Research on Multimodal Fusion of Temporal Electronic Medical Records
Abstract
1. Introduction
- We propose an integrative approach that combines time series clinical note data, time series tabular data, static clinical note data, and static tabular data, resulting in improved performance on two types of predictive tasks;
- Addressing the irregularity and non-uniformity of medical time series data, we employ a time window to mitigate these challenges. Simultaneously, the integration of an attention-backtracking module enhances our model’s ability to capture long-term dependencies;
- By comparing two types of prediction models, utilizing LSTM and a deep neural network (DNN), we demonstrate that neglecting the temporal sequence information embedded in time series data can have detrimental effects on the predictive performance of the model.
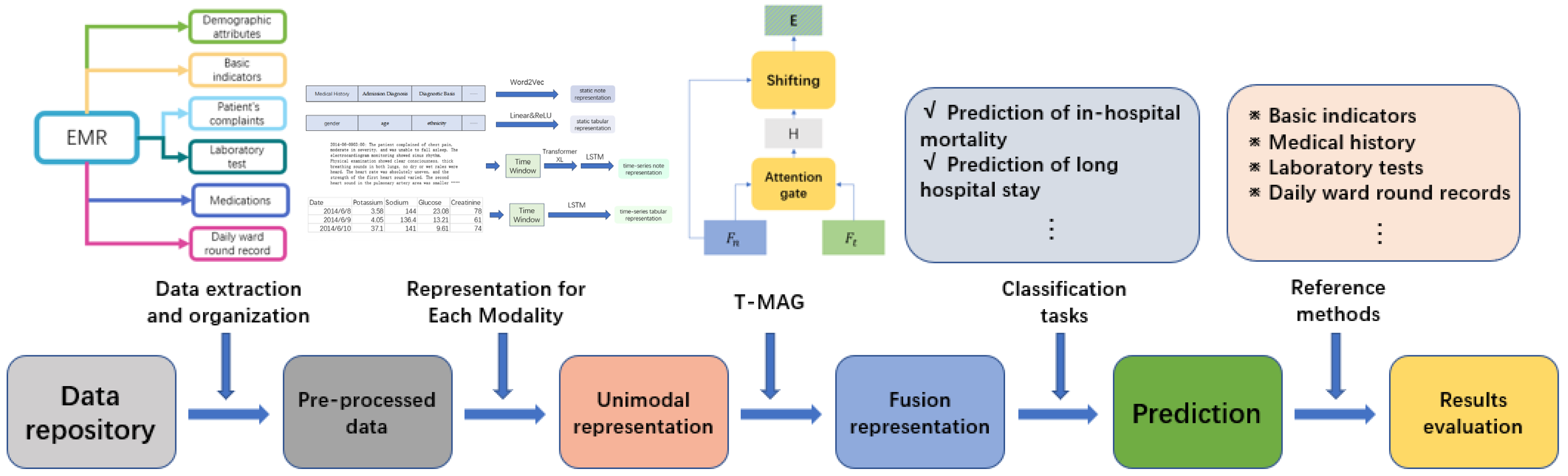
2. Materials and Methods
2.1. Dataset and Data Preprocessing
- Static tabular data: This is obtained from the admission records and includes 6 demographic attributes of the patient, as well as 7 basic physical examination parameters;
- Static note data: Also obtained from admission records, this category encompasses 15 types of information, including the patient’s complaints, medical history, specialized examinations, admission diagnoses, and confirmed diagnoses. Additionally, due to the absence of punctuation in some text data, such as in-patient diagnoses, we utilized the Jieba segmentation tool for tokenization [28]. This tool is a specialized Chinese tokenization tool that automatically identifies new words and proper nouns based on Chinese text;
- Temporal tabular data: Extracted from progress notes, this contains laboratory test results and vital signs measured at two or more time points during the hospital stay. It includes a total of 95 parameters;
- Temporal note data: Derived from progress notes, this category consists of daily ward round records for each day of the patient’s hospitalization. In cases where multiple rounds occur on the same day, only the first-round record of the day is selected.
