Deep-Learning-Based Automated Anomaly Detection of EEGs in Intensive Care Units
Abstract
1. Introduction
- The automated anomaly detection targets patients who are heavily ill and taken care of intensively in the intensive care unit of the Taichung Veterans General Hospital (TCVGH), a national-level medical center, not patients taking some routine and/or physical examinations.
- We attempt to detect anomaly brainwaves before their occurrence so that we can consider possible follow-up treatments in advance. The developed early detection models have promising performance and show great potential in clinical applications.
2. Related Works
3. Materials and Methods
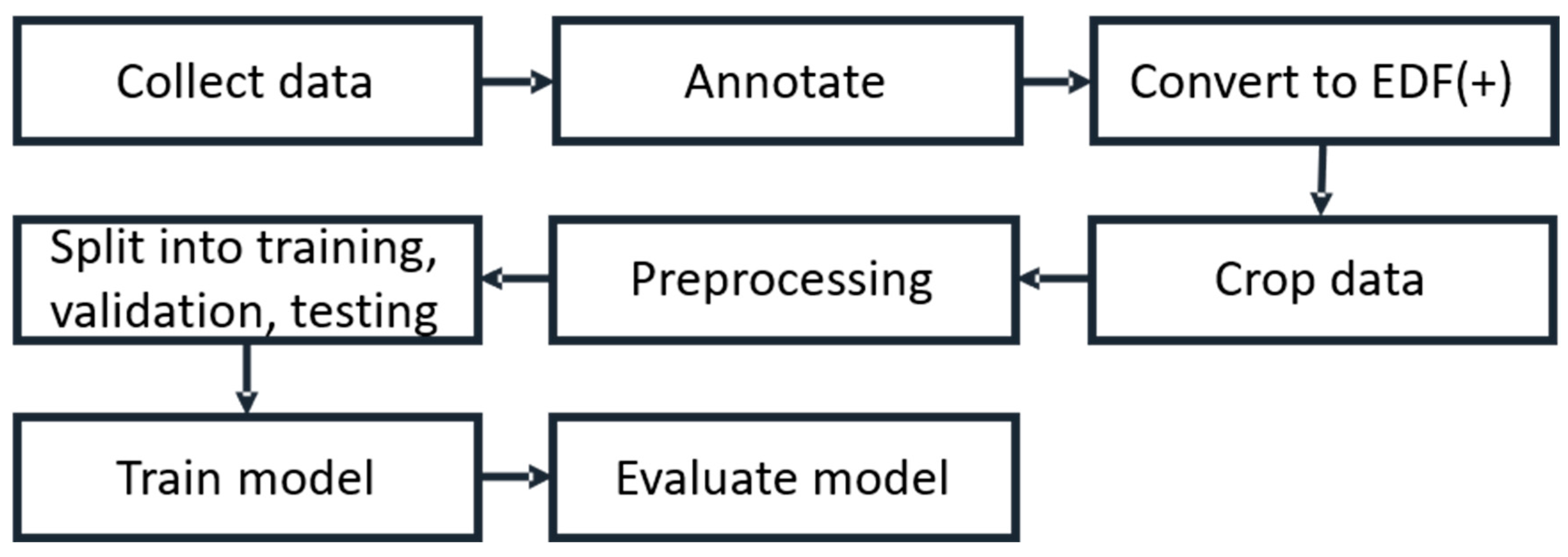
3.1. Working Flow
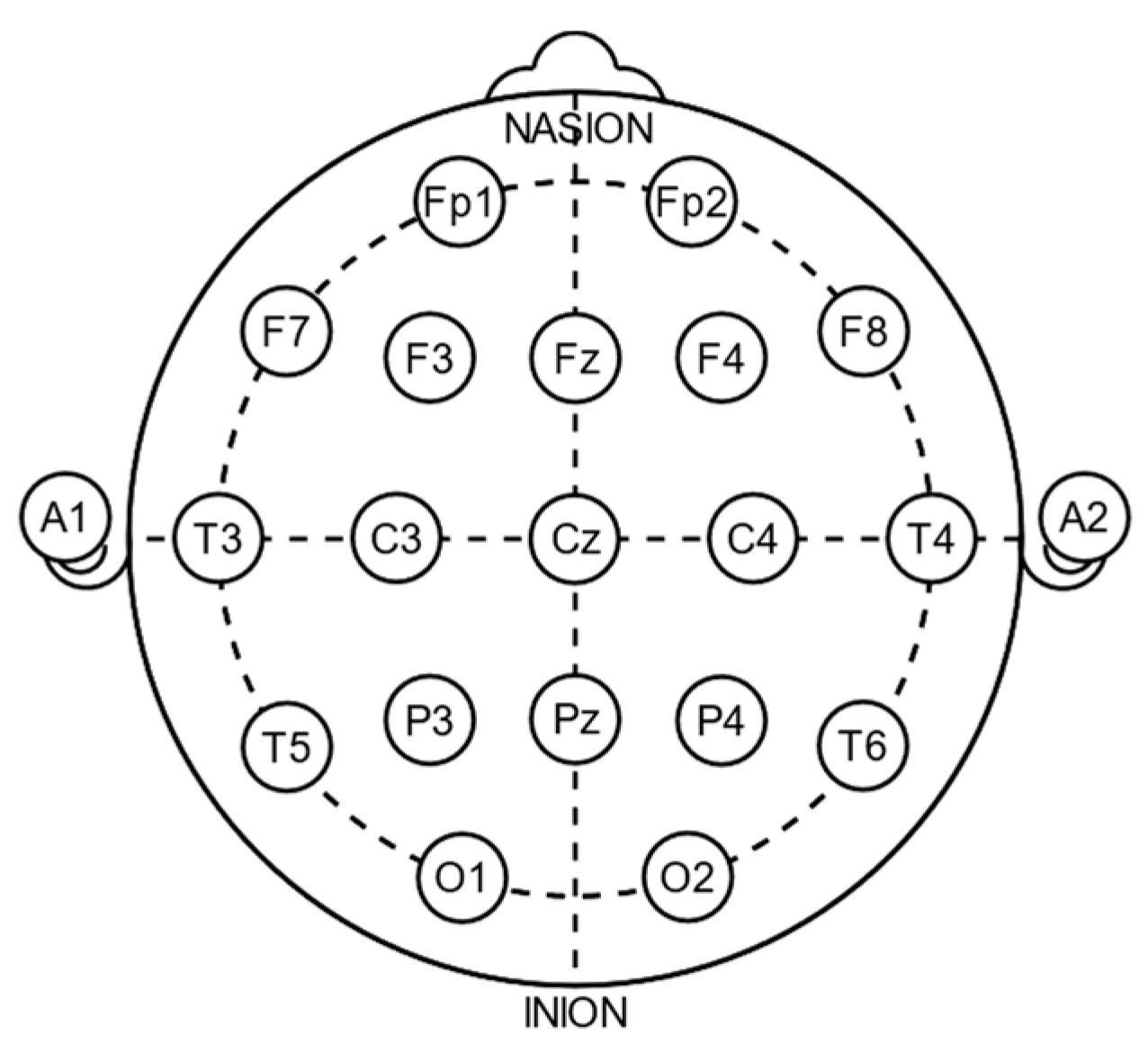
3.2. ICU Data
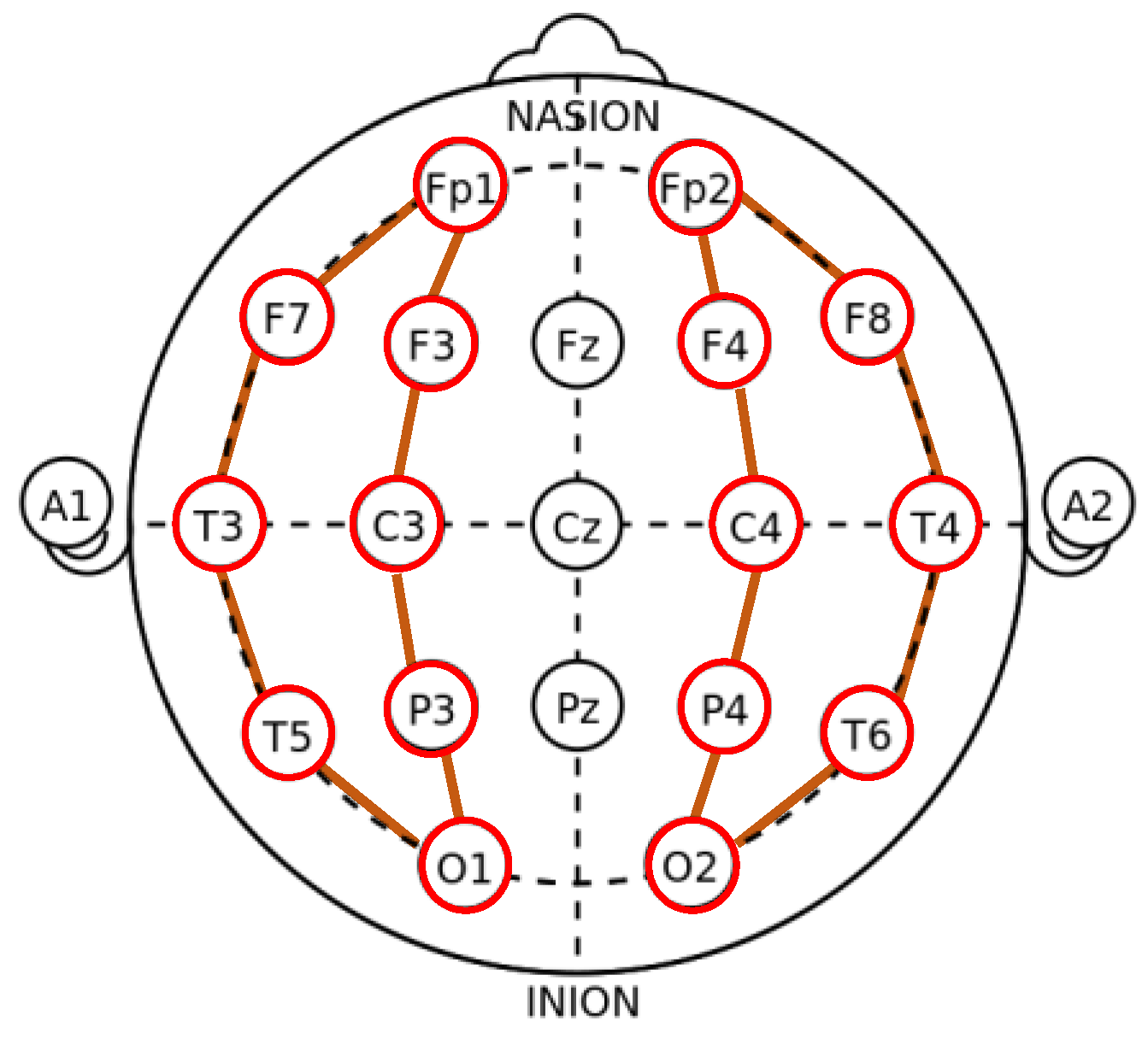
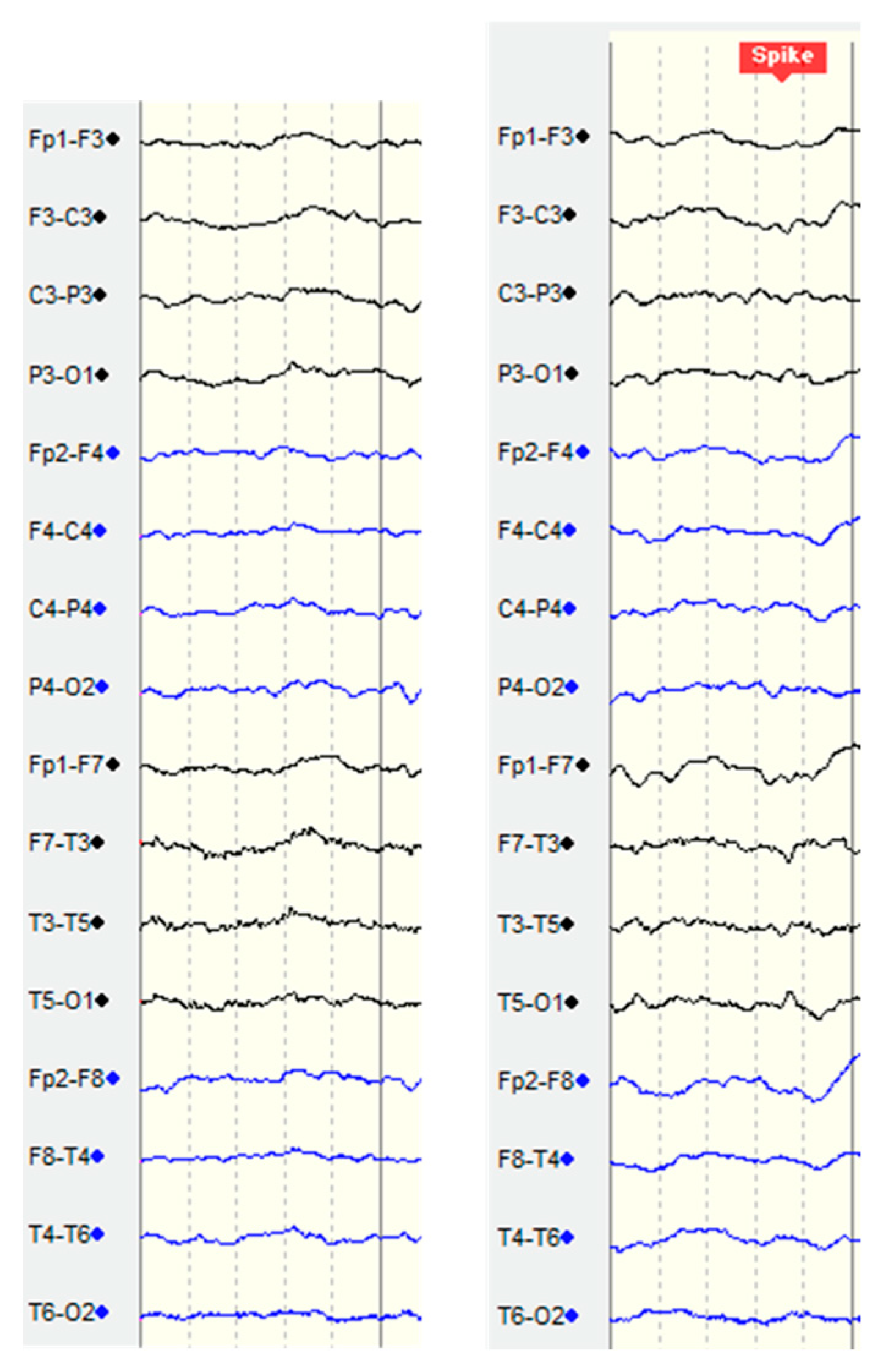
3.3. Preprocessing
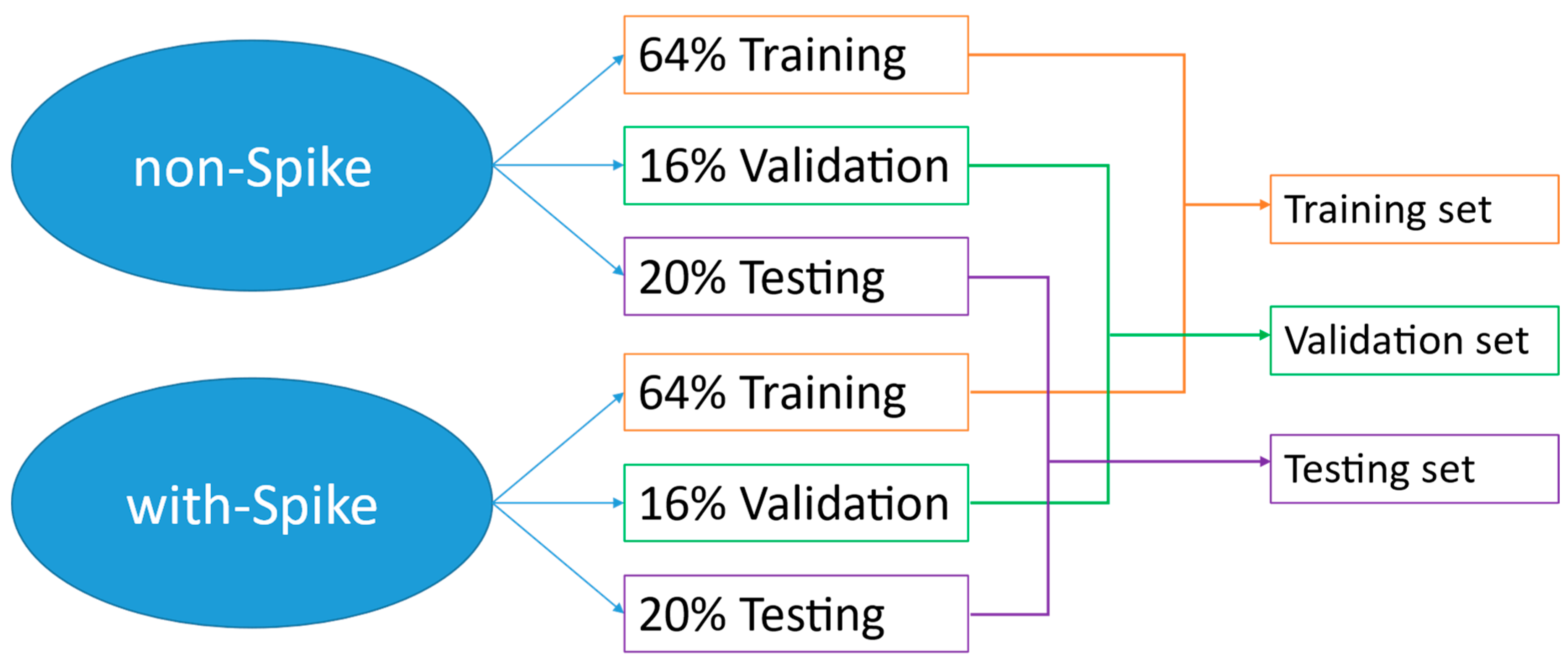
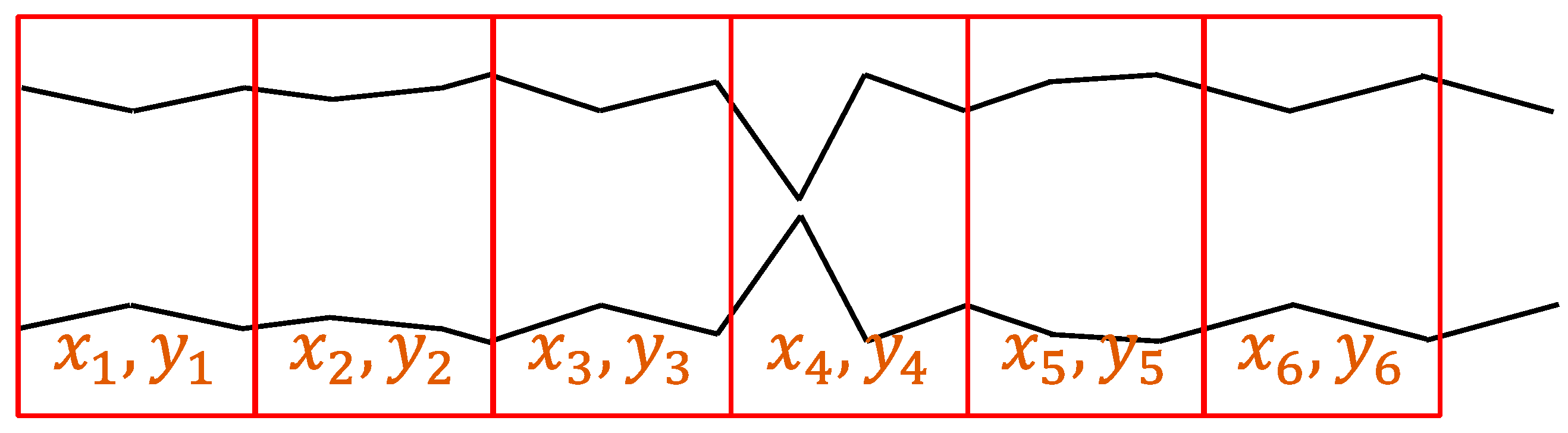
3.4. Sampling Method
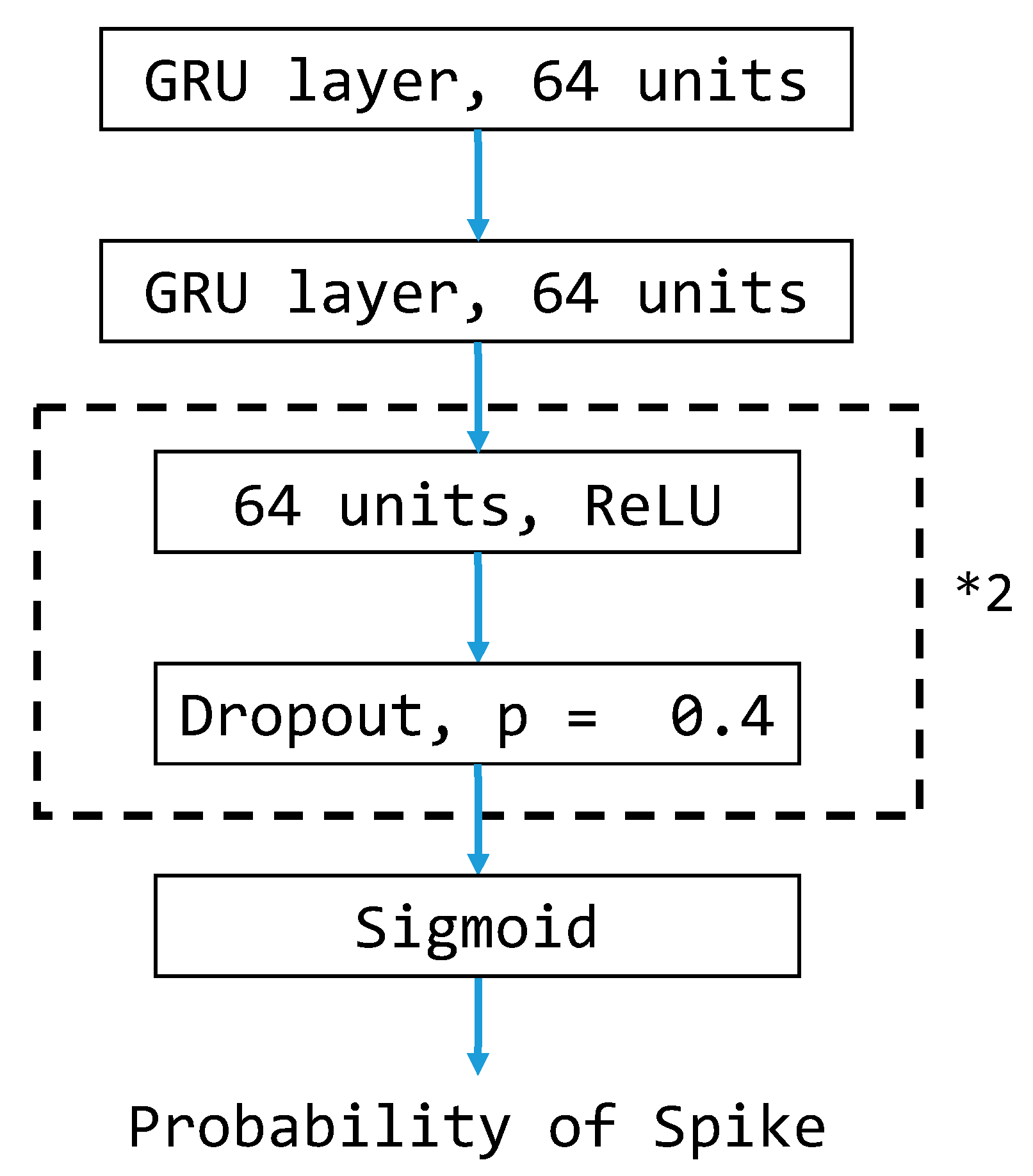
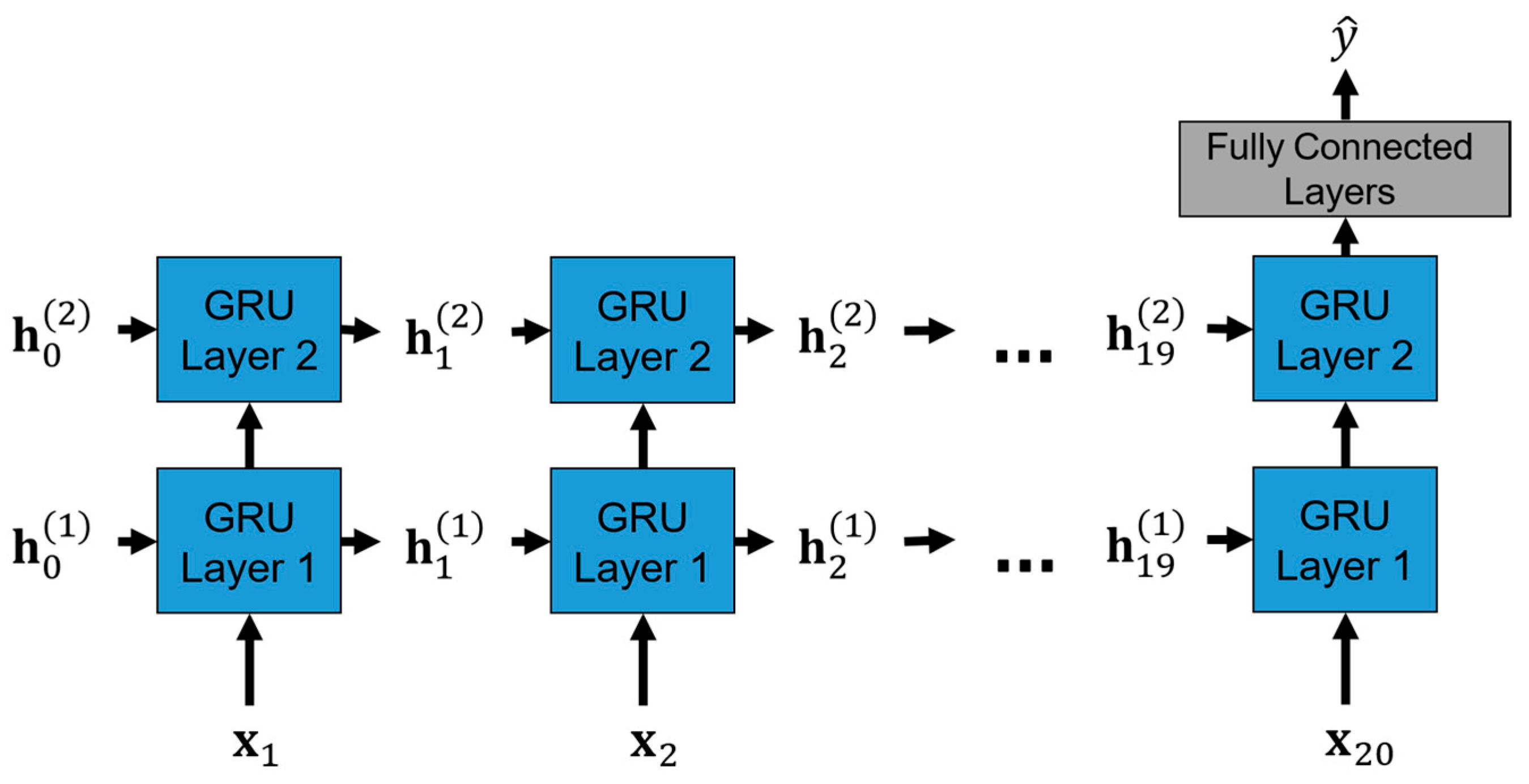
3.5. GRU-Based Model Architecture
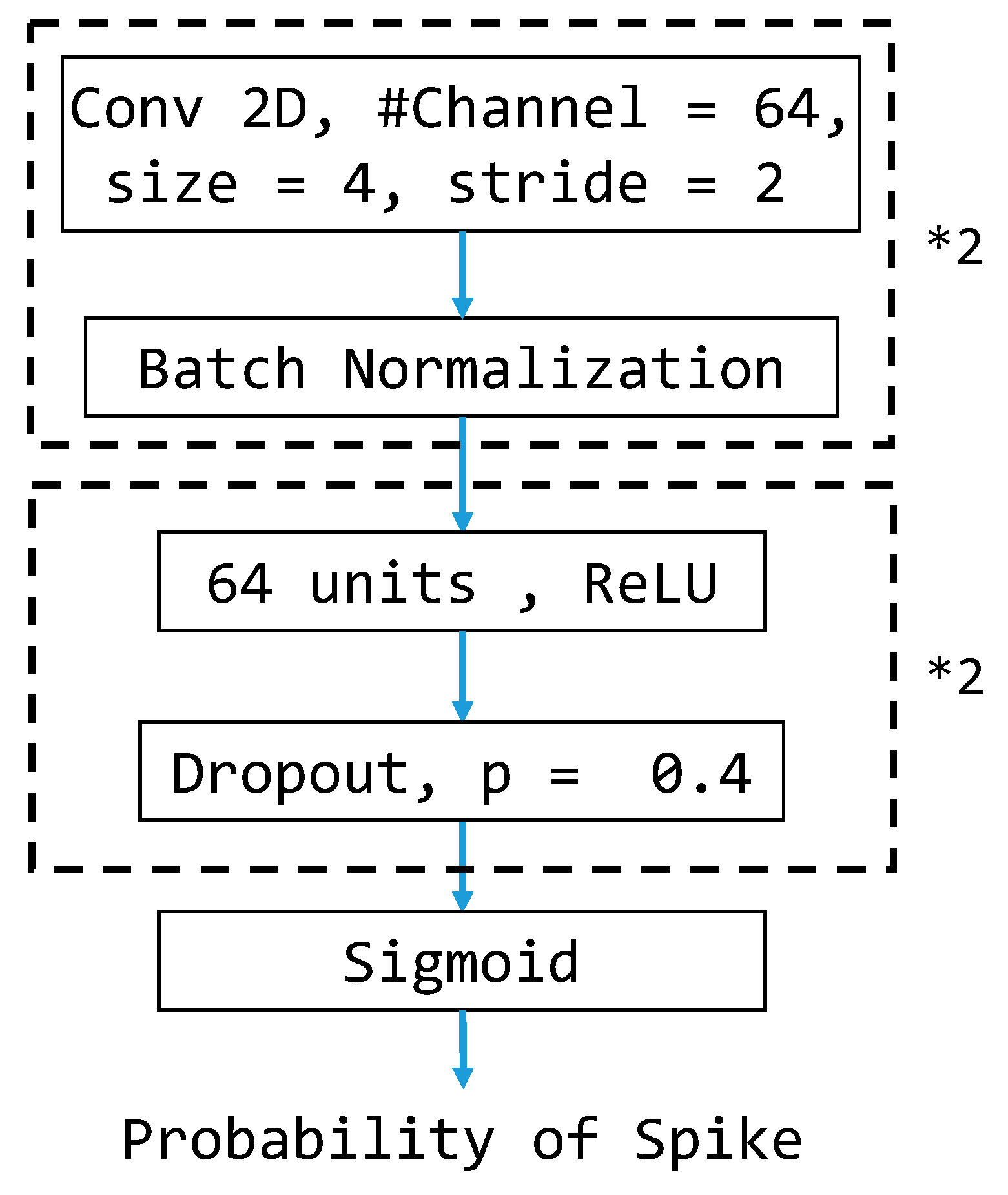
3.6. CNN-Based Model Architecture
3.7. Class Weight
3.8. Performance Metrics
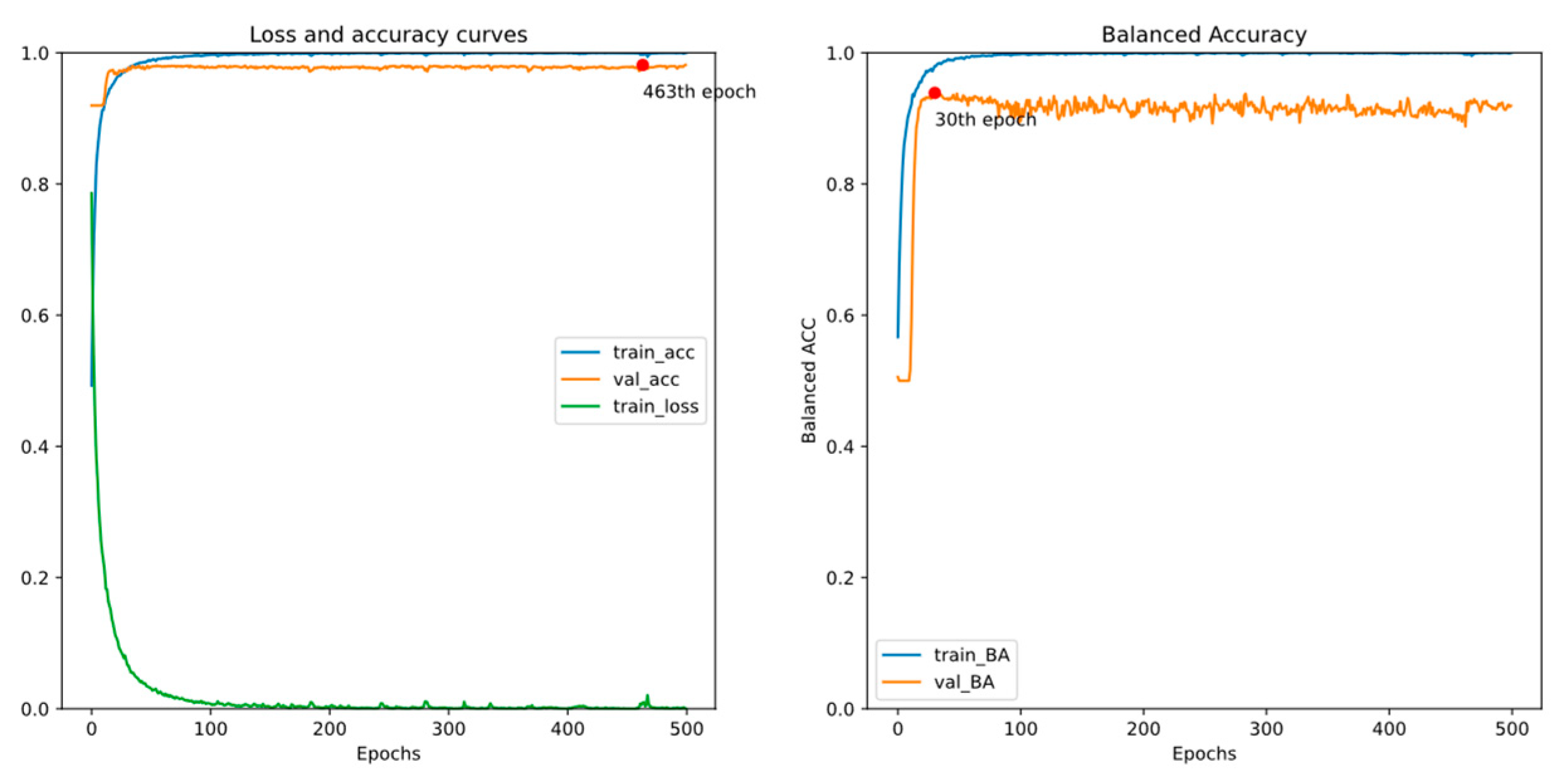
4. Experiments and Results
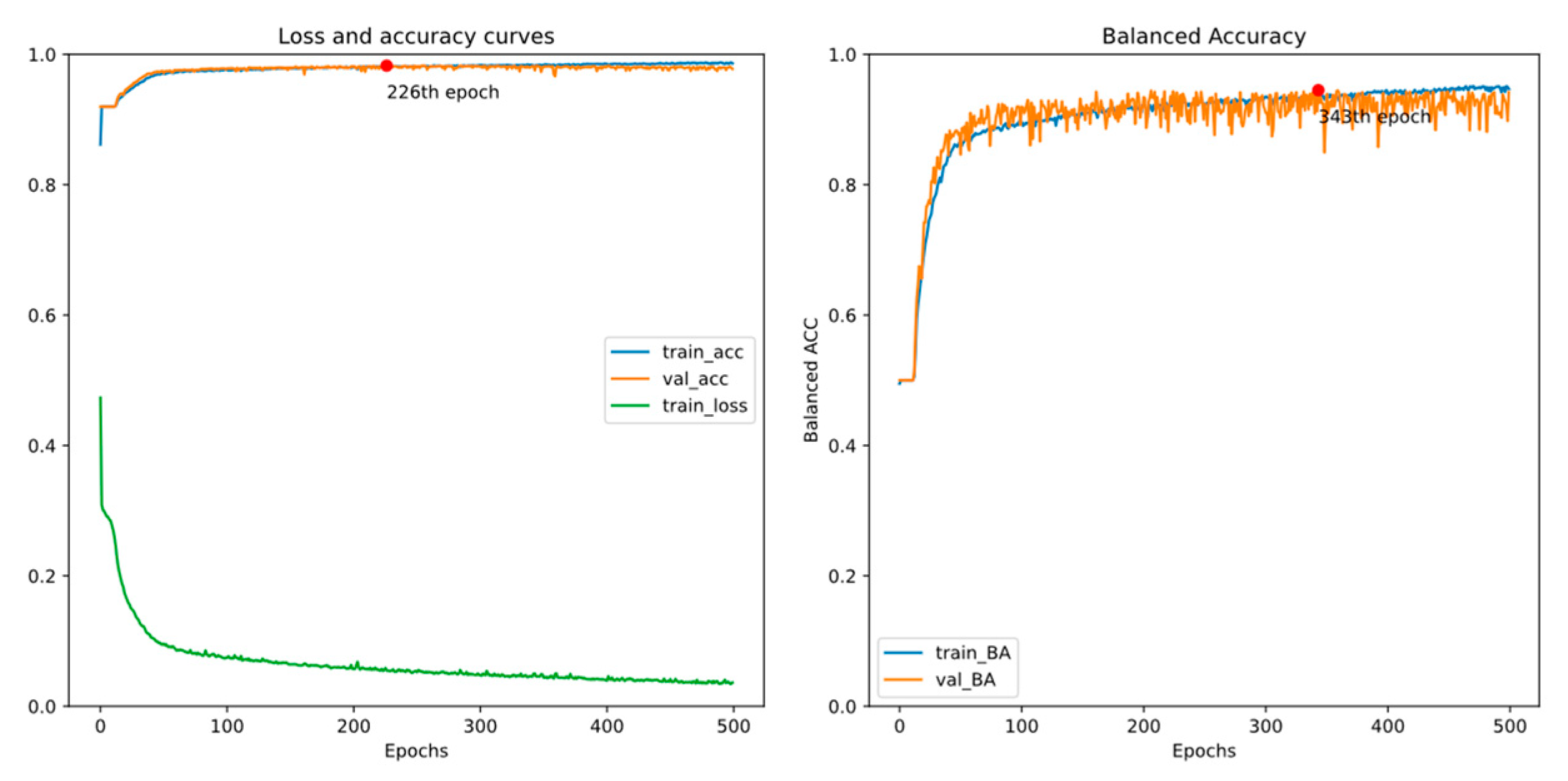
4.1. Experiment 1
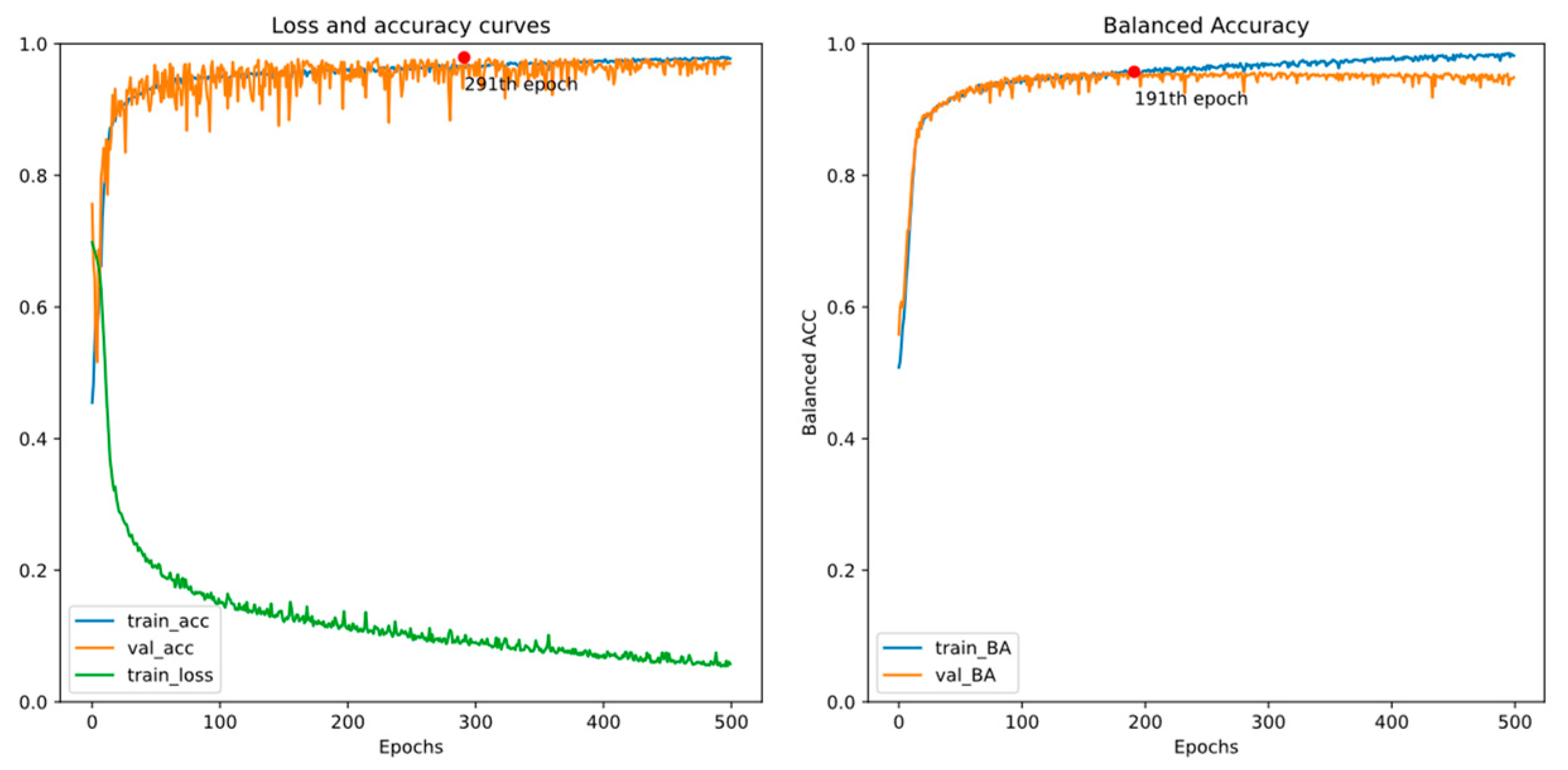
4.2. Experiment 2
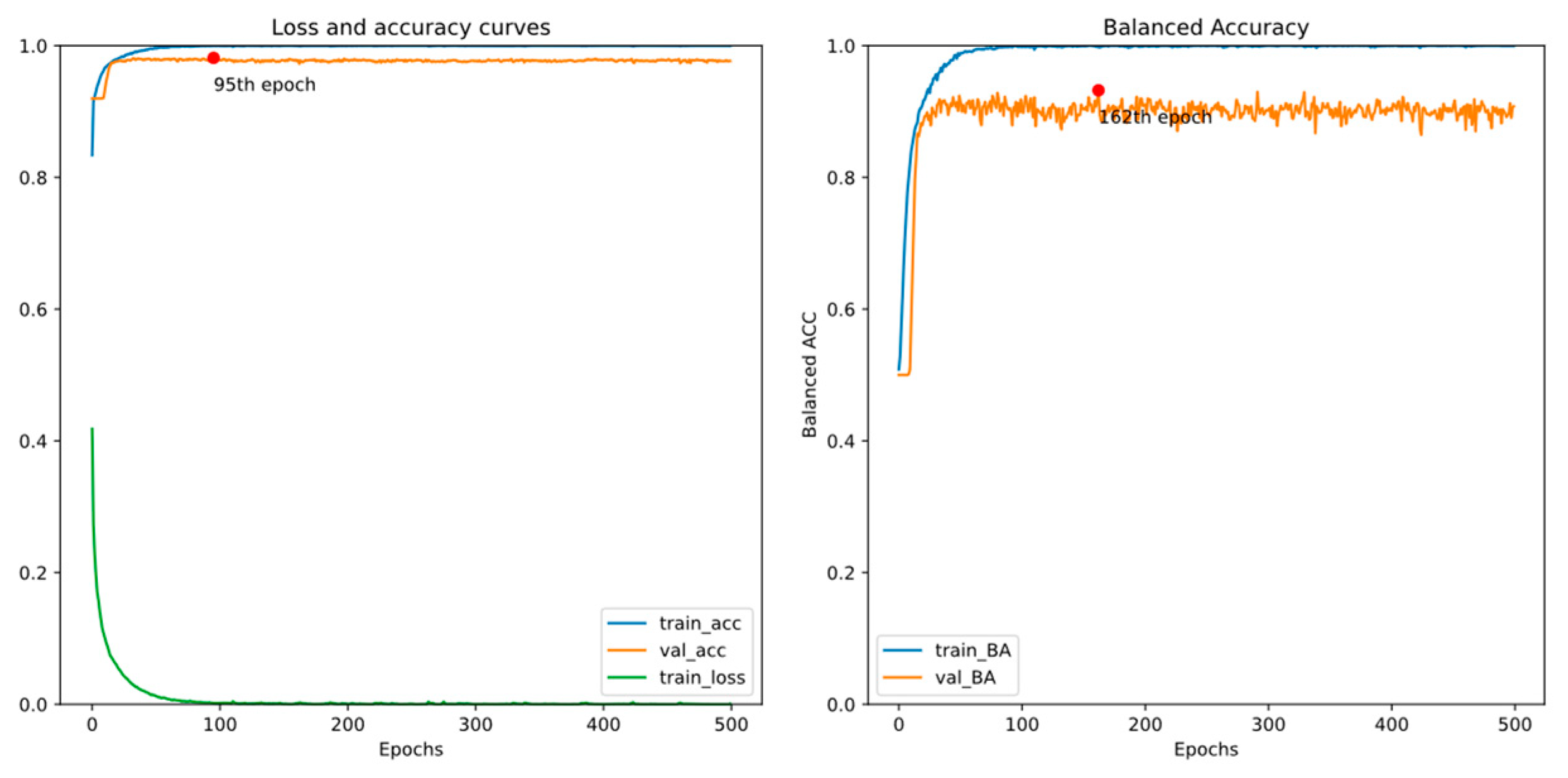
4.3. Early Detection
