RNA Sequencing Revealed a Weak Response of Gingival Fibroblasts Exposed to Hyaluronic Acid
Abstract
1. Introduction
2. Materials and Methods
2.1. Gingival Fibroblasts and Macrophages
2.2. Total RNA Isolation, Sequencing, and Data Analysis
2.3. Reverse Transcription Quantitative Real-Time PCR (RT-qPCR)
2.4. Volcano Plot, Heat Map, and Gene Set Enrichment Analysis
| ATAD2B-F: ATTCATGCGCTAAAGGAAATGGT | ATAD2B-R: AGGAGGGCCATAAAACAAACAG [49] |
| CXCL1-F: TCCTGCATCCCCCATAGTTA | CXCL1-R: CTTCAGGAACAGCCACCAGT [50] |
| CXCL10-F TGCCATTCTGATTTGCTGCC | CXCL10-R: TGCAGGTACAGCGTACAGTT [51] |
| CXCL2-F: CCCATGGTTAAGAAAATCATCG | CXCL2-R: CTTCAGGAACAGCCACCAAT [52] |
| DGKE-F: GACGGGCACCTGATCTTGTG | DGKE-R: CTGGAGGCTACACCAGAAGG [53] |
| DUSP4-F: GGGGTCCTGTGGAGATCCTT | DUSP4-R: GGCAGTCCGAGGAGACATTC [54] |
| FOXC2-F: CCTCCTGGTATCTCAACCACA | FOXC2-R: GAGGGTCGAGTTCTCAATCCC [55] |
| GAPDH-F: AGCCACATCGCTCAGACAC | GAPDH-R: GCCCAATACGACCAAATCC [56] |
| HBEGF-F: ATCGTGGGGCTTCTCATGTTT | HBEGF-R: TTAGTCATGCCCAACTTCACTTT [57] |
| IL8/CXCL8-F: AACTTCTCCACAACCCTCTG | IL8/CXCL8-R: TTGGCAGCCTTCCTGATTTC [58] |
| PLK3-F: TTTTCGCACCACTTTGAGGAC | PLK3-R: GAGGCCAGAAAGGATCTGCC [59] |
| PTGS2-F: CCTGTGCCTGATGATTGC | PTGS2-R: CTGATGCGTGAAGTGCTG [60] |
| SLC16A6-F: CGCTGTGTTTGCTTTCGCACCA | SLC16A6-R: TTTTCGGTGACGCTGGTCCTCT [61] |
3. Results
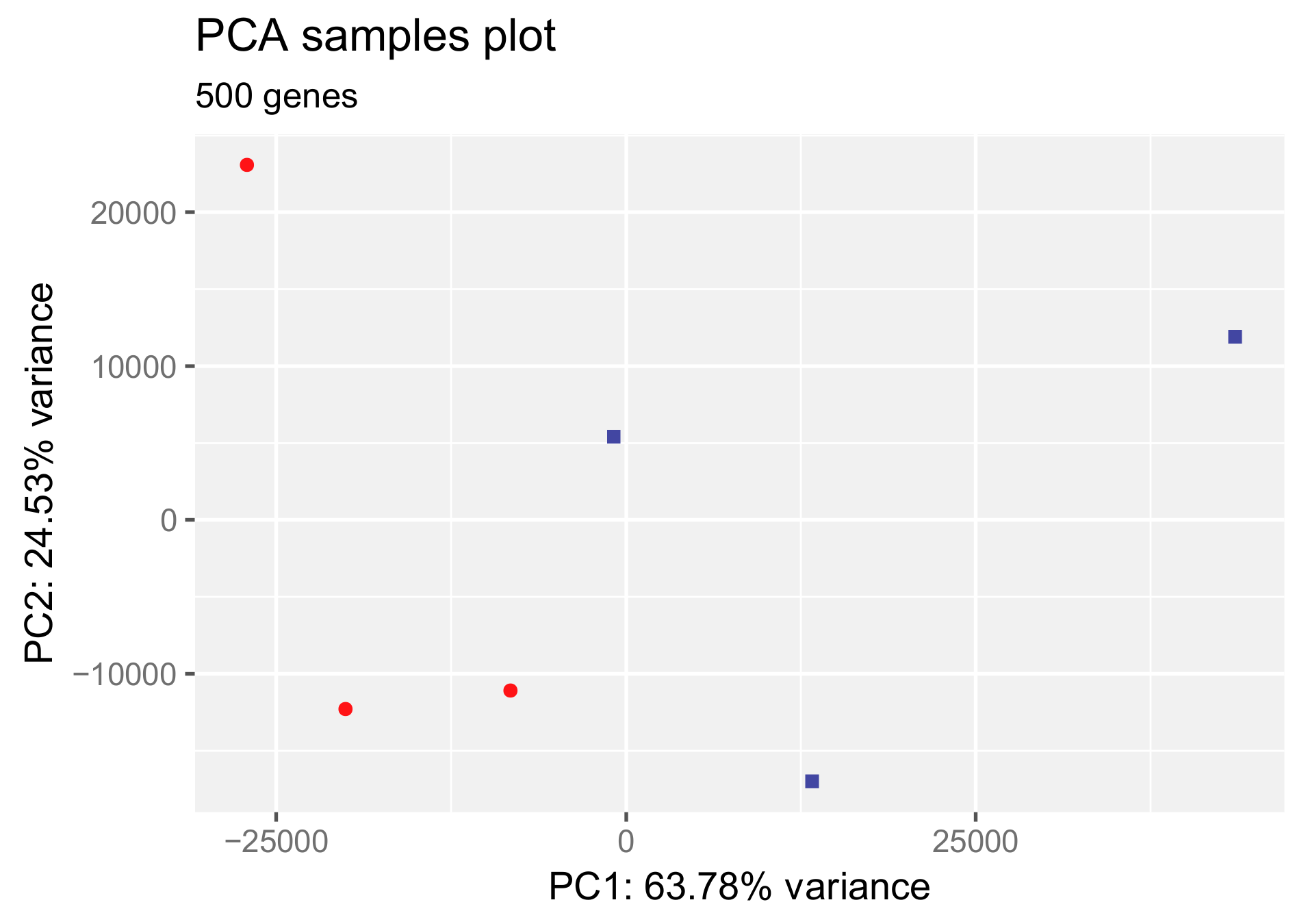
3.1. Principal Component Analysis and Heat Map of Gene Expression Changes by HAc
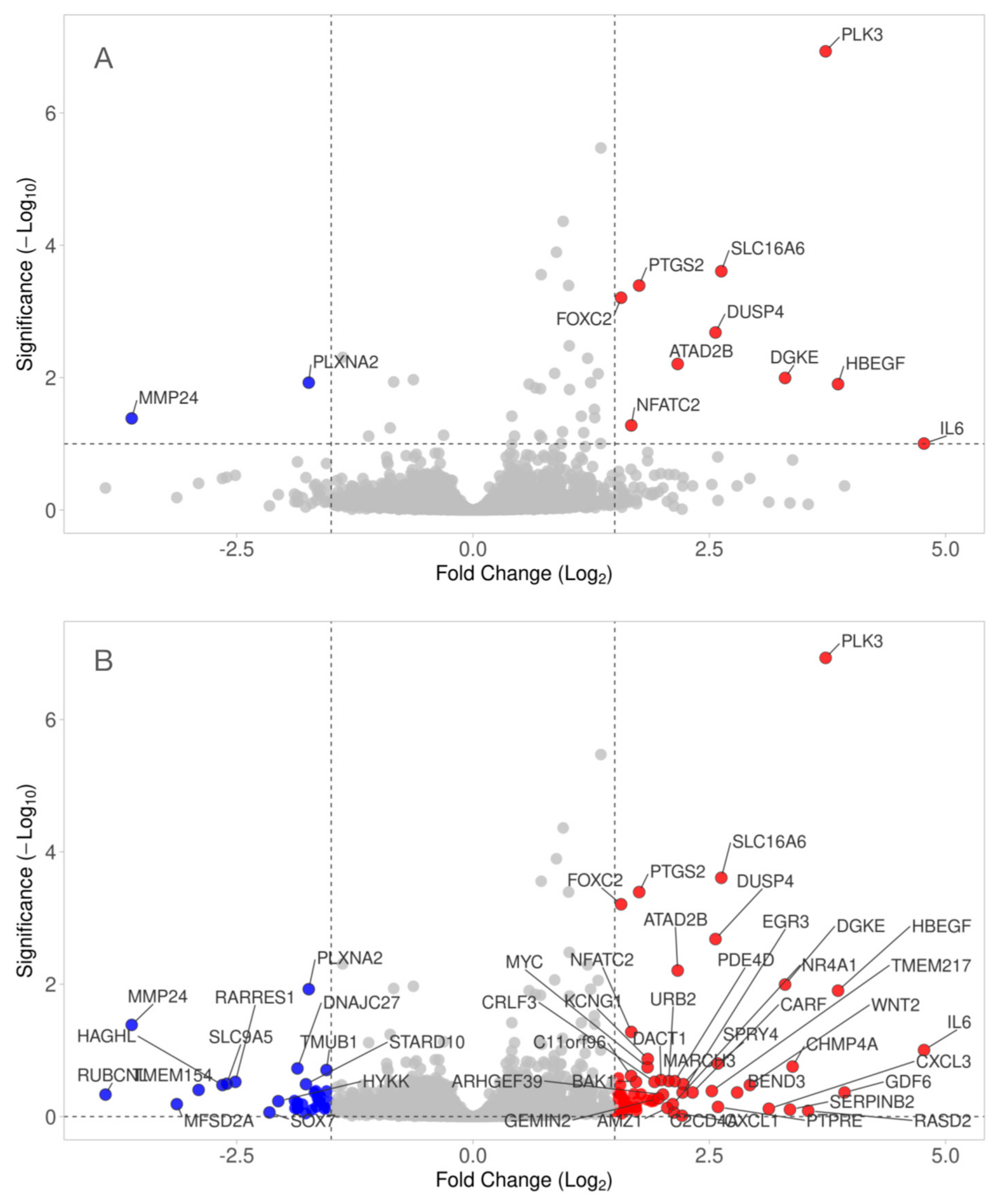
3.2. Volcano Blot of Gene Expression Changes by HAc
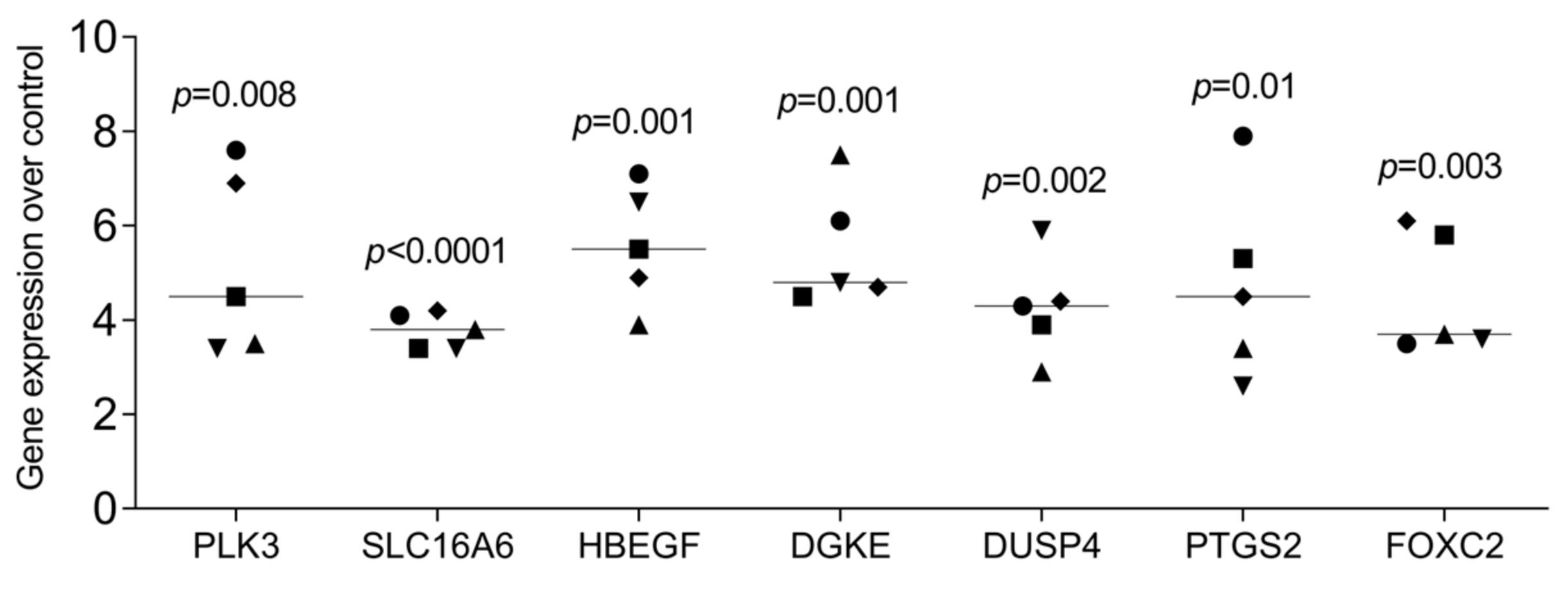
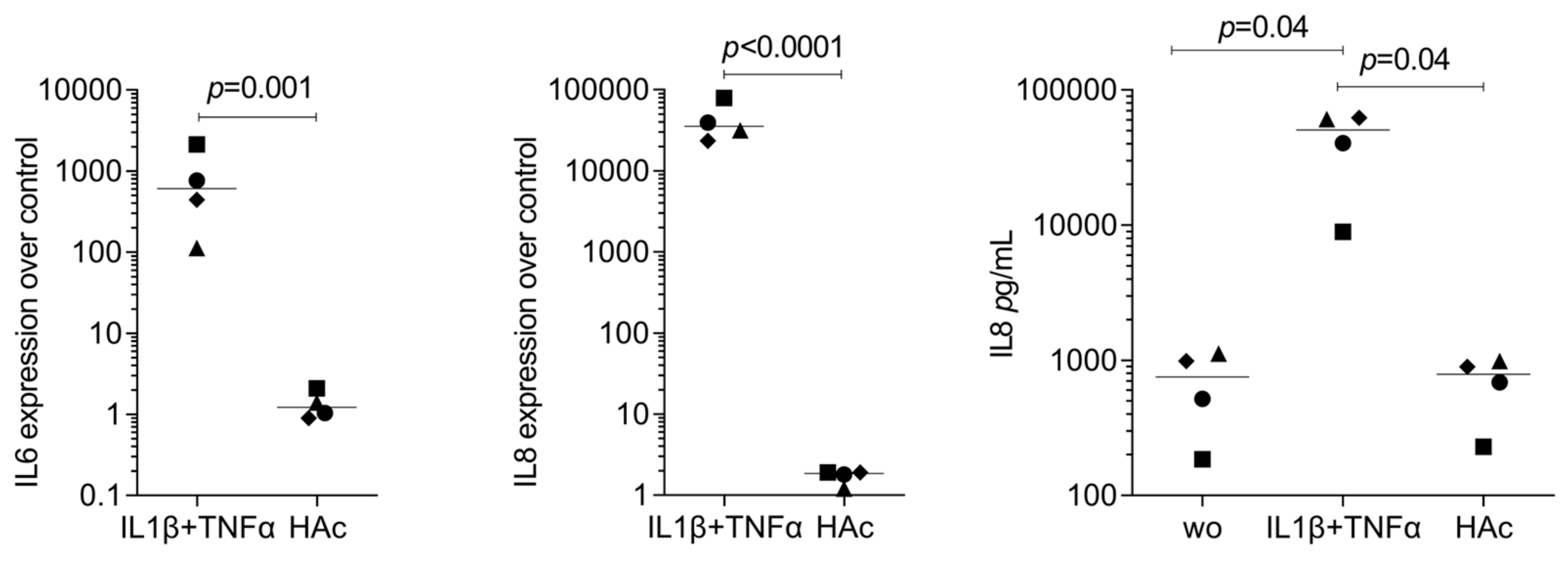
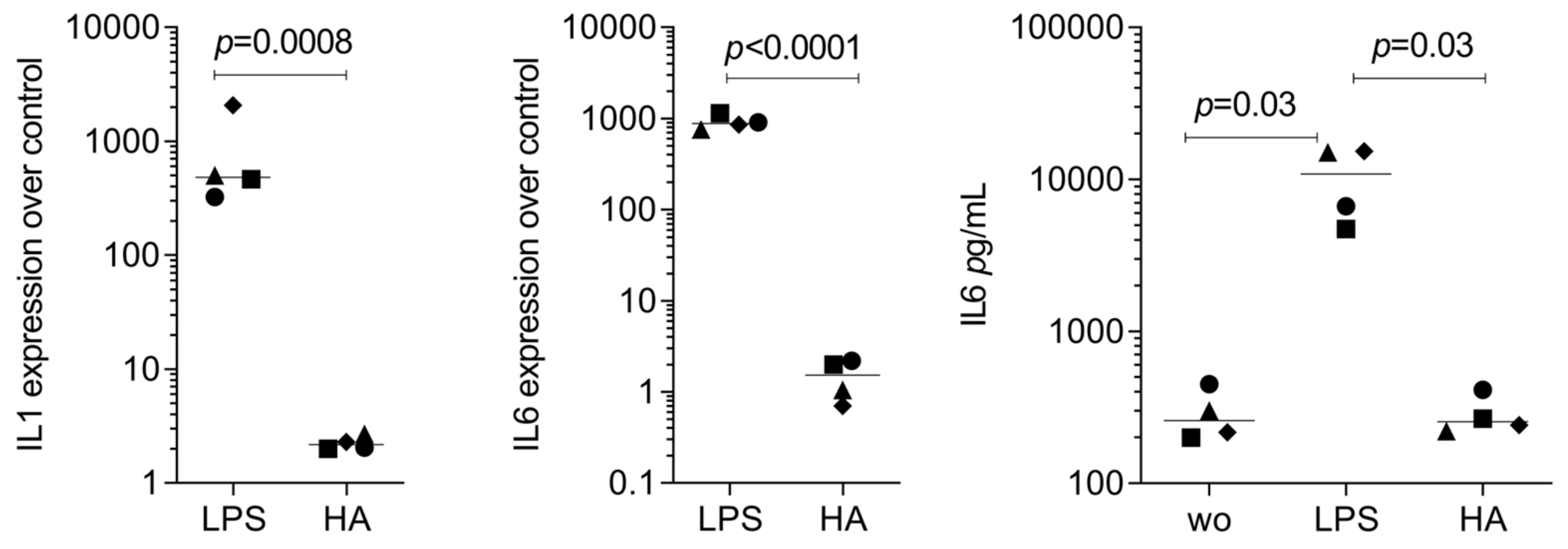
3.3. RT-PCR Gene Expression Changes by HAc
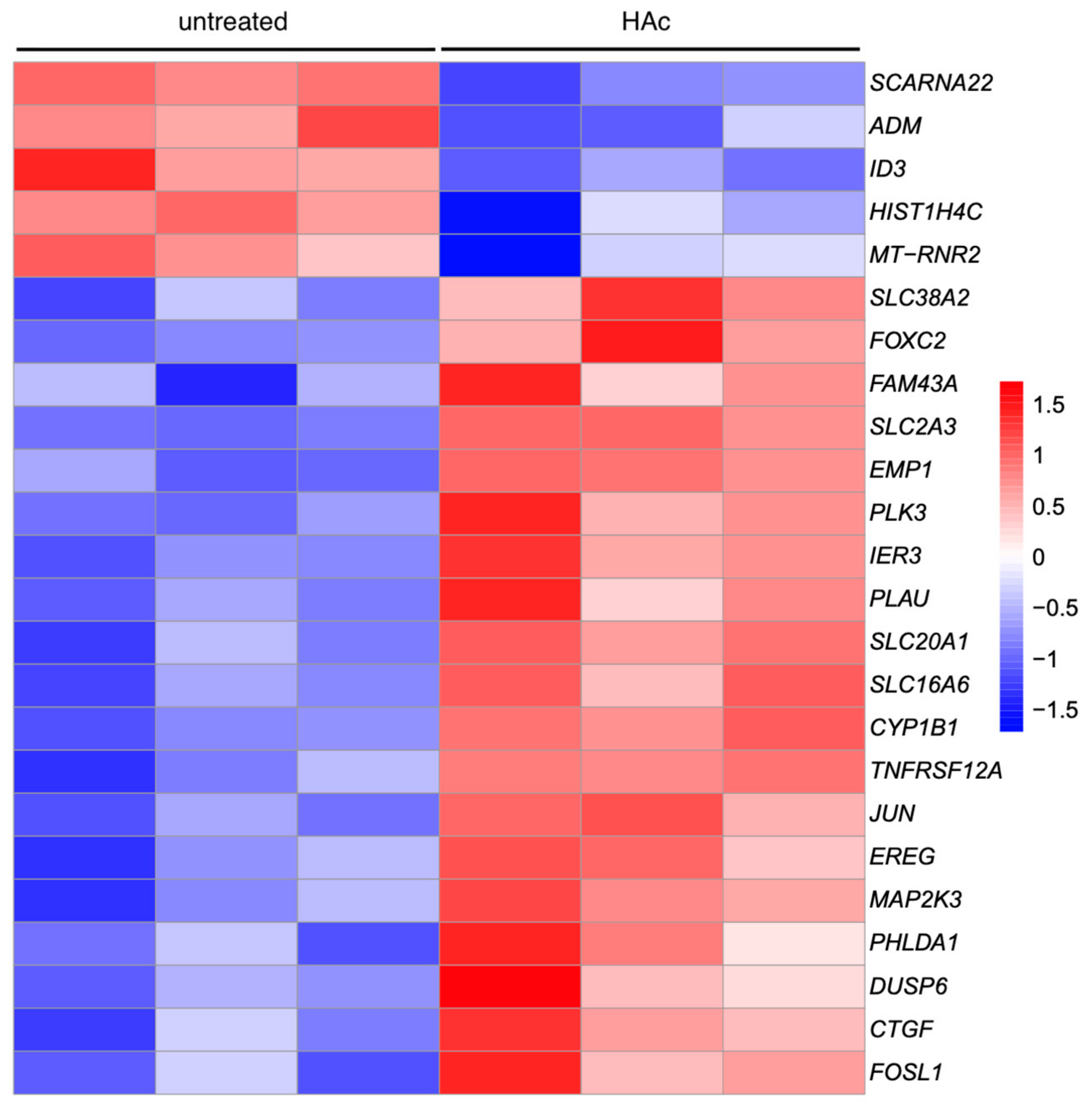
3.4. Heat Map of Gene Expression Changes by HAc
3.5. STRING Cluster Analysis of Differentially Expressed Genes by HAc
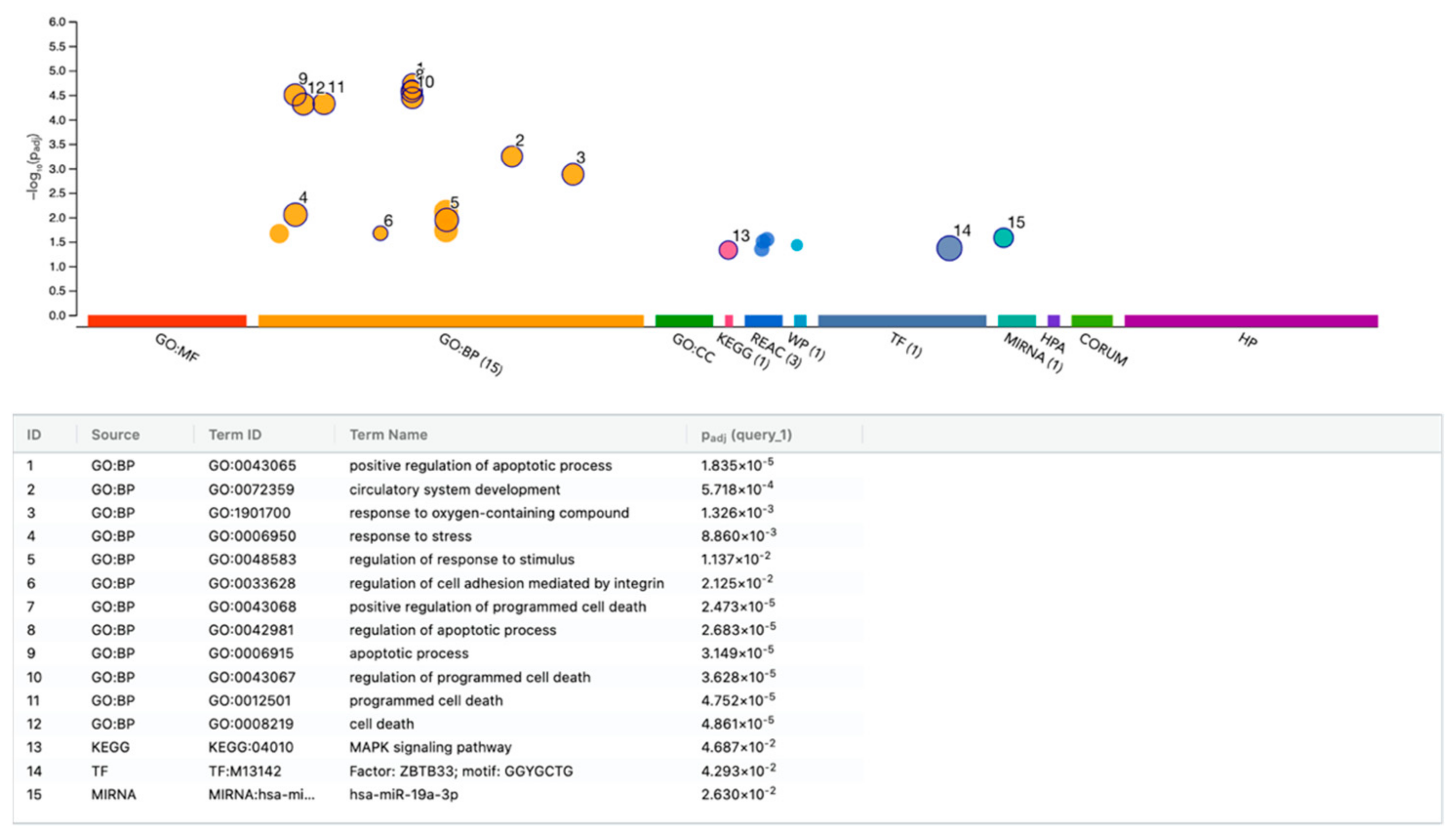
3.6. g:Profiler Analysis of Gene Expression Changes by HAc
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fundaro, S.P.; Salti, G.; Malgapo, D.M.H.; Innocenti, S. The Rheology and Physicochemical Characteristics of Hyaluronic Acid Fillers: Their Clinical Implications. Int. J. Mol. Sci. 2022, 23, 10518. [Google Scholar] [CrossRef] [PubMed]
- Tamer, T.M. Hyaluronan and synovial joint: Function, distribution and healing. Interdiscip. Toxicol. 2013, 6, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Papakonstantinou, E.; Roth, M.; Karakiulakis, G. Hyaluronic acid: A key molecule in skin aging. Dermato-Endocrinology 2012, 4, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Fagerholm, P.; Fitzsimmons, T.; Harfstrand, A.; Schenholm, M. Reactive formation of hyaluronic acid after small and large lens injury. Acta Ophthalmol. 1992, 70, 58–64. [Google Scholar] [CrossRef]
- Hynnekleiv, L.; Magno, M.; Vernhardsdottir, R.R.; Moschowits, E.; Tonseth, K.A.; Dartt, D.A.; Vehof, J.; Utheim, T.P. Hyaluronic acid in the treatment of dry eye disease. Acta Ophthalmol. 2022, 100, 844–860. [Google Scholar] [CrossRef]
