Magnetic Field Intervention Enhances Cellular Migration Rates in Biological Scaffolds
Abstract
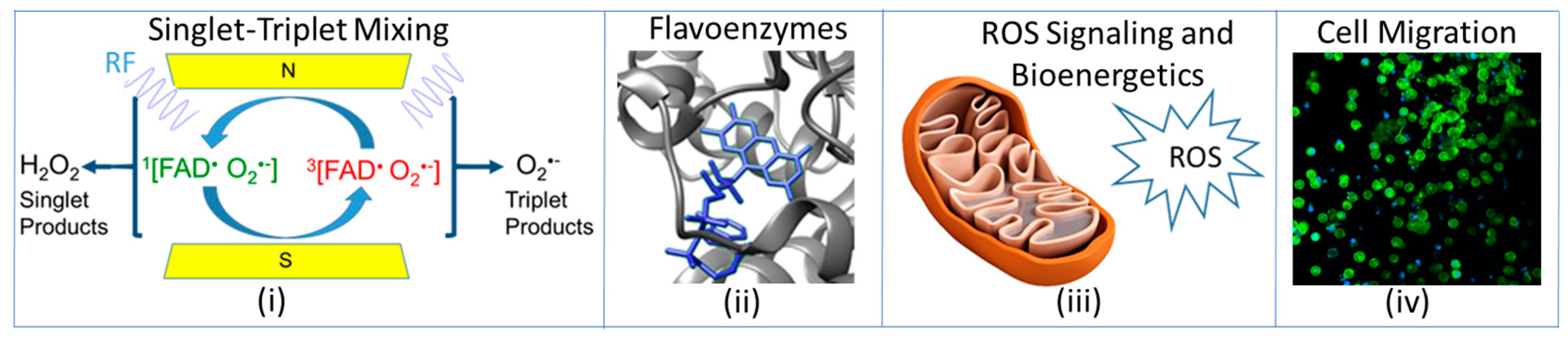
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Bioink Synthesis
2.3. Bioprinting-Based Construct Formation
2.4. Magnetic Field Instrumentation
2.5. DAPI-Phalloidin
2.6. Scanning Electron Microscopy
2.7. Image Processing
2.8. Data Analysis
3. Results
3.1. Nuclei and Cytoskeleton Imaging
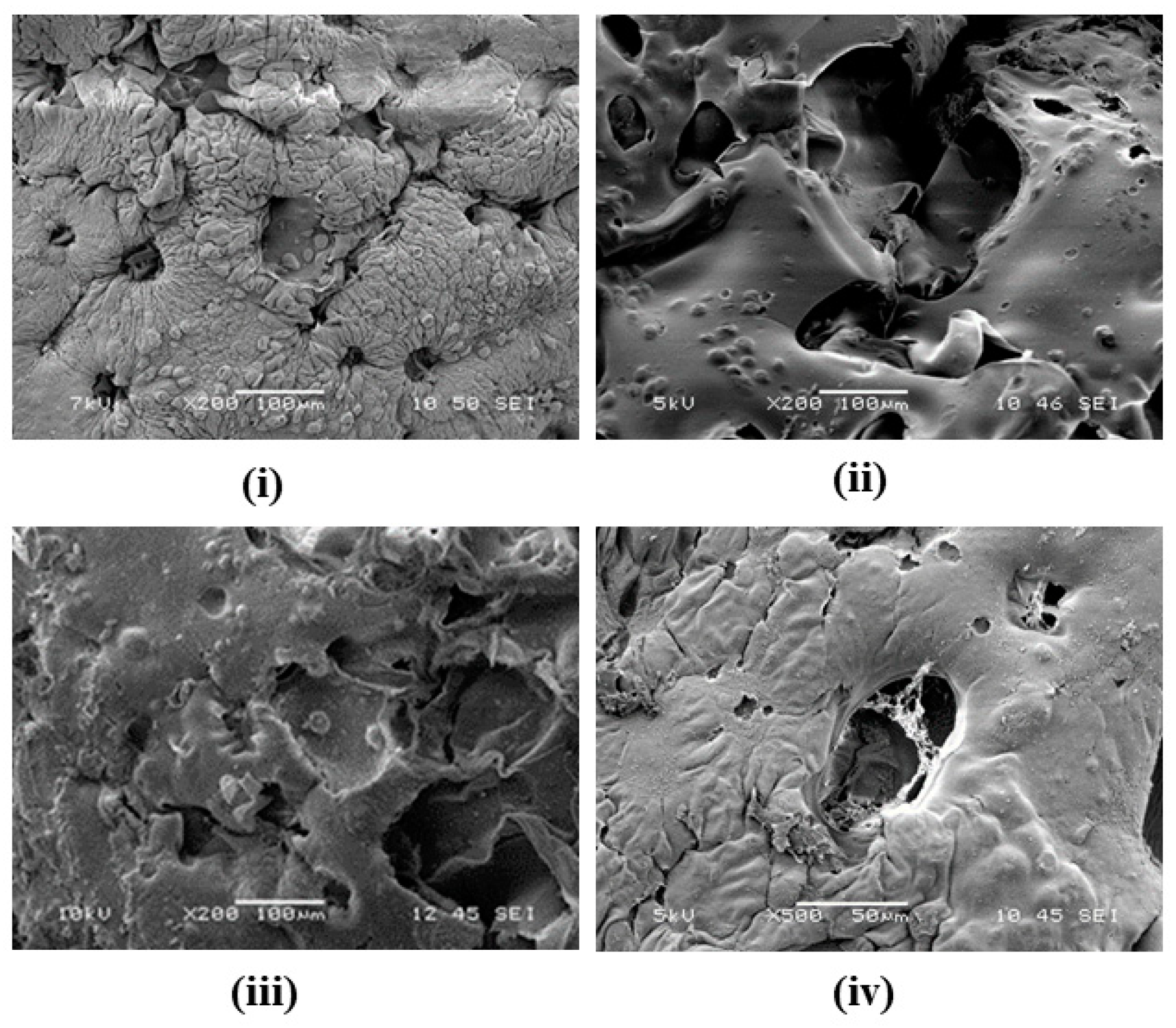
3.2. Surface Topography and CDM Formation
3.3. Change in Cell Density
4. Discussion
Study Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Usselman, R.; Hill, I.; Singel, D.; Martino, C. Spin Biochemistry Modulates Reactive Oxygen Species (ROS) Production by Radio Frequency Magnetic Fields. PLoS ONE 2014, 9, e93065. [Google Scholar] [CrossRef] [PubMed]
- Usselman, R.J.; Chavarriaga, C.; Castello, P.R.; Procopio, M.; Ritz, T.; Dratz, E.A.; Singel, D.J.; Martino, C.F. The Quantum Biology of Reactive Oxygen Species Partitioning Impacts Cellular Bioenergetics. Sci. Rep. 2016, 6, 38543. [Google Scholar] [CrossRef] [PubMed]
- Yap, J.L.Y.; Tai, Y.K.; Frohlich, J.; Fong, C.H.H.; Yin, J.N.; Foo, Z.L.; Ramanan, S.; Beyer, C.; Toh, S.J.; Casarosa, M.; et al. Ambient and supplemental magnetic fields promote myogenesis via a TRPC1-mitochondrial axis: Evidence of a magnetic mitohormetic mechanism. FASEB J. 2019, 33, 12853–12872. [Google Scholar] [CrossRef] [PubMed]
- Franco-Obregon, A. Magnetic mitohormesis: A non-invasive therapy for inflammatory disorders? Biocell 2023, 47, 239–244. [Google Scholar] [CrossRef]
- Arthaut, L.D.; Jourdan, N.; Mteyrek, A.; Procopio, M.; El-Esawi, M.; d’Harlingue, A.; Bouchet, P.E.; Witczak, J.; Ritz, T.; Klarsfeld, A.; et al. Blue-light induced accumulation of reactive oxygen species is a consequence of the Drosophila cryptochrome photocycle. PLoS ONE 2017, 12, e0171836. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Bertagna, F.; D’Souza, E.M.; Heyes, D.J.; Johannissen, L.O.; Nery, E.T.; Pantelias, A.; Sanchez-Pedreño Jimenez, A.; Slocombe, L.; Spencer, M.G.; et al. Quantum Biology: An Update and Perspective. Quantum Rep. 2021, 3, 80–126. [Google Scholar] [CrossRef]
