef]
- Wang, R.X.; Fan, J.P.; Li, Y. Deep Multi-Scale Fusion Neural Network for Multi-Class Arrhythmia Detection. IEEE J. Biomed. Health Inform. 2020, 24, 2461–2472. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Kwon, J.M.; Kim, K.H.; Medina-Inojosa, J.R.; Jeon, K.H.; Cho, S.; Lee, S.Y.; Park, J.; Oh, B.H. Artificial intelligence algorithm for detecting myocardial infarction using six-lead electrocardiography. Sci. Rep. 2020, 10, 10. [Google Scholar] [CrossRef]
- Kumar, P.S.; Ramasamy, M.; Kallur, K.R.; Rai, P.; Varadan, V.K. Personalized LSTM Models for ECG Lead Transformations Led to Fewer Diagnostic Errors Than Generalized Models: Deriving 12-Lead ECG from Lead II, V2, and V6. Sensors 2023, 23, 19. [Google Scholar] [CrossRef]
- Li, Z.; Zhou, D.S.; Wan, L.; Li, J.; Mou, W.F. Heartbeat classification using deep residual convolutional neural network from 2-lead electrocardiogram. J. Electrocardiol. 2020, 58, 105–112. [Google Scholar] [CrossRef]
- Gibson, C.M.; Mehta, S.; Ceschim, M.R.S.; Frauenfelder, A.; Vieira, D.; Botelho, R.; Fernandez, F.; Villagran, C.; Niklitschek, S.; Matheus, C.I.; et al. Evolution of single-lead ECG for STEMI detection using a deep learning approach. Int. J. Cardiol. 2022, 346, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Serrano, S.; Rodrigo, M.; Calvo, C.J.; Millet, J.; Castells, F. From 12 to 1 ECG lead: Multiple cardiac condition detection mixing a hybrid machine learning approach with a one-versus-rest classification strategy. Physiol. Meas. 2022, 43, 17. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.F.; Liu, C.Y.; Zhao, L.N.; Zhang, X.Y.; Wu, X.L.; Xu, X.Y.; Liu, Y.L.; Ma, C.Y.; Wei, S.S.; He, Z.Q.; et al. An Open Access Database for Evaluating the Algorithms of Electrocardiogram Rhythm and Morphology Abnormality Detection. J. Med. Imaging Health Inform. 2018, 8, 1368–1373. [Google Scholar] [CrossRef]