- Tammi, R.; Tammi, M.; Hakkinen, L.; Larjava, H. Histochemical localization of hyaluronate in human oral epithelium using a specific hyaluronate-binding probe. Arch. Oral Biol. 1990, 35, 219–224. [Google Scholar] [CrossRef]
- Ijuin, C.; Ohno, S.; Tanimoto, K.; Honda, K.; Tanne, K. Regulation of hyaluronan synthase gene expression in human periodontal ligament cells by tumour necrosis factor-alpha, interleukin-1beta and interferon-gamma. Arch. Oral Biol. 2001, 46, 767–772. [Google Scholar] [CrossRef]
- Stern, R.; Jedrzejas, M.J. Hyaluronidases: Their genomics, structures, and mechanisms of action. Chem. Rev. 2006, 106, 818–839. [Google Scholar] [CrossRef]
- Entwistle, J.; Hall, C.L.; Turley, E.A. HA receptors: Regulators of signalling to the cytoskeleton. J. Cell Biochem. 1996, 61, 569–577. [Google Scholar] [CrossRef]
- Peach, R.J.; Hollenbaugh, D.; Stamenkovic, I.; Aruffo, A. Identification of hyaluronic acid binding sites in the extracellular domain of CD44. J. Cell Biol. 1993, 122, 257–264. [Google Scholar] [CrossRef]
- Aruffo, A.; Stamenkovic, I.; Melnick, M.; Underhill, C.B.; Seed, B. CD44 is the principal cell surface receptor for hyaluronate. Cell 1990, 61, 1303–1313. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Yang, B.L.; Savani, R.C.; Turley, E.A. Identification of a common hyaluronan binding motif in the hyaluronan binding proteins RHAMM, CD44 and link protein. EMBO J. 1994, 13, 286–296. [Google Scholar] [CrossRef]
- McCourt, P.A.; Gustafson, S. On the adsorption of hyaluronan and ICAM-1 to modified hydrophobic resins. Int. J. Biochem. Cell Biol. 1997, 29, 1179–1189. [Google Scholar] [CrossRef] [PubMed]
- Knudson, W.; Chow, G.; Knudson, C.B. CD44-mediated uptake and degradation of hyaluronan. Matrix Biol. 2002, 21, 15–23. [Google Scholar] [CrossRef]
- Zhang, Y.W.; Mess, J.; Aizarani, N.; Mishra, P.; Johnson, C.; Romero-Mulero, M.C.; Rettkowski, J.; Schonberger, K.; Obier, N.; Jacklein, K.; et al. Hyaluronic acid-GPRC5C signalling promotes dormancy in haematopoietic stem cells. Nat. Cell Biol. 2022, 24, 1038–1048. [Google Scholar] [CrossRef]
- Moutsopoulos, N.M.; Moutsopoulos, H.M. The oral mucosa: A barrier site participating in tissue-specific and systemic immunity. Oral Dis. 2018, 24, 22–25. [Google Scholar] [CrossRef]
- Dutzan, N.; Abusleme, L.; Bridgeman, H.; Greenwell-Wild, T.; Zangerle-Murray, T.; Fife, M.E.; Bouladoux, N.; Linley, H.; Brenchley, L.; Wemyss, K.; et al. On-going Mechanical Damage from Mastication Drives Homeostatic Th17 Cell Responses at the Oral Barrier. Immunity 2017, 46, 133–147. [Google Scholar] [CrossRef]
- Larjava, H.; Hakkinen, L.; Rahemtulla, F. A biochemical analysis of human periodontal tissue proteoglycans. Biochem. J. 1992, 284 Pt 1, 267–274. [Google Scholar] [CrossRef]
- Arumuganainar, D.; Krishna Naik, V.; Ramadoss, R.; Balasundaram, A.; Victor, D.J. Evaluation of CD44 antigen in type 2 diabetic patients with periodontitis: An immunohistochemical study. Dent. Med. Probl. 2024, 61, 225–231. [Google Scholar] [CrossRef]