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kennedy, J.D.; Gerard, E.E. Continuous EEG Monitoring in the Intensive Care Unit. Curr. Neurol. Neurosci. Rep. 2012, 12, 419–428. [Google Scholar] [CrossRef]
- Craik, A.; He, Y.; Contreras-Vidal, J.L. Deep learning for electroencephalogram (EEG) classification tasks: A review. J. Neural Eng. 2019, 16, 031001. [Google Scholar] [CrossRef]
- Chambon, S.; Galtier, M.N.; Arnal, P.J.; Wainrib, G.; Gramfort, A. A Deep Learning Architecture for Temporal Sleep Stage Classification Using Multivariate and Multimodal Time Series. IEEE Trans. Neural Syst. Rehabil. Eng. 2018, 26, 758–769. [Google Scholar] [CrossRef]
- Biswal, S.; Kulas, J.; Sun, H.; Goparaju, B.; Westover, M.B.; Bianchi, M.T.; Sun, J. SLEEPNET: Automated Sleep Staging System via Deep Learning. arXiv 2017, arXiv:1707.08262. [Google Scholar] [CrossRef]
- Eldele, E.; Chen, Z.; Liu, C.; Wu, M.; Kwoh, C.K.; Li, X.; Guan, C. An Attention-Based Deep Learning Approach for Sleep Stage Classification With Single-Channel EEG. IEEE Trans. Neural Syst. Rehabil. Eng. 2021, 29, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Jirayucharoensak, S.; Pan-Ngum, S.; Israsena, P. EEG-Based Emotion Recognition Using Deep Learning Network with Principal Component Based Covariate Shift Adaptation. Sci. World J. 2014, 2014, 627892. [Google Scholar] [CrossRef]
- Chen, J.X.; Zhang, P.W.; Mao, Z.J.; Huang, Y.F.; Jiang, D.M.; Zhang, Y.N. Accurate EEG-Based Emotion Recognition on Combined Features Using Deep Convolutional Neural Networks. IEEE Access 2019, 7, 44317–44328. [Google Scholar] [CrossRef]