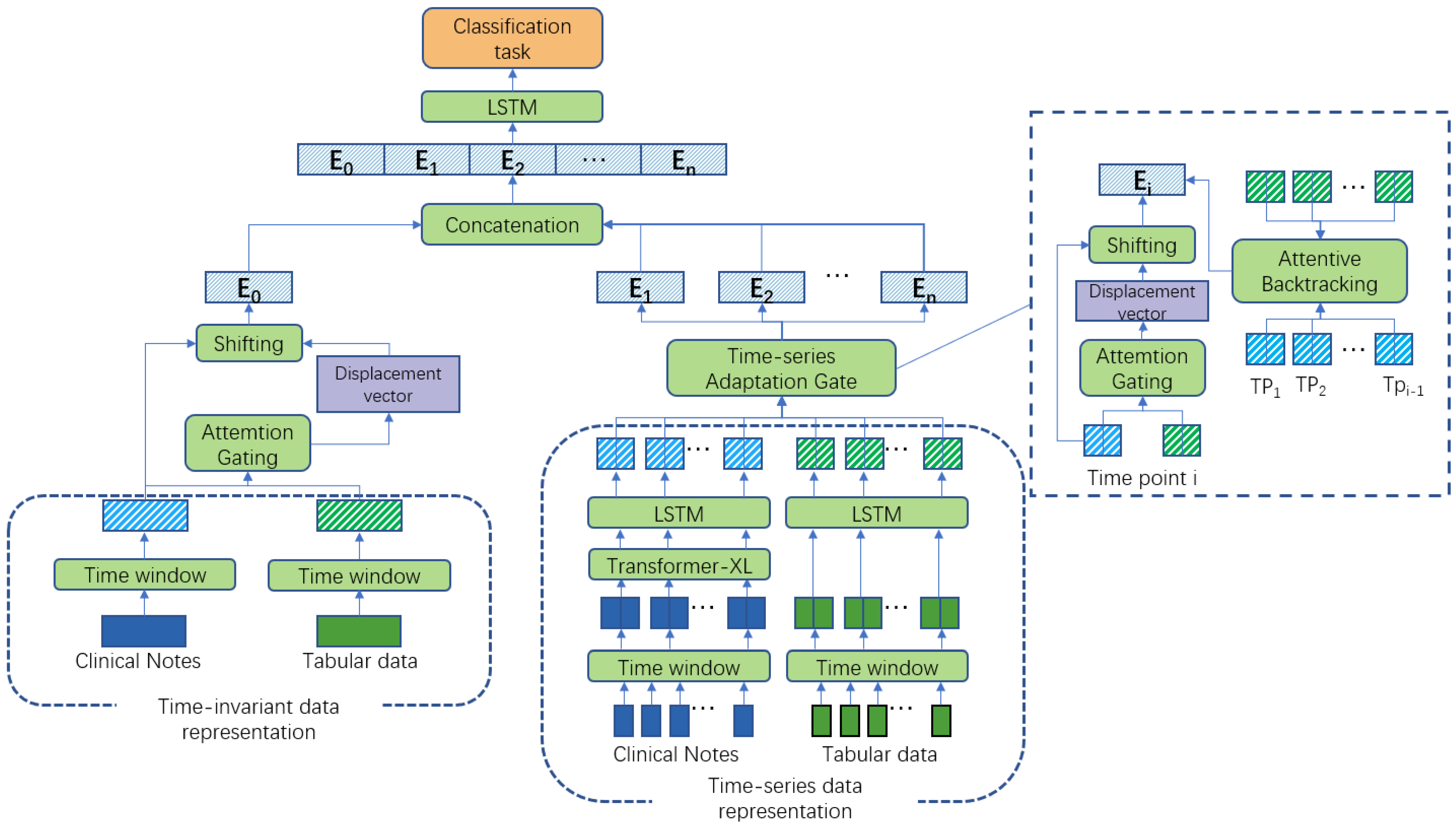
2.2. The T-MAG Model
2.2.1. Representation for Each Modality
- Feature embedding for static tabular data
- Feature Embedding for static note data
- Feature Embedding for temporal tabular data
- Feature Embedding for temporal note data
2.2.2. Fusion for Each Modality
2.3. Evaluation of the T-MAG-Based Multimodal Fusion Model
2.3.1. Tasks and Indexes for the Performance Evaluation
2.3.2. Evaluation Experiments
2.3.3. Comparative Models
- Models using a simple fusion method—fusion-convolutional neural network (Fusion-CNN) and fusion-LSTM [33];
- MAG-based fusion models—MAG-LSTM and MAG-DNN.