- Jin, S.H.; Lee, J.E.; Yun, J.H.; Kim, I.; Ko, Y.; Park, J.B. Isolation and characterization of human mesenchymal stem cells from gingival connective tissue. J. Periodontal Res. 2015, 50, 461–467. [Google Scholar] [CrossRef]
- Lin, N.H.; Menicanin, D.; Mrozik, K.; Gronthos, S.; Bartold, P.M. Putative stem cells in regenerating human periodontium. J. Periodontal Res. 2008, 43, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Trubiani, O.; Di Primio, R.; Traini, T.; Pizzicannella, J.; Scarano, A.; Piattelli, A.; Caputi, S. Morphological and cytofluorimetric analysis of adult mesenchymal stem cells expanded ex vivo from periodontal ligament. Int. J. Immunopathol. Pharmacol. 2005, 18, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Takigawa, M.; Takashiba, S.; Nagai, A.; Miyamoto, M.; Kurihara, H.; Murayama, Y. Role of cytokine in the induction of adhesion molecules on cultured human gingival fibroblasts. J. Periodontol. 1994, 65, 230–235. [Google Scholar] [CrossRef]
- Zhu, X.; von Werdt, L.; Zappala, G.; Sculean, A.; Eick, S.; Stahli, A. In vitro activity of hyaluronic acid and human serum on periodontal biofilm and periodontal ligament fibroblasts. Clin. Oral Investig. 2023, 27, 5021–5029. [Google Scholar] [CrossRef]
- Casale, M.; Moffa, A.; Vella, P.; Sabatino, L.; Capuano, F.; Salvinelli, B.; Lopez, M.A.; Carinci, F.; Salvinelli, F. Hyaluronic acid: Perspectives in dentistry. A systematic review. Int. J. Immunopathol. Pharmacol. 2016, 29, 572–582. [Google Scholar] [CrossRef]
- Shirakata, Y.; Imafuji, T.; Nakamura, T.; Shinohara, Y.; Iwata, M.; Setoguchi, F.; Noguchi, K.; Sculean, A. Cross-linked hyaluronic acid gel with or without a collagen matrix in the treatment of class III furcation defects: A histologic and histomorphometric study in dogs. J. Clin. Periodontol. 2022, 49, 1079–1089. [Google Scholar] [CrossRef]
- Shirakata, Y.; Nakamura, T.; Kawakami, Y.; Imafuji, T.; Shinohara, Y.; Noguchi, K.; Sculean, A. Healing of buccal gingival recessions following treatment with coronally advanced flap alone or combined with a cross-linked hyaluronic acid gel. An experimental study in dogs. J. Clin. Periodontol. 2021, 48, 570–580. [Google Scholar] [CrossRef]
- Eliezer, M.; Sculean, A.; Miron, R.J.; Nemcovsky, C.; Weinberg, E.; Weinreb, M.; Zoabi, H.; Bosshardt, D.D.; Fujioka-Kobayashi, M.; Moses, O. Hyaluronic acid slows down collagen membrane degradation in uncontrolled diabetic rats. J. Periodontal Res. 2019, 54, 644–652. [Google Scholar] [CrossRef]
- Vela, O.C.; Boariu, M.; Rusu, D.; Iorio-Siciliano, V.; Sculean, A.; Stratul, S.I. Clinical and Radiographic Evaluation of Intrabony Periodontal Defects Treated with Hyaluronic Acid or Enamel Matrix Proteins: A 6-Month Prospective Study. Oral Health Prev. Dent. 2024, 22, 257–270. [Google Scholar] [CrossRef]
- Bilhan, H.; Friedmann, A. The “Tunneled Sandwich” Technique for Preserving the Buccal Tissue Volume After Immediate Implantation: A Retrospective Report of 10 Cases. Int. J. Periodontics Restor. Dent. 2023, 43, s53–s64. [Google Scholar] [CrossRef]