- Tabar, Y.R.; Halici, U. A novel deep learning approach for classification of EEG motor imagery signals. J. Neural Eng. 2017, 14, 016003. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.U.; Alsulaiman, M.; Muhammad, G.; Mekhtiche, M.A.; Hossain, M.S. Deep Learning for EEG motor imagery classification based on multi-layer CNNs feature fusion. Future Gener. Comput. Syst. 2019, 101, 542–554. [Google Scholar] [CrossRef]
- Yin, Z.; Zhang, J. Cross-session classification of mental workload levels using EEG and an adaptive deep learning model. Biomed. Signal Process. Control. 2017, 33, 30–47. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, X.; Zhang, W.; Chen, J. Learning Spatial–Spectral–Temporal EEG Features With Recurrent 3D Convolutional Neural Networks for Cross-Task Mental Workload Assessment. IEEE Trans. Neural Syst. Rehabil. Eng. 2019, 27, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Hussein, R.; Palangi, H.; Ward, R.; Wang, Z.J. Epileptic Seizure Detection: A Deep Learning Approach. arXiv 2018, arXiv:1803.09848. [Google Scholar] [CrossRef]
- Cecotti, H.; Eckstein, M.P.; Giesbrecht, B. Single-Trial Classification of Event-Related Potentials in Rapid Serial Visual Presentation Tasks Using Supervised Spatial Filtering. IEEE Trans. Neural Netw. Learn. Syst. 2014, 25, 2030–2042. [Google Scholar] [CrossRef] [PubMed]
- Vahid, A.; Bluschke, A.; Roessner, V.; Stober, S.; Beste, C. Deep Learning Based on Event-Related EEG Differentiates Children with ADHD from Healthy Controls. J. Clin. Med. 2019, 8, 1055. [Google Scholar] [CrossRef] [PubMed]