2.3.4. Ablation Experiments
2.3.5. Experimental Setup
3. Results
3.1. Impact of the Main Modality on the Model Performance
3.2. Impact of Different Subsets on Model Performance
3.3. Results of Comparative Experiments
3.4. Results of Ablation Experiments
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AMI | Acute myocardial infarction |
| AUPRC | Area under the precision-recall curve |
| AUROC | Area under the receiver operating characteristic curve |
| BERT | Bidirectional encoder representations from transformers |
| BiLSTM | Bi-directional long short-term memory |
| CNN | Convolutional neural network |
| DNN | Deep neural networks |
| EMR | Electronic medical record |
| FIDDLE | Flexible data-driven pipeline |
| ICD | International classification of diseases |
| ICU | Intensive care unit |
| LSTM | Long short-term memory |
| MAG | Multimodal adaptation gate |
| PatchTST | Patch time series transformer |
| RNN | Recurrent neural network |
Appendix A. Tumbling Time Window
| Size of Time Window | Number of Subsequences | Prediction of In-Hospital Mortality | Prediction of Long Hospital Stay | ||||
|---|---|---|---|---|---|---|---|
| AUROC | AUPRC | F1 | AUROC | AUPRC | F1 | ||
| 1 | 30 | 0.907 | 0.323 | 0.495 | 0.825 | 0.543 | 0.397 |
| 2 | 15 | 0.924 | 0.356 | 0.519 | 0.867 | 0.601 | 0.448 |
| 3 | 10 | 0.928 | 0.363 | 0.535 | 0.881 | 0.632 | 0.478 |
| 5 | 6 | 0.923 | 0.351 | 0.525 | 0.844 | 0.588 | 0.425 |
| 10 | 3 | 0.877 | 0.303 | 0.485 | 0.813 | 0.524 | 0.376 |
| 15 | 2 | 0.868 | 0.298 | 0.488 | 0.815 | 0.519 | 0.374 |
References
- Charles, D.; Gabriel, M.; Furukawa, M.F. Adoption of electronic health record systems among US non-federal acute care hospitals: 2008–2012. ONC Data Brief 2013, 9, 9. [Google Scholar]
- Esther Omolara, A.; Jantan, A.; Abiodun, O.I.; Arshad, H.; Dada, K.V.; Emmanuel, E. HoneyDetails: A prototype for ensuring patient’s information privacy and thwarting electronic health record threats based on decoys. Health Inform. J. 2020, 26, 2083–2104. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Li, Y.; Zhang, Z.; Shen, D.; Xu, J.; Zheng, X.; Wang, T.; Tang, B.; Lei, J.; Zhang, J. Adoption of electronic health records (EHRs) in China during the past 10 years: Consecutive survey data analysis and comparison of Sino-American challenges and experiences. J. Med. Internet Res. 2021, 23, e24813. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Pouyanfar, S.; Yang, Y.; Chen, S.C.; Iyengar, S.S. Computational health informatics in the big data age: A survey. ACM Comput. Surv. (CSUR) 2016, 49, 12. [Google Scholar] [CrossRef]
- Tang, S.; Davarmanesh, P.; Song, Y.; Koutra, D.; Sjoding, M.W.; Wiens, J. Democratizing EHR analyses with FIDDLE: A flexible data-driven preprocessing pipeline for structured clinical data. J. Am. Med. Inform. Assoc. 2020, 27, 1921–1934. [Google Scholar] [CrossRef] [PubMed]
- Noraziani, K.; Nurul’Ain, A.; Azhim, M.Z.; Eslami, S.R.; Drak, B.; Sharifa Ezat, W.; Siti Nurul Akma, A. An overview of electronic medical record implementation in healthcare system: Lesson to learn. World Appl. Sci. J. 2013, 25, 323–332. [Google Scholar]
- An, Q.; Rahman, S.; Zhou, J.; Kang, J.J. A Comprehensive Review on Machine Learning in Healthcare Industry: Classification, Restrictions, Opportunities and Challenges. Sensors 2023, 23, 4178. [Google Scholar] [CrossRef]