- Martis, R.J.; Acharya, U.R.; Min, L.C. ECG beat classification using PCA, LDA, ICA and Discrete Wavelet Transform. Biomed. Signal Process. Control 2013, 8, 437–448. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Pan, J.; Tompkins, W.J. A Real-Time QRS Detection Algorithm. IEEE Trans. Biomed. Eng. BME 1985, 32, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning Deep Features for Discriminative Localization. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; IEEE: Seattle, WA, USA, 2016. [Google Scholar]
- Lin, M.; Chen, Q.; Yan, S. Network in network. arXiv 2013, arXiv:1312.4400. [Google Scholar]
- Yao, Q.H.; Wang, R.X.; Fan, X.M.; Liu, J.K.; Li, Y. Multi-class Arrhythmia detection from 12-lead varied-length ECG using Attention-based Time-Incremental Convolutional Neural Network. Inf. Fusion 2020, 53, 174–182. [Google Scholar] [CrossRef]
- Kaiming, H.; Xiangyu, Z.; Shaoqing, R.; Jian, S. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. In Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015; pp. 1026–1034. [Google Scholar]
- Saxe, A.M.; McClelland, J.L.; Ganguli, S. Exact solutions to the nonlinear dynamics of learning in deep linear neural networks. arXiv 2013, arXiv:1312.6120. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Geng, Q.C.; Liu, H.; Gao, T.L.; Liu, R.S.; Chen, C.; Zhu, Q.; Shu, M.L. An ECG Classification Method Based on Multi-Task Learning and CoT Attention Mechanism. Healthcare 2023, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.M.; Huang, C.H.; Shih, E.S.C.; Hu, Y.F.; Hwang, M.J. Detection and Classification of Cardiac Arrhythmias by a Challenge-Best Deep Learning Neural Network Model. Iscience 2020, 23, 26. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Yau, C.Y. Fitting time series models for longitudinal surveys with nonignorable missing data. J. Stat. Plan. Inference 2021, 214, 1–12. [Google Scholar] [CrossRef]
- Le, K.H.; Pham, H.H.; Nguyen, T.B.; Nguyen, T.A.; Thanh, T.N.; Do, C.D. Enhancing deep learning-based 3-lead ecg classification with heartbeat counting and demographic data integration. In Proceedings of the 2022 IEEE-EMBS Conference on Biomedical Engineering and Sciences (IECBES), Kuala Lumpur, Malaysia, 7–9 December 2022. [Google Scholar]