- Wilson, S.B.; Emerson, R. Spike detection: A review and comparison of algorithms. Clin. Neurophysiol. 2002, 113, 1873–1881. [Google Scholar] [CrossRef] [PubMed]
- Halford, J.J. Computerized epileptiform transient detection in the scalp electroencephalogram: Obstacles to progress and the example of computerized ECG interpretation. Clin. Neurophysiol. 2009, 120, 1909–1915. [Google Scholar] [CrossRef] [PubMed]
- Gotman, J.; Gloor, P. Automatic recognition and quantification of interictal epileptic activity in the human scalp EEG. Electroencephalogr. Clin. Neurophysiol. 1976, 41, 513–529. [Google Scholar] [CrossRef]
- Exarchos, T.P.; Tzallas, A.T.; Fotiadis, D.I.; Konitsiotis, S.; Giannopoulos, S. EEG transient event detection and classification using association rules. IEEE Trans. Inf. Technol. Biomed. 2006, 10, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Ji, Z.; Sugi, T.; Goto, S.; Wang, X.; Ikeda, A.; Nagamine, T.; Shibasaki, H.; Nakamura, M. An automatic spike detection system based on elimination of false positives using the large-area context in the scalp EEG. IEEE Trans. Biomed. Eng. 2011, 58, 2478–2488. [Google Scholar] [CrossRef]
- Adjouadi, M.; Cabrerizo, M.; Ayala, M.; Sanchez, D.; Yaylali, I.; Jayakar, P.; Barreto, A. A new mathematical approach based on orthogonal operators for the detection of interictal spikes in epileptogenic data. Biomed. Sci. Instrum. 2004, 40, 175–180. [Google Scholar]