- Kamal, U.; Zunaed, M.; Nizam, N.B.; Hasan, T. Anatomy-xnet: An anatomy aware convolutional neural network for thoracic disease classification in chest X-rays. IEEE J. Biomed. Health Inform. 2022, 26, 5518–5528. [Google Scholar] [CrossRef]
- Kamal, S.A.; Yin, C.; Qian, B.; Zhang, P. An interpretable risk prediction model for healthcare with pattern attention. BMC Med. Inform. Decis. Mak. 2020, 20, 307. [Google Scholar] [CrossRef]
- Kemp, J.; Rajkomar, A.; Dai, A.M. Improved hierarchical patient classification with language model pretraining over clinical notes. arXiv 2019, arXiv:1909.03039. [Google Scholar]
- Min, X.; Yu, B.; Wang, F. Predictive modeling of the hospital readmission risk from patients’ claims data using machine learning: A case study on COPD. Sci. Rep. 2019, 9, 2362. [Google Scholar] [CrossRef] [PubMed]
- Harutyunyan, H.; Khachatrian, H.; Kale, D.C.; Ver Steeg, G.; Galstyan, A. Multitask learning and benchmarking with clinical time series data. Sci. Data 2019, 6, 96. [Google Scholar] [CrossRef] [PubMed]
- Suo, Q.; Ma, F.; Yuan, Y.; Huai, M.; Zhong, W.; Gao, J.; Zhang, A. Deep patient similarity learning for personalized healthcare. IEEE Trans. Nanobiosci. 2018, 17, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Wang, F.; Zhang, P.; Hu, J. Risk prediction with electronic health records: A deep learning approach. In Proceedings of the 2016 SIAM International Conference on Data Mining, Miami, FL, USA, 5–7 May 2016; Society for Industrial and Applied Mathematics: Philadelphia, PA, USA, 2016; pp. 432–440. [Google Scholar]
- Lee, J.; Maslove, D.M.; Dubin, J.A. Personalized mortality prediction driven by electronic medical data and a patient similarity metric. PLoS ONE 2015, 10, e0127428. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; McDermott, M.B.A.; Chauhan, G.; Ghassemi, M.; Hughes, M.C.; Naumann, T. Mimic-extract: A data extraction, preprocessing, and representation pipeline for mimic-iii. In Proceedings of the ACM Conference on Health, Inference, and Learning, Toronto, ON, Canada, 2–4 February 2020; pp. 222–235. [Google Scholar]
- Shao, C.; Fang, F.; Bai, F.; Wang, B. An interpolation method combining Snurbs with window interpolation adjustment. In Proceedings of the 2014 4th IEEE International Conference on Information Science and Technology, Shenzhen, China, 26–28 April 2014; IEEE: Piscataway, NJ, USA, 2014; pp. 176–179. [Google Scholar]
- Shickel, B.; Tighe, P.J.; Bihorac, A.; Rashidi, P. Deep EHR: A survey of recent advances in deep learning techniques for electronic health record (EHR) analysis. IEEE J. Biomed. Health Inform. 2017, 22, 1589–1604. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Xi, X.; Chen, J.; Sheng, V.S.; Ma, J.; Cui, Z. A survey of deep learning for electronic health records. Appl. Sci. 2022, 12, 11709. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N. Attention Is All You Need. In Proceedings of the 31st Conference on Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017; pp. 5998–6008. [Google Scholar]
- Kirchberger, I.; Heier, M.; Kuch, B.; von Scheidt, W.; Meisinger, C. Presenting symptoms of myocardil infarction predict short-and long-term mortality: The MONICA/KORA Myocardial Infarction Registry. Am. Heart J. 2012, 164, 856–861. [Google Scholar] [CrossRef]