- Husseini, B.; Friedmann, A.; Wak, R.; Ghosn, N.; Khoury, G.; El Ghoul, T.; Abboud, C.K.; Younes, R. Clinical and radiographic assessment of cross-linked hyaluronic acid addition in demineralized bovine bone based alveolar ridge preservation: A human randomized split-mouth pilot study. J. Stomatol. Oral Maxillofac. Surg. 2023, 124, 101426. [Google Scholar] [CrossRef] [PubMed]
- Kalimeri, E.; Roccuzzo, A.; Stahli, A.; Oikonomou, I.; Berchtold, A.; Sculean, A.; Kloukos, D. Adjunctive use of hyaluronic acid in the treatment of gingival recessions: A systematic review and meta-analysis. Clin. Oral Investig. 2024, 28, 329. [Google Scholar] [CrossRef] [PubMed]
- Onisor, F.; Bran, S.; Mester, A.; Voina-Tonea, A. Efficiency of Hyaluronic Acid in Infrabony Defects: A Systematic Review of Human Clinical Trials. Medicina 2022, 58, 580. [Google Scholar] [CrossRef] [PubMed]
- Joshi, V.M.; Kandaswamy, E.; Germain, J.S.; Schiavo, J.H.; Fm, H.S. Effect of hyaluronic acid on palatal wound healing: A systematic review. Clin. Oral Investig. 2024, 28, 565. [Google Scholar] [CrossRef]
- Makdisi, J.; Akbari, S.; Zayeri, F.; AslRoosta, H.; Yaghobee, S. Application of Hyaluronic Acid for Treatment of Interdental Papillary Deficiency: A Systematic Review and Meta-Analysis. Front. Dent. 2023, 20, 19. [Google Scholar] [CrossRef]
- Fujioka-Kobayashi, M.; Muller, H.D.; Mueller, A.; Lussi, A.; Sculean, A.; Schmidlin, P.R.; Miron, R.J. In vitro effects of hyaluronic acid on human periodontal ligament cells. BMC Oral Health 2017, 17, 44. [Google Scholar] [CrossRef]
- Asparuhova, M.B.; Chappuis, V.; Stahli, A.; Buser, D.; Sculean, A. Role of hyaluronan in regulating self-renewal and osteogenic differentiation of mesenchymal stromal cells and pre-osteoblasts. Clin. Oral Investig. 2020, 24, 3923–3937. [Google Scholar] [CrossRef]
- Asparuhova, M.B.; Kiryak, D.; Eliezer, M.; Mihov, D.; Sculean, A. Activity of two hyaluronan preparations on primary human oral fibroblasts. J. Periodontal Res. 2019, 54, 33–45. [Google Scholar] [CrossRef]
- Kessler-Becker, D.; Krieg, T.; Eckes, B. Expression of pro-inflammatory markers by human dermal fibroblasts in a three-dimensional culture model is mediated by an autocrine interleukin-1 loop. Biochem. J. 2004, 379, 351–358. [Google Scholar] [CrossRef]
- Froget, S.; Barthelemy, E.; Guillot, F.; Soler, C.; Coudert, M.C.; Benbunan, M.; Dosquet, C. Wound healing mediator production by human dermal fibroblasts grown within a collagen-GAG matrix for skin repair in humans. Eur. Cytokine Netw. 2003, 14, 60–64. [Google Scholar]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Imani, A.; Panahipour, L.; Kuhtreiber, H.; Mildner, M.; Gruber, R. RNAseq of Gingival Fibroblasts Exposed to PRF Membrane Lysates and PRF Serum. Cells 2024, 13, 1308. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Gerst, R.; Hölzer, M. PCAGO: An interactive tool to analyze RNA-Seq data with principal component analysis. bioRxiv 2019, 1–4. [Google Scholar] [CrossRef]