- Indiradevi, K.P.; Elias, E.; Sathidevi, P.S.; Dinesh Nayak, S.; Radhakrishnan, K. A multi-level wavelet approach for automatic detection of epileptic spikes in the electroencephalogram. Comput. Biol. Med. 2008, 38, 805–816. [Google Scholar] [CrossRef]
- Acir, N.; Oztura, I.; Kuntalp, M.; Baklan, B.; Guzelis, C. Automatic detection of epileptiform events in EEG by a three-stage procedure based on artificial neural networks. IEEE Trans. Biomed. Eng. 2005, 52, 30–40. [Google Scholar] [CrossRef]
- Tzallas, A.T.; Karvelis, P.S.; Katsis, C.D.; Fotiadis, D.I.; Giannopoulos, S.; Konitsiotis, S. A method for classification of transient events in EEG recordings: Application to epilepsy diagnosis. Methods Inf. Med. 2006, 45, 610–621. [Google Scholar] [CrossRef]
- Güler, I.; Übeyli, E.D. Adaptive neuro-fuzzy inference system for classification of EEG signals using wavelet coefficients. J. Neurosci. Methods 2005, 148, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Abi-Chahla, F. Nvidia’s CUDA: The End of the CPU? Tom’s Hardware. 2008. Available online: https://www.tomshardware.com/reviews/nvidia-cuda-gpu,1954.html (accessed on 26 March 2024).
- Rácz, M.; Liber, C.; Németh, E.; Fiáth, R.; Rokai, J.; Harmati, I.; Ulbert, I.; Márton, G. Spike detection and sorting with deep learning. J. Neural Eng. 2020, 17, 016038. [Google Scholar] [CrossRef]