- Chen, H.; Shi, L.; Xue, M.; Wang, N.; Dong, X.; Cai, Y.; Chen, J.; Zhu, W.; Xu, H.; Meng, Q. Geographic variations in In-Hospital mortality and use of percutaneous coronary intervention following acute myocardial infarction in China: A nationwide Cross-Sectional analysis. J. Am. Heart Assoc. 2018, 7, e008131. [Google Scholar] [CrossRef]
- Lee, H.C.; Park, J.S.; Choe, J.C.; Ahn, J.H.; Lee, H.W.; Oh, J.-H.; Choi, J.H.; Cha, K.S.; Hong, T.J.; Jeong, M.H. Prediction of 1-year mortality from acute myocardial infarction using machine learning. Am. J. Cardiol. 2020, 133, 23–31. [Google Scholar] [CrossRef]
- Gyárfás, I.; Keltai, M.; Salim, Y. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries in a case-control study based on the INTERHEART study. Orvosi Hetil. 2006, 147, 675–686. [Google Scholar]
- Braunwald, E.; Antman, E.M.; Beasley, J.W.; Califf, R.M.; Cheitlin, M.D.; Hochman, J.S.; Jones, R.H.; Kereiakes, D.; Kupersmith, J.; Levin, T.N.; et al. ACC/AHA 2002 guideline update for the management of patients with unstable angina and non–ST-segment elevation myocardial infarction—Summary article: A report of the American College of Cardiology/American Heart Association task force on practice guidelines (Committee on the Management of Patients with Unstable Angina). J. Am. Coll. Cardiol. 2002, 40, 1366–1374. [Google Scholar] [PubMed]
- Boag, W.; Doss, D.; Naumann, T.; Szolovits, P. What’s in a note? unpacking predictive value in clinical note representations. AMIA Summits Transl. Sci. Proc. 2018, 2018, 26. [Google Scholar]
- Yang, B.; Wu, L. How to leverage multimodal EHR data for better medical predictions? arXiv 2021, arXiv:2110.15763. [Google Scholar]
- Jieba. Available online: https://github.com/fxsjy/jieba (accessed on 15 February 2020).
- Dai, Z.; Yang, Z.; Yang, Y.; Carbonell, J.; Le, Q.; Salakhutdinov, R. Transformer-xl: Attentive language models beyond a fixed-length context. arXiv 2019, arXiv:1901.02860. [Google Scholar]
- Wang, Y.; Shen, Y.; Liu, Z.; Liang, P.P.; Zadeh, A.; Morency, L.-P. Words can shift: Dynamically adjusting word representations using nonverbal behaviors. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; Volume 33, pp. 7216–7223. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Ahsan, M.M.; Luna, S.A.; Siddique, Z. Machine-learning-based disease diagnosis: A comprehensive review. Proc. Healthc. 2022, 10, 541. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Yin, C.; Zeng, J.; Yuan, X.; Zhang, P. Combining structured and unstructured data for predictive models: A deep learning approach. BMC Med. Inform. Decis. Mak. 2020, 20, 280. [Google Scholar] [CrossRef]
- Tsai, Y.H.H.; Bai, S.; Liang, P.P.; Kolter, J.Z.; Morency, L.-P.; Salakhutdinov, R. Multimodal transformer for unaligned multimodal language sequences. In Proceedings of the Association for Computational Linguistics Meeting, Florence, Italy, 28 July–2 August 2019; NIH Public Access: Bethesda, MD, USA, 2019; p. 6558. [Google Scholar]
- Zhang, Y.; Yan, J. Crossformer: Transformer utilizing cross-dimension dependency for multivariate time series forecasting. In Proceedings of the 11th International Conference on Learning Representations (ICLR 2023), Kigali, Rwanda, 1–5 May 2023. [Google Scholar]