- Goedhart, J.; Luijsterburg, M.S. VolcaNoseR is a web app for creating, exploring, labeling and sharing volcano plots. Sci. Rep. 2020, 10, 20560. [Google Scholar] [CrossRef]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Adler, P.; Vilo, J.; Peterson, H. g:Profiler-interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic Acids Res. 2023, 51, W207–W212. [Google Scholar] [CrossRef]
- Dudakovic, A.; Camilleri, E.T.; Xu, F.; Riester, S.M.; McGee-Lawrence, M.E.; Bradley, E.W.; Paradise, C.R.; Lewallen, E.A.; Thaler, R.; Deyle, D.R.; et al. Epigenetic Control of Skeletal Development by the Histone Methyltransferase Ezh2. J. Biol. Chem. 2015, 290, 27604–27617. [Google Scholar] [CrossRef]
- Luo, Q.; Liu, Y.; Shi, K.; Shen, X.; Yang, Y.; Liang, X.; Lu, L.; Qiao, W.; Chen, A.; Hong, D.; et al. An autonomous activation of interleukin-17 receptor signaling sustains inflammation and promotes disease progression. Immunity 2023, 56, 2006–2020.e2006. [Google Scholar] [CrossRef]
- Kong, W.; Zhao, G.; Chen, H.; Wang, W.; Shang, X.; Sun, Q.; Guo, F.; Ma, X. Analysis of therapeutic targets and prognostic biomarkers of CXC chemokines in cervical cancer microenvironment. Cancer Cell Int. 2021, 21, 399. [Google Scholar] [CrossRef]
- Wiley, C.D.; Velarde, M.C.; Lecot, P.; Liu, S.; Sarnoski, E.A.; Freund, A.; Shirakawa, K.; Lim, H.W.; Davis, S.S.; Ramanathan, A.; et al. Mitochondrial Dysfunction Induces Senescence with a Distinct Secretory Phenotype. Cell Metab. 2016, 23, 303–314. [Google Scholar] [CrossRef]
- Wang, Z.; Zhou, Z.; Zhang, Y.; Zuo, F.; Du, J.; Wang, M.; Hu, M.; Sun, Y.; Wang, X.; Liu, M.; et al. Diacylglycerol kinase epsilon protects against renal ischemia/reperfusion injury in mice through Kruppel-like factor 15/klotho pathway. Ren. Fail. 2022, 44, 902–913. [Google Scholar] [CrossRef] [PubMed]
- Kosinsky, R.L.; Gonzalez, M.M.; Saul, D.; Barros, L.L.; Sagstetter, M.R.; Fedyshyn, Y.; Nair, A.; Sun, Z.; Hamdan, F.H.; Gibbons, H.R.; et al. The FOXP3(+) Pro-Inflammatory T Cell: A Potential Therapeutic Target in Crohn’s Disease. Gastroenterology 2024, 166, 631–644. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Li, J.; Luo, Z.; Zhang, S.; Xue, S.; Wang, K.; Shi, Y.; Zhang, C.; Chen, H.; Li, Z. Roles of dopamine receptors and their antagonist thioridazine in hepatoma metastasis. Onco Targets Ther. 2015, 8, 1543–1552. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Arenas-Hernandez, M.; Vega-Sanchez, R. Housekeeping gene expression stability in reproductive tissues after mitogen stimulation. BMC Res. Notes 2013, 6, 285. [Google Scholar] [CrossRef]
- Hu, C.; Wang, S.; Wang, J.; Ruan, X.; Wu, L.; Zhang, Z.; Wang, X.; Zhang, J.; Liu, Y.; Li, Y.; et al. B7-H3 enhances colorectal cancer progression by regulating HB-EGF via HIF-1alpha. J. Gastrointest. Oncol. 2024, 15, 1035–1049. [Google Scholar] [CrossRef]