- Zhang, X.B.; Jiang, M.Z.; Polat, K.; Alhudhaif, A.; Hemanth, J.; Wu, W.Q. Detection of Atrial Fibrillation from Variable-Duration ECG Signal Based on Time-Adaptive Densely Network and Feature Enhancement Strategy. IEEE J. Biomed. Health Inform. 2023, 27, 944–955. [Google Scholar] [PubMed]
- Zhang, Y.T.; Li, J.Y.; Wei, S.S.; Zhou, F.Y.; Li, D. Heartbeats Classification Using Hybrid Time-Frequency Analysis and Transfer Learning Based on ResNet. IEEE J. Biomed. Health Inform. 2021, 25, 4175–4184. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Holzinger, A.; Biemann, C.; Pattichis, C.S.; Kell, D.B. What do we need to build explainable AI systems for the medical domain? arXiv 2017, arXiv:1712.09923. [Google Scholar]
- Hurst, J.W. Methods used to interpret the 12-lead electrocardiogram: Pattern memorization versus the use of vector concepts. Clin. Cardiol. 2000, 23, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Bruce, K.; Moore, A.K.; Mello, I.; Hilliard, T.; Day, M. Basics of the 12-lead ECG. Nursing 2023, 53, 20–25. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017. [Google Scholar]



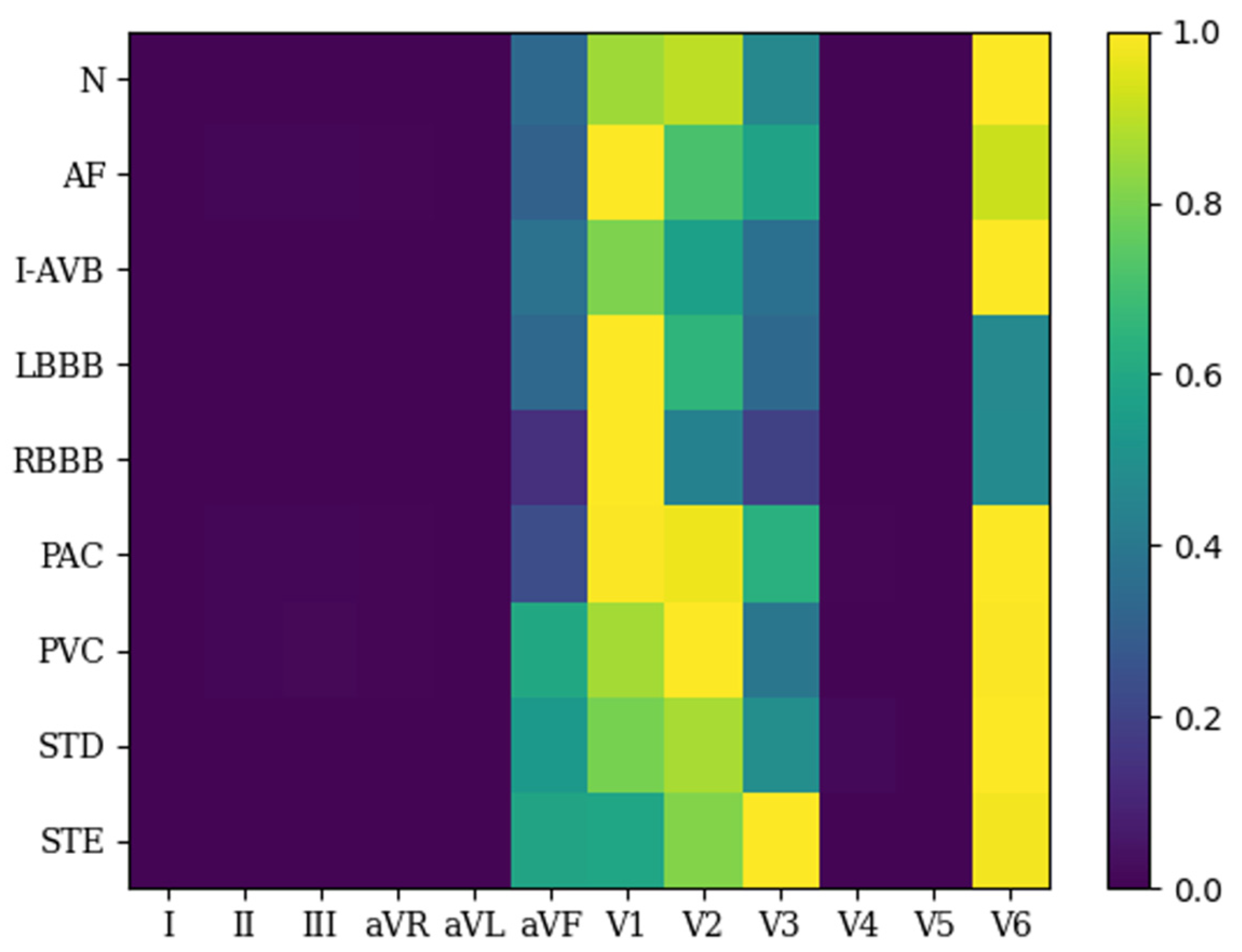
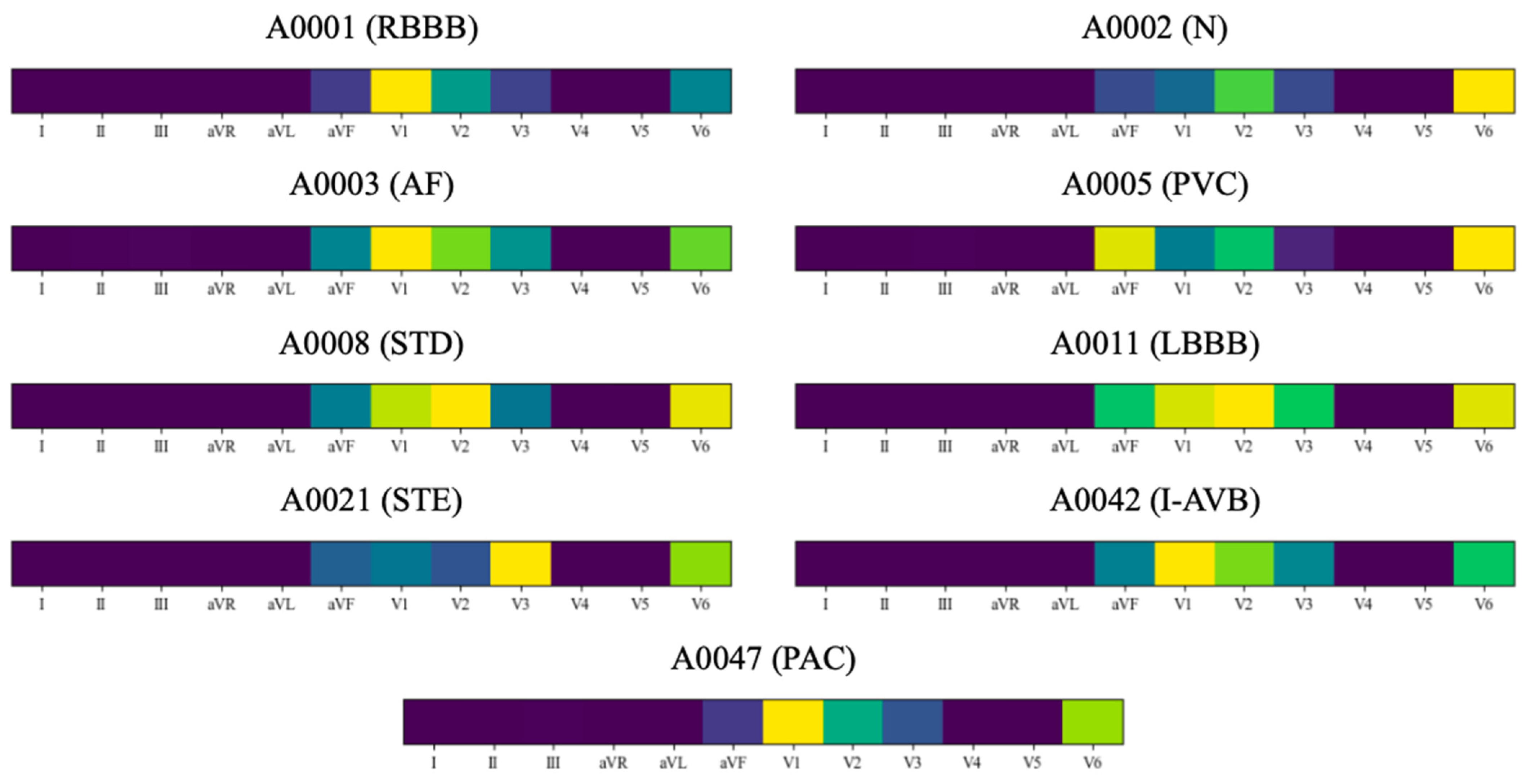
| Categories | Number of Signals | Number of Heartbeats | ||||
|---|---|---|---|---|---|---|
| Training Set | Test Set | Total Number | ||||
| Before | After | Before | After | |||
| N | 918 | 13,027 | 13,027 | 3314 | 16,341 | 16,341 |
| AF | 1098 | 12,784 | 12,784 | 3168 | 15,952 | 15,925 |
| IAVB | 704 | 8248 | 8248 | 2159 | 10,407 | 10,407 |
| LBBB | 207 | 2356 | 7068 | 566 | 2922 | 7634 |
| RBBB | 1695 | 19,512 | 19,512 | 4959 | 24,471 | 24,471 |
| PAC | 556 | 9201 | 9201 | 2293 | 11,494 | 11,494 |
| PAV | 672 | 11,491 | 11,491 | 2842 | 14,333 | 14,333 |
| STD | 825 | 10,939 | 10,939 | 2653 | 13,592 | 13,592 |
| STE | 202 | 2733 | 8199 | 619 | 3352 | 8818 |
| Total | 6877 | 90,291 | 100,469 | 22,573 | 112,864 | 123,069 |
| Layer Name | Output Size | Kernel Size/Stride |