- Nie, Y.; Nguyen, N.H.; Sinthong, P.; Kalagnanam, J. A time series is worth 64 words: Long-term forecasting with transformers. In Proceedings of the 11th International Conference on Learning Representations (ICLR 2023), Kigali, Rwanda, 1–5 May 2023. [Google Scholar]
- Zhang, X.; Li, S.; Chen, Z.; Yan, X.; Petzold, L.R. Improving medical predictions by irregular multimodal electronic health records modeling. In Proceedings of the International Conference on Machine Learning, PMLR, Honolulu, HI, USA, 23–29 July 2023; pp. 41300–41313. [Google Scholar]
- Jalamangala Shivananjaiah, S.K.; Kumari, S.; Majid, I.; Wang, S.Y. Predicting near-term glaucoma progression: An artificial intelligence approach using clinical free-text notes and data from electronic health records. Front. Med. 2023, 10, 371. [Google Scholar] [CrossRef]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An imperative style, high-performance deep learning library. In Proceedings of the 33rd International Conference on Neural Information Processing Systems, Vancouver, BC, Canada, 8–14 December 2019; pp. 8026–8037. [Google Scholar]
- Grant, D.; Papież, B.W.; Parsons, G.; Tarassenko, L.; Mahdi, A. Deep learning classification of cardiomegaly using combined imaging and non-imaging ICU data. In Proceedings of the Medical Image Understanding and Analysis: 25th Annual Conference, MIUA 2021, Oxford, UK, 12–14 July 2021; Springer International Publishing: Berlin/Heidelberg, Germany, 2021; pp. 547–558. [Google Scholar]
- Liu, J.; Zhang, Z.; Razavian, N. Deep ehr: Chronic disease prediction using medical notes. In Proceedings of the Machine Learning for Healthcare Conference, PMLR, Palo Alto, CA, USA, 17–18 August 2018; pp. 440–464. [Google Scholar]
- Searle, T.; Ibrahim, Z.; Teo, J.; Dobson, R.J. Discharge summary hospital course summarisation of in-patient Electronic Health Record text with clinical concept guided deep pre-trained Transformer models. J. Biomed. Inform. 2023, 141, 104358. [Google Scholar] [CrossRef]
- Ning, W.; Lei, S.; Yang, J.; Cao, Y.; Jiang, P.; Yang, Q.; Zhang, J.; Wang, X.; Chen, F.; Geng, Z.; et al. Open resource of clinical data from patients with pneumonia for the prediction of COVID-19 outcomes via deep learning. Nat. Biomed. Eng. 2020, 4, 1197–1207. [Google Scholar] [CrossRef]
- Xu, Y.; Biswal, S.; Deshpande, S.R.; Maher, K.O.; Sun, J. Raim: Recurrent attentive and intensive model of multimodal patient monitoring data. In Proceedings of the 24th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, London, UK, 19–23 August 2018; pp. 2565–2573. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Jacenków, G.; O’Neil, A.Q.; Tsaftaris, S.A. Indication as prior knowledge for multimodal disease classification in chest radiographs with transformers. In Proceedings of the 2022 IEEE 19th International Symposium on Biomedical Imaging (ISBI), Kolkata, India, 28–31 March 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 1–5. [Google Scholar]
- Zeng, A.; Chen, M.; Zhang, L.; Xu, Q. Are transformers effective for time series forecasting? In Proceedings of the AAAI Conference on Artificial Intelligence, Washington, DC, USA, 7–14 February 2023; Volume 37, pp. 11121–11128. [Google Scholar]