- Fukumori, K.; Nguyen, H.T.T.; Yoshida, N.; Tanaka, T. Fully Data-driven Convolutional Filters with Deep Learning Models for Epileptic Spike Detection. In Proceedings of the ICASSP 2019—2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 2772–2776. [Google Scholar]
- Johansen, A.R.; Jin, J.; Maszczyk, T.; Dauwels, J.; Cash, S.S.; Westover, M.B. Epileptiform spike detection via convolutional neural networks. In Proceedings of the 2016 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Shanghai, China, 20–25 March 2016; pp. 754–758. [Google Scholar] [CrossRef][Green Version]
- Saif-ur-Rehman, M.; Lienkämper, R.; Parpaley, Y.; Wellmer, J.; Liu, C.; Lee, B.; Kellis, S.; Andersen, R.; Iossifidis, I.; Glasmachers, T.; et al. SpikeDeeptector: A deep-learning based method for detection of neural spiking activity. J. Neural Eng. 2019, 16, 056003. [Google Scholar] [CrossRef]
- Thirunavukarasu, R.; Gnanasambandan, R.; Gopikrishnan, M.; Palanisamy, V. Towards computational solutions for precision medicine based big data healthcare system using deep learning models: A review. Comput. Biol. Med. 2022, 149, 106020. [Google Scholar] [CrossRef] [PubMed]
- Miotto, R.; Wang, F.; Wang, S.; Jiang, X.; Dudley, J.T. Deep learning for healthcare: Review, opportunities and challenges. Brief. Bioinform. 2017, 19, 1236–1246. [Google Scholar] [CrossRef]
- Kotei, E.; Thirunavukarasu, R. Computational techniques for the automated detection of mycobacterium tuberculosis from digitized sputum smear microscopic images: A systematic review. Prog. Biophys. Mol. Biol. 2022, 171, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Rajkomar, A.; Oren, E.; Chen, K.; Dai, A.M.; Hajaj, N.; Hardt, M.; Liu, P.J.; Liu, X.; Marcus, J.; Sun, M.; et al. Scalable and accurate deep learning with electronic health records. npj Digit. Med. 2018, 1, 18. [Google Scholar] [CrossRef]
- Ting, D.S.W.; Peng, L.; Varadarajan, A.V.; Keane, P.A.; Burlina, P.M.; Chiang, M.F.; Schmetterer, L.; Pasquale, L.R.; Bressler, N.M.; Webster, D.R.; et al. Deep learning in ophthalmology: The technical and clinical considerations. Prog. Retin. Eye Res. 2019, 72, 100759. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Tan, J.; Han, D.; Zhu, H. From machine learning to deep learning: Progress in machine intelligence for rational drug discovery. Drug Discov. Today 2017, 22, 1680–1685. [Google Scholar] [CrossRef]
- Singh, R.; Lanchantin, J.; Robins, G.; Qi, Y. DeepChrome: Deep-learning for predicting gene expression from histone modifications. Bioinformatics 2016, 32, i639–i648. [Google Scholar] [CrossRef]
- Niedermeyer, E.; Lopes da Silva, F.H. Electroencephalography: Basic Principles, Clinical Applications, and Related Fields; Lippincott Williams & Wilkins: Baltimore, MD, USA, 2004. [Google Scholar]
- Tatum, W.O. Handbook of EEG Interpretation; Demos Medical Publishing: New York, NY, USA, 2014. [Google Scholar]
- トマトン124. Electrode Locations of International 10-20 System for EEG (Electroencephalography) Recording. 2010. Available online: https://commons.wikimedia.org/wiki/File:21_electrodes_of_International_10-20_system_for_EEG.svg (accessed on 26 March 2024).