|---|---|---|
| Input | (12, 1, 325) | - |
| Cov1 | (12, 12, 325) | 2/1 |
| Cov2 | (12, 12, 325) | 1/1 |
| Cov3 | (12, 24, 325) | 24/2 |
| Maxpooling | (12, 24, 163) | 3/1 |
| Cov4 | (12, 24, 163) | 1/1 |
| Cov5 | (12, 32, 163) | 32/1 |
| Maxpooling | (12, 32, 163) | 3/1 |
| Cov6 | (12, 32, 163) | 1/1 |
| Cov7 | (12, 48, 163) | 32/1 |
| Maxpooling | (12, 48, 163) | 3/1 |
| Cov8 | (12, 48, 163) | 1/1 |
| Cov9 | (12, 60, 163) | 48/1 |
| Maxpooling | (12, 60, 163) | 3/1 |
| GAP | (12, 60) | - |
| Linear | 9 | - |
| True Labels | Predicted | Precision (%) | Recall (%) | F1-Score | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | AF | IAVB | LBBB | RBBB | PAC | PVC | STD | STE | ||||
| N | 3189 | 9 | 9 | 0 | 18 | 30 | 26 | 26 | 7 | 95.79 | 96.23 | 0.9601 |
| AF | 20 | 2823 | 47 | 4 | 62 | 81 | 86 | 40 | 5 | 88.85 | 89.11 | 0.8883 |
| IAVB | 6 | 49 | 2043 | 0 | 12 | 14 | 24 | 9 | 2 | 93.50 | 94.63 | 0.9406 |
| LBBB | 0 | 9 | 1 | 544 | 1 | 1 | 6 | 4 | 0 | 97.32 | 96.11 | 0.9671 |
| RBBB | 7 | 53 | 10 | 1 | 4818 | 24 | 37 | 9 | 0 | 96.57 | 97.16 | 0.9686 |
| PAC | 34 | 119 | 35 | 1 | 21 | 1974 | 55 | 52 | 2 | 88.88 | 86.09 | 0.8746 |
| PVC | 47 | 96 | 25 | 9 | 35 | 56 | 2506 | 63 | 5 | 89.66 | 88.18 | 0.8891 |
| STD | 19 | 26 | 12 | 0 | 20 | 38 | 53 | 2485 | 0 | 92.34 | 93.67 | 0.9300 |
| STE | 7 | 4 | 3 | 0 | 2 | 3 | 2 | 3 | 595 | 96.59 | 96.12 | 0.9636 |
| Average | - | - | - | - | - | - | - | - | - | 93.25 | 93.03 | 0.9313 |
| Authors | Year | Themes | Database | Methods | Precision (%) | Recall (%) | F1-Score |
|---|---|---|---|---|---|---|---|
| Chen et al. [26] | 2020 | 9-class | The CPSC 2018 DB | CNN | - | - | 0.8400 |
| Wang et al. [9] | 2020 | 9-class | The CPSC 2018 DB | Multi-scale CNN | 83.80 | 82.20 | 0.8280 |
| Liu et al. [27] | 2021 | 9-class | The CPSC 2018 DB | NAS-TCAM-S | - | - | 0.7813 |
| Li et al. [6] | 2022 | 9-class | The CPSC 2018 DB | CNN + Channel Attention + ensemble model | 84.47 | 80.31 | 0.8170 |
| Le et al. [28] | 2022 | 9-class | The CPSC 2018 DB | X3ECG w/HC + DDI | - | - | 0.8140 |