- Woo, G.; Liu, C.; Sahoo, D.; Kumar, A.; Hoi, S. Learning deep time-index models for time series forecasting. In Proceedings of the International Conference on Machine Learning, PMLR, Honolulu, HI, USA, 23–29 July 2023; pp. 37217–37237. [Google Scholar]
- Tang, F.; Xiao, C.; Wang, F.; Zhou, J. Predictive modeling in urgent care: A comparative study of machine learning approaches. Jamia Open 2018, 1, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Ghassemi, M.; Celi, L.A.; Stone, D.J. State of the art review: The data revolution in critical care. Crit. Care 2015, 19, 118. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Huang, F.; Lv, J.; Duan, Y.; Qin, Z.; Li, G.; Tian, G. Do RNN and LSTM have long memory? In Proceedings of the International Conference on Machine Learning, PMLR, Virtual, 13–18 July 2023; pp. 11365–11375. [Google Scholar]
- Schaefer, A.M.; Udluft, S.; Zimmermann, H.-G. Learning long-term dependencies with recurrent neural networks. Neurocomputing 2008, 71, 2481–2488. [Google Scholar] [CrossRef]
- Ke, N.R.; Goyal, A.; Bilaniuk, O.; Binas, J.; Mozer, M.C.; Pal, C.; Bengio, Y. Sparse attentive backtracking: Temporal credit assignment through reminding. In Proceedings of the 32nd International Conference on Neural Information Processing Systems, Montréal, QC, Canada, 3–8 December 2018; pp. 7651–7662. [Google Scholar]
- Goyal, A.; Lamb, A.; Hoffmann, J.; Sodhani, S.; Levine, S.; Bengio, Y.; Schölkopf, B. Recurrent independent mechanisms. arXiv 2019, arXiv:1909.10893. [Google Scholar]

| Modal | Feature Category | AMI Dataset | Stroke Dataset | Example |
|---|---|---|---|---|
| Number of Features | ||||
| Static tabular | Gender | 2 | 2 | male, female |
| Age | 2 | 2 | >60 years, ≤60 years | |
| Ethnicity | 12 | 16 | Han ethnicity, Hui ethnicity | |
| Marital status | 3 | 3 | married, divorced, unmarried | |
| Department | 7 | 18 | neurosurgery, vascular surgery | |
| Admission method | 2 | 2 | emergency department | |
| Basic indicators | 7 | 7 | height, weight, temperature | |
| Static note | Chief Complaint | 1 | 1 | chief complaint |
| Medical History | 5 | 5 | current medical history, past medical history | |
| Specialized Examination | 2 | 2 | specialist examination, auxiliary examination | |
| Admission Diagnosis | 3 | 3 | confirm diagnosis, supplementary diagnosis | |
| Characteristics | 1 | 1 | patient characteristics | |
| Diagnostic Basis | 2 | 2 | diagnostic basis, differential diagnosis | |
| Treatment Plan | 1 | 1 | treatment plan | |
| Temporal tabular | Laboratory tests | 73 | 71 | serum triglyceride, serum creatinine |
| Medications | 17 | 21 | angiotensin-converting enzyme inhibitor, heparin | |
| Vital signs measured | 5 | 5 | respiratory rate, pulse rate | |
| Temporal note | Daily ward round records | 1 | 1 | daily ward round records |
| Data Set | Selection of Main Modality | Prediction of In-Hospital Mortality | Prediction of Long Hospital Stay | ||||
|---|---|---|---|---|---|---|---|
| AUROC | AUPRC | F1 | AUROC | AUPRC | F1 | ||
| AMI | Static notes and temporal notes | 0.928 | 0.363 | 0.535 | 0.881 | 0.632 | 0.478 |
| Static tabular data and temporal notes | 0.923 | 0.351 | 0.520 | 0.879 | 0.630 | 0.473 | |
| Static notes and temporal tabular data | 0.925 | 0.359 | 0.528 | 0.877 | 0.626 | 0.466 | |