- Mao, P.; Wu, S.; Li, J.; Fu, W.; He, W.; Liu, X.; Slutsky, A.S.; Zhang, H.; Li, Y. Human alveolar epithelial type II cells in primary culture. Physiol. Rep. 2015, 3, e12288. [Google Scholar] [CrossRef]
- Li, C.; Park, S.; Zhang, X.; Dai, W.; Xu, D. Mutual regulation between Polo-like kinase 3 and SIAH2 E3 ubiquitin ligase defines a regulatory network that fine-tunes the cellular response to hypoxia and nickel. J. Biol. Chem. 2017, 292, 11431–11444. [Google Scholar] [CrossRef]
- Lan, J.; Deng, Z.; Wang, Q.; Li, D.; Fan, K.; Chang, J.; Ma, Y. Neuropeptide substance P attenuates colitis by suppressing inflammation and ferroptosis via the cGAS-STING signaling pathway. Int. J. Biol. Sci. 2024, 20, 2507–2531. [Google Scholar] [CrossRef]
- Thiecke, M.J.; Wutz, G.; Muhar, M.; Tang, W.; Bevan, S.; Malysheva, V.; Stocsits, R.; Neumann, T.; Zuber, J.; Fraser, P.; et al. Cohesin-Dependent and -Independent Mechanisms Mediate Chromosomal Contacts between Promoters and Enhancers. Cell Rep. 2020, 32, 107929. [Google Scholar] [CrossRef] [PubMed]
- Ang, Z.; Er, J.Z.; Tan, N.S.; Lu, J.; Liou, Y.C.; Grosse, J.; Ding, J.L. Human and mouse monocytes display distinct signalling and cytokine profiles upon stimulation with FFAR2/FFAR3 short-chain fatty acid receptor agonists. Sci. Rep. 2016, 6, 34145. [Google Scholar] [CrossRef] [PubMed]
- Kozlowska, A.K.; Florczak, A.; Smialek, M.; Dondajewska, E.; Mackiewicz, A.; Kortylewski, M.; Dams-Kozlowska, H. Functionalized bioengineered spider silk spheres improve nuclease resistance and activity of oligonucleotide therapeutics providing a strategy for cancer treatment. Acta Biomater. 2017, 59, 221–233. [Google Scholar] [CrossRef]
- Amirrasouli, M.M.; Shamsara, M. Comparing the in vivo and in vitro effects of hypoxia (3% O2) on directly derived cells from murine cardiac explants versus murine cardiosphere derived cells. J. Stem Cells Regen. Med. 2017, 13, 35–44. [Google Scholar] [CrossRef]
- Blattler, A.; Yao, L.; Wang, Y.; Ye, Z.; Jin, V.X.; Farnham, P.J. ZBTB33 binds unmethylated regions of the genome associated with actively expressed genes. Epigenetics Chromatin 2013, 6, 13. [Google Scholar] [CrossRef]
- Feng, S.; Zhu, X.; Fan, B.; Xie, D.; Li, T.; Zhang, X. miR-19a-3p targets PMEPA1 and induces prostate cancer cell proliferation, migration and invasion. Mol. Med. Rep. 2016, 13, 4030–4038. [Google Scholar] [CrossRef]
- David-Raoudi, M.; Tranchepain, F.; Deschrevel, B.; Vincent, J.C.; Bogdanowicz, P.; Boumediene, K.; Pujol, J.P. Differential effects of hyaluronan and its fragments on fibroblasts: Relation to wound healing. Wound Repair. Regen. 2008, 16, 274–287. [Google Scholar] [CrossRef]
- Olsson, M.; Bremer, L.; Aulin, C.; Harris, H.E. Fragmented hyaluronan has no alarmin function assessed in arthritis synovial fibroblast and chondrocyte cultures. Innate Immun. 2018, 24, 131–141. [Google Scholar] [CrossRef]