- Kemp, B.; Värri, A.; Rosa, A.C.; Nielsen, K.D.; Gade, J. A simple format for exchange of digitized polygraphic recordings. Electroencephalogr. Clin. Neurophysiol. 1992, 82, 391–393. [Google Scholar] [CrossRef] [PubMed]
- Kemp, B.; Olivan, J. European data format ‘plus’ (EDF+), an EDF alike standard format for the exchange of physiological data. Clin. Neurophysiol. 2003, 114, 1755–1761. [Google Scholar] [CrossRef] [PubMed]
- Louis, E.K.S.; Frey, L.C.; Britton, J.W.; Hopp, J.L.; Korb, P.; Koubeissi, M.Z.; Lievens, W.E.; Pestana-Knight, E.M.; Foundation, C.C.C. Electroencephalography (EEG): An Introductory Text and Atlas of Normal and Abnormal Findings in Adults, Children, and Infants; Louis, E.K.S., Frey, L.C., Eds.; American Epilepsy Society: Chicago, IL, USA, 2016. [Google Scholar]
- Chatrian, G.E. A glossary of terms most commonly used by clinical electroencephalographers. Electroenceph. Clin. Neurophysiol. 1974, 37, 538–548. [Google Scholar]
- Wu, J.C.; Yu, H.W.; Tsai, T.H.; Lu, H.H. Dynamically Synthetic Images for Federated Learning of medical images. Comput. Methods Programs Biomed. 2023, 242, 107845. [Google Scholar] [CrossRef]


| Objective | Method | ICU Patients | Montage | Window Size | Performance |
|---|---|---|---|---|---|
| transient event classification [18] | mimetic analysis, power spectral analysis | No | bipolar | 355 ms | 87.38% accuracy |
| spike detection [19] | template matching | No | average reference, bipolar | 5.12 s | 92.6% selectivity |
| IED detection [21] | wavelet analysis | No | average reference | 3 s | 90.5% accuracy |
| epileptic activity classification [23] | artificial neural network | No | bipolar | 355 ms | 84.48% accuracy |
| spike detection [28] | deep learning | No | average reference | 0.5 s | 0.947 AUC |
| spike detection (ours) | deep learning | Yes | bipolar | 160 ms | 94.66% balanced accuracy |
| Predicted Class | |||
|---|---|---|---|
| Positive | Negative | ||
| True Class | Positive | True Positive (TP) | False Negative (FN) |
| Negative | False Positive (FP) | True Negative (TN) | |
| Model | By | Accuracy | Sensitivity | Specificity | BA | |
|---|---|---|---|---|---|---|
| No Class Weight | GRU | Acc | 98.19 | 85.24 | 99.33 | 92.28 |
| BA | 97.32 | 91.54 | 97.83 | 94.68 | ||
| CNN | Acc | 97.93 | 81.30 | 99.38 | 90.34 | |
| BA | 97.21 | 87.80 | 98.04 | 92.92 | ||
| With Class Weight | GRU | Acc | 97.83 | 87.99 | 98.69 | 93.34 |
| BA | 95.95 | 93.11 | 96.19 | 94.65 | ||
| CNN | Acc | 97.61 | 81.10 | 99.05 | 90.08 | |
| BA | 97.15 | 90.16 | 97.76 | 93.96 | ||
| Zero-rule Baseline | 91.95 | 0.00 | 100.00 | 50.00 | ||
| Model | By | Accuracy | Sensitivity | Specificity | BA | |
|---|---|---|---|---|---|---|
| No Class Weight | GRU | Acc | 98.02 | 82.12 | 99.41 | 90.77 |
| BA | 97.58 | 90.18 | 98.23 | 94.20 | ||
| CNN | Acc | 97.80 | 85.27 | 98.90 | 92.08 | |
| BA | 97.13 | 88.21 | 97.92 | 93.06 | ||
| With Class Weight | GRU | Acc | 97.93 | 85.27 | 99.04 | 92.15 |
| BA | 95.95 | 93.12 | 96.19 | 94.66 | ||
| CNN | Acc | 97.94 | 83.50 | 99.21 | 91.35 | |
| BA | 97.61 | 89.98 | 98.28 | 94.13 | ||
| Zero-rule Baseline | 91.94 | 0.00 | 100.00 | 50.00 | ||
| # of Windows Early | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| Accuracy | 95.95 | 93.38 | 93.65 | 94.26 | 93.15 | 94.18 | 94.96 | 93.65 | 94.97 | 96.57 |
| Sensitivity | 93.12 | 93.59 | 90.82 | 93.22 | 92.37 | 91.56 | 89.36 | 92.33 | 85.54 | 82.04 |
| Specificity | 96.19 | 93.36 | 93.83 | 94.34 | 93.21 | 94.35 | 95.31 | 93.74 | 95.61 | 97.49 |
| Balanced Accuracy | 94.66 | 93.48 | 92.33 | 93.78 | 92.79 | 92.95 | 92.34 | 93.03 | 90.57 | 89.76 |
| Zero-rule Accuracy | 91.94 | 93.82 | 93.79 | 93.69 | 93.77 | 93.99 | 94.04 | 93.80 | 93.64 | 94.08 |
| Zero-rule Baseline | Sensitivity: 0.00 | Specificity: 100.00 | Balanced Accuracy: 50.00 | |||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.C.-H.; Liao, N.-C.; Yang, T.-H.; Hsieh, C.-C.; Huang, J.-A.; Pai, Y.-W.; Huang, Y.-J.; Wu, C.-L.; Lu, H.H.-S. Deep-Learning-Based Automated Anomaly Detection of EEGs in Intensive Care Units. Bioengineering 2024, 11, 421. https://doi.org/10.3390/bioengineering11050421
Wu JC-H, Liao N-C, Yang T-H, Hsieh C-C, Huang J-A, Pai Y-W, Huang Y-J, Wu C-L, Lu HH-S. Deep-Learning-Based Automated Anomaly Detection of EEGs in Intensive Care Units. Bioengineering. 2024; 11(5):421. https://doi.org/10.3390/bioengineering11050421
Chicago/Turabian StyleWu, Jacky Chung-Hao, Nien-Chen Liao, Ta-Hsin Yang, Chen-Cheng Hsieh, Jin-An Huang, Yen-Wei Pai, Yi-Jhen Huang, Chieh-Liang Wu, and Henry Horng-Shing Lu. 2024. "Deep-Learning-Based Automated Anomaly Detection of EEGs in Intensive Care Units" Bioengineering 11, no. 5: 421. https://doi.org/10.3390/bioengineering11050421
APA StyleWu, J. C.-H., Liao, N.-C., Yang, T.-H., Hsieh, C.-C., Huang, J.-A., Pai, Y.-W., Huang, Y.-J., Wu, C.-L., & Lu, H. H.-S. (2024). Deep-Learning-Based Automated Anomaly Detection of EEGs in Intensive Care Units. Bioengineering, 11(5), 421. https://doi.org/10.3390/bioengineering11050421