| Static tabular data and temporal tabular data | 0.919 | 0.346 | 0.516 | 0.874 | 0.621 | 0.454 | |
| Stroke | Static notes and temporal notes | 0.954 | 0.455 | 0.671 | 0.847 | 0.508 | 0.376 |
| Static tabular data and temporal notes | 0.951 | 0.447 | 0.665 | 0.836 | 0.486 | 0.359 | |
| Static notes and temporal tabular data | 0.945 | 0.438 | 0.651 | 0.834 | 0.479 | 0.352 | |
| Static tabular data and temporal tabular data | 0.933 | 0.425 | 0.644 | 0.818 | 0.443 | 0.334 | |
| Model | In-Hospital Mortality | Long Length of Stay | |||||
|---|---|---|---|---|---|---|---|
| AUROC | AUPRC | F1 | AUROC | AUPRC | F1 | ||
| T-MAG | T-MAG | 0.928 | 0.363 | 0.535 | 0.881 | 0.632 | 0.478 |
| Neural Network | DNN | 0.748 | 0.228 | 0.313 | 0.726 | 0.423 | 0.318 |
| LSTM | 0.769 | 0.243 | 0.328 | 0.758 | 0.528 | 0.413 | |
| Fusion Methods | Fusion-CNN | 0.816 | 0.267 | 0.403 | 0.716 | 0.513 | 0.405 |
| Fusion-LSTM | 0.828 | 0.287 | 0.435 | 0.818 | 0.544 | 0.436 | |
| Fusion Methods | MulT | 0.913 | 0.323 | 0.502 | 0.856 | 0.593 | 0.468 |
| Crossformer | 0.893 | 0.319 | 0.491 | 0.855 | 0.597 | 0.461 | |
| PatchTST | 0.838 | 0.296 | 0.447 | 0.825 | 0.537 | 0.422 | |
| MISTS-fusion | 0.917 | 0.341 | 0.508 | 0.866 | 0.609 | 0.438 | |
| Glaucoma-fusion | 0.821 | 0.277 | 0.415 | 0.805 | 0.565 | 0.461 | |
| MAG-DNN | 0.844 | 0.312 | 0.481 | 0.838 | 0.557 | 0.445 | |
| MAG-LSTM | 0.916 | 0.339 | 0.511 | 0.874 | 0.619 | 0.451 | |
| Model | In-Hospital Mortality | Long Length of Stay | |||||
|---|---|---|---|---|---|---|---|
| AUROC | AUPRC | F1 | AUROC | AUPRC | F1 | ||
| T-MAG | T-MAG | 0.954 | 0.455 | 0.671 | 0.847 | 0.508 | 0.376 |
| Neural Network | DNN | 0.849 | 0.333 | 0.505 | 0.736 | 0.375 | 0.245 |
| LSTM | 0.856 | 0.349 | 0.511 | 0.746 | 0.378 | 0.255 | |
| Fusion Methods | Fusion-CNN | 0.879 | 0.388 | 0.573 | 0.767 | 0.408 | 0.286 |
| Fusion-LSTM | 0.887 | 0.401 | 0.595 | 0.822 | 0.430 | 0.317 | |
| Fusion Methods | MulT | 0.933 | 0.435 | 0.641 | 0.830 | 0.441 | 0.332 |
| Crossformer | 0.945 | 0.450 | 0.658 | 0.844 | 0.498 | 0.370 | |
| PatchTST | 0.927 | 0.423 | 0.633 | 0.825 | 0.435 | 0.323 | |
| MISTS-fusion | 0.949 | 0.451 | 0.660 | 0.833 | 0.445 | 0.339 | |
| Glaucoma-fusion | 0.891 | 0.405 | 0.611 | 0.815 | 0.425 | 0.310 | |
| MAG-DNN | 0.914 | 0.411 | 0.625 | 0.799 | 0.419 | 0.301 | |
| MAG-LSTM | 0.938 | 0.445 | 0.647 | 0.836 | 0.488 | 0.358 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, M.; Wang, M.; Gao, B.; Li, Y.; Huang, J.; Chen, H. Research on Multimodal Fusion of Temporal Electronic Medical Records. Bioengineering 2024, 11, 94. https://doi.org/10.3390/bioengineering11010094
Ma M, Wang M, Gao B, Li Y, Huang J, Chen H. Research on Multimodal Fusion of Temporal Electronic Medical Records. Bioengineering. 2024; 11(1):94. https://doi.org/10.3390/bioengineering11010094
Chicago/Turabian StyleMa, Moxuan, Muyu Wang, Binyu Gao, Yichen Li, Jun Huang, and Hui Chen. 2024. "Research on Multimodal Fusion of Temporal Electronic Medical Records" Bioengineering 11, no. 1: 94. https://doi.org/10.3390/bioengineering11010094
APA StyleMa, M., Wang, M., Gao, B., Li, Y., Huang, J., & Chen, H. (2024). Research on Multimodal Fusion of Temporal Electronic Medical Records. Bioengineering, 11(1), 94. https://doi.org/10.3390/bioengineering11010094