- Williams, D.W.; Greenwell-Wild, T.; Brenchley, L.; Dutzan, N.; Overmiller, A.; Sawaya, A.P.; Webb, S.; Martin, D.; Genomics, N.N.; Computational Biology, C.; et al. Human oral mucosa cell atlas reveals a stromal-neutrophil axis regulating tissue immunity. Cell 2021, 184, 4090–4104.e4015. [Google Scholar] [CrossRef]
- Li, J.; Ye, L.J.; Dai, Y.W.; Wang, H.W.; Gao, J.; Shen, Y.H.; Wang, F.; Dai, Q.G.; Wu, Y.Q. Single-cell analysis reveals a unique microenvironment in peri-implantitis. J. Clin. Periodontol. 2024, 51, 1665–1676. [Google Scholar] [CrossRef]
- Mo, J.J.; Lai, Y.R.; Huang, Q.R.; Li, Y.R.; Zhang, Y.J.; Chen, R.Y.; Qian, S.J. Single-cell sequencing identifies inflammation-promoting fibroblast-neutrophil interaction in peri-implantitis. J. Clin. Periodontol. 2024, 51, 196–208. [Google Scholar] [CrossRef] [PubMed]
- Cozza, G.; Salvi, M. The Acidophilic Kinases PLK2 and PLK3: Structure, Substrate Targeting and Inhibition. Curr. Protein Pept. Sci. 2018, 19, 728–745. [Google Scholar] [CrossRef] [PubMed]
- Uebanso, T.; Fukui, M.; Naito, C.; Shimohata, T.; Mawatari, K.; Takahashi, A. SLC16a6, mTORC1, and Autophagy Regulate Ketone Body Excretion in the Intestinal Cells. Biology 2023, 12, 1467. [Google Scholar] [CrossRef]
- Felmlee, M.A.; Jones, R.S.; Rodriguez-Cruz, V.; Follman, K.E.; Morris, M.E. Monocarboxylate Transporters (SLC16): Function, Regulation, and Role in Health and Disease. Pharmacol. Rev. 2020, 72, 466–485. [Google Scholar] [CrossRef]
- Almubarak, A.; Zhang, Q.; Zhang, C.H.; Abdelwahab, N.; Kume, T.; Lassar, A.B.; Berry, F.B. FOXC1 and FOXC2 regulate growth plate chondrocyte maturation towards hypertrophy in the embryonic mouse limb skeleton. Development 2024, 151, dev202798. [Google Scholar] [CrossRef]
- Li, P.; Deng, Q.; Liu, J.; Yan, J.; Wei, Z.; Zhang, Z.; Liu, H.; Li, B. Roles for HB-EGF in Mesenchymal Stromal Cell Proliferation and Differentiation During Skeletal Growth. J. Bone Miner. Res. 2019, 34, 295–309. [Google Scholar] [CrossRef]
- Zhu, J.; Chaki, M.; Lu, D.; Ren, C.; Wang, S.S.; Rauhauser, A.; Li, B.; Zimmerman, S.; Jun, B.; Du, Y.; et al. Loss of diacylglycerol kinase epsilon in mice causes endothelial distress and impairs glomerular Cox-2 and PGE2 production. Am. J. Physiol. Renal Physiol. 2016, 310, F895–F908. [Google Scholar] [CrossRef]
- Liu, D.; Ding, Q.; Dai, D.F.; Padhy, B.; Nayak, M.K.; Li, C.; Purvis, M.; Jin, H.; Shu, C.; Chauhan, A.K.; et al. Loss of diacylglycerol kinase epsilon causes thrombotic microangiopathy by impairing endothelial VEGFA signaling. JCI Insight 2021, 6, e146959. [Google Scholar] [CrossRef]
- Ito, T.; Young, M.J.; Li, R.; Jain, S.; Wernitznig, A.; Krill-Burger, J.M.; Lemke, C.T.; Monducci, D.; Rodriguez, D.J.; Chang, L.; et al. Paralog knockout profiling identifies DUSP4 and DUSP6 as a digenic dependence in MAPK pathway-driven cancers. Nat. Genet. 2021, 53, 1664–1672. [Google Scholar] [CrossRef]
- Lloyd, J.T.; Glass, K.C. Biological function and histone recognition of family IV bromodomain-containing proteins. J. Cell Physiol. 2018, 233, 1877–1886. [Google Scholar] [CrossRef]