| Zhang et al. [29] | 2023 | 3-class | The CPSC 2018 DB | 1-D CNN + Fine-tuning | - | - | 0.8200 |
| Jiang et al. [5] | 2023 | 9-class | The CPSC 2018 DB | Multi-scale + Multi-model CNN | 84.91 | 82.64 | 0.8352 |
| Geng et al. [25] | 2023 | 9-class | The CPSC 2018 DB | SE-ResNet + task-specific model | 85.20 | 80.00 | 0.8270 |
| This work | 2024 | 9-class | The CPSC 2018 DB | Lead selection + ResBiTime | 93.25 | 93.03 | 0.9313 |
| Neural Network | Precision (%) | Recall (%) | F1-Score |
|---|---|---|---|
| VGG [17] | 81.52 | 81.16 | 0.8108 |
| ResNet [31] | 86.52 | 87.58 | 0.8688 |
| This work (i.e., lead-wise network + ResBiTime network) | 93.25 | 93.03 | 0.9313 |
| Neural Network | Precision (%) | Recall (%) | F1-Score | Memory Usage (MB) |
|---|---|---|---|---|
| ResBiTime network | 92.85 | 93.13 | 0.9302 | 674.52 |
| This work (i.e., lead-wise network + ResBiTime network) | 93.25 | 93.03 | 0.9313 | 396.61 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Shen, T.; Jiang, S.; Wang, J.; Ma, Y.; Zhang, Y. Visualized Lead Selection for Arrhythmia Classification Based on a Lead Activation Heatmap Using Multi-Lead ECGs. Bioengineering 2024, 11, 578. https://doi.org/10.3390/bioengineering11060578
Wang H, Shen T, Jiang S, Wang J, Ma Y, Zhang Y. Visualized Lead Selection for Arrhythmia Classification Based on a Lead Activation Heatmap Using Multi-Lead ECGs. Bioengineering. 2024; 11(6):578. https://doi.org/10.3390/bioengineering11060578
Chicago/Turabian StyleWang, Heng, Tengqun Shen, Shoufen Jiang, Jilin Wang, Yijun Ma, and Yatao Zhang. 2024. "Visualized Lead Selection for Arrhythmia Classification Based on a Lead Activation Heatmap Using Multi-Lead ECGs" Bioengineering 11, no. 6: 578. https://doi.org/10.3390/bioengineering11060578
APA StyleWang, H., Shen, T., Jiang, S., Wang, J., Ma, Y., & Zhang, Y. (2024). Visualized Lead Selection for Arrhythmia Classification Based on a Lead Activation Heatmap Using Multi-Lead ECGs. Bioengineering, 11(6), 578. https://doi.org/10.3390/bioengineering11060578