- Oh, J.E.; Kim, H.J.; Kim, W.S.; Lee, Z.H.; Ryoo, H.M.; Hwang, S.J.; Lee, Y.; Kim, H.H. PlexinA2 mediates osteoblast differentiation via regulation of Runx2. J. Bone Miner. Res. 2012, 27, 552–562. [Google Scholar] [CrossRef] [PubMed]
- Robertson, G.; Xie, C.; Chen, D.; Awad, H.; Schwarz, E.M.; O’Keefe, R.J.; Guldberg, R.E.; Zhang, X. Alteration of femoral bone morphology and density in COX-2−/− mice. Bone 2006, 39, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Simon, A.M.; Manigrasso, M.B.; O’Connor, J.P. Cyclo-oxygenase 2 function is essential for bone fracture healing. J. Bone Miner. Res. 2002, 17, 963–976. [Google Scholar] [CrossRef] [PubMed]
- Luo, N.; Knudson, W.; Askew, E.B.; Veluci, R.; Knudson, C.B. CD44 and hyaluronan promote the bone morphogenetic protein 7 signaling response in murine chondrocytes. Arthritis Rheumatol. 2014, 66, 1547–1558. [Google Scholar] [CrossRef]
- Nedvetzki, S.; Gonen, E.; Assayag, N.; Reich, R.; Williams, R.O.; Thurmond, R.L.; Huang, J.F.; Neudecker, B.A.; Wang, F.S.; Turley, E.A.; et al. RHAMM, a receptor for hyaluronan-mediated motility, compensates for CD44 in inflamed CD44-knockout mice: A different interpretation of redundancy. Proc. Natl. Acad. Sci. USA 2004, 101, 18081–18086. [Google Scholar] [CrossRef]
- Snetkov, P.; Zakharova, K.; Morozkina, S.; Olekhnovich, R.; Uspenskaya, M. Hyaluronic Acid: The Influence of Molecular Weight on Structural, Physical, Physico-Chemical, and Degradable Properties of Biopolymer. Polymers 2020, 12, 1800. [Google Scholar] [CrossRef]
- Stern, R.; Asari, A.A.; Sugahara, K.N. Hyaluronan fragments: An information-rich system. Eur. J. Cell Biol. 2006, 85, 699–715. [Google Scholar] [CrossRef]
- Krejcova, D.; Pekarova, M.; Safrankova, B.; Kubala, L. The effect of different molecular weight hyaluronan on macrophage physiology. Neuro Endocrinol. Lett. 2009, 30 (Suppl. S1), 106–111. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panahipour, L.; Imani, A.; dos Santos Sanches, N.; Kühtreiber, H.; Mildner, M.; Gruber, R. RNA Sequencing Revealed a Weak Response of Gingival Fibroblasts Exposed to Hyaluronic Acid. Bioengineering 2024, 11, 1307. https://doi.org/10.3390/bioengineering11121307
Panahipour L, Imani A, dos Santos Sanches N, Kühtreiber H, Mildner M, Gruber R. RNA Sequencing Revealed a Weak Response of Gingival Fibroblasts Exposed to Hyaluronic Acid. Bioengineering. 2024; 11(12):1307. https://doi.org/10.3390/bioengineering11121307
Chicago/Turabian StylePanahipour, Layla, Atefe Imani, Natália dos Santos Sanches, Hannes Kühtreiber, Michael Mildner, and Reinhard Gruber. 2024. "RNA Sequencing Revealed a Weak Response of Gingival Fibroblasts Exposed to Hyaluronic Acid" Bioengineering 11, no. 12: 1307. https://doi.org/10.3390/bioengineering11121307
APA StylePanahipour, L., Imani, A., dos Santos Sanches, N., Kühtreiber, H., Mildner, M., & Gruber, R. (2024). RNA Sequencing Revealed a Weak Response of Gingival Fibroblasts Exposed to Hyaluronic Acid. Bioengineering, 11(12), 1307. https://doi.org/10.3390/bioengineering11121307