- Zadeh-Haghighi, H.; Simon, C. Magnetic field effects in biology from the perspective of the radical pair mechanism. J. R. Soc. Interface 2022, 19, 20220325. [Google Scholar] [CrossRef]
- Hore, P.J.; Mouritsen, H. The Radical-Pair Mechanism of Magnetoreception. Annu. Rev. Biophys. 2016, 45, 299–344. [Google Scholar] [CrossRef]
- Ikeya, N.; Woodward, J.R. Cellular autofluorescence is magnetic field sensitive. Proc. Natl. Acad. Sci. USA 2021, 118, e2018043118. [Google Scholar] [CrossRef]
- Parate, D.; Franco-Obregon, A.; Frohlich, J.; Beyer, C.; Abbas, A.A.; Kamarul, T.; Hui, J.H.P.; Yang, Z. Enhancement of mesenchymal stem cell chondrogenesis with short-term low intensity pulsed electromagnetic fields. Sci. Rep. 2017, 7, 9421. [Google Scholar] [CrossRef]
- Wong, C.J.K.; Tai, Y.K.; Yap, J.L.Y.; Fong, C.H.H.; Loo, L.S.W.; Kukumberg, M.; Frohlich, J.; Zhang, S.; Li, J.Z.; Wang, J.W.; et al. Brief exposure to directionally-specific pulsed electromagnetic fields stimulates extracellular vesicle release and is antagonized by streptomycin: A potential regenerative medicine and food industry paradigm. Biomaterials 2022, 287, 121658. [Google Scholar] [CrossRef] [PubMed]
- Thoni, V.; Mauracher, D.; Ramalingam, A.; Fiechtner, B.; Sandbichler, A.M.; Egg, M. Quantum based effects of therapeutic nuclear magnetic resonance persistently reduce glycolysis. iScience 2022, 25, 105536. [Google Scholar] [CrossRef] [PubMed]
- Van Huizen, A.V.; Morton, J.M.; Kinsey, L.J.; Von Kannon, D.G.; Saad, M.A.; Birkholz, T.R.; Czajka, J.M.; Cyrus, J.; Barnes, F.S.; Beane, W.S. Weak magnetic fields alter stem cell-mediated growth. Sci. Adv. 2019, 5, eaau7201. [Google Scholar] [CrossRef] [PubMed]
- Kinsey, L.J.; Van Huizen, A.V.; Beane, W.S. Weak magnetic fields modulate superoxide to control planarian regeneration. Front. Phys. 2023, 10, 1086809. [Google Scholar] [CrossRef]
- Carter, C.S.; Huang, S.C.; Searby, C.C.; Cassaidy, B.; Miller, M.J.; Grzesik, W.J.; Piorczynski, T.B.; Pak, T.K.; Walsh, S.A.; Acevedo, M.; et al. Exposure to Static Magnetic and Electric Fields Treats Type 2 Diabetes. Cell Metab. 2020, 32, 561–574.E7. [Google Scholar] [CrossRef] [PubMed]
- Tai, Y.K.; Chan, K.K.W.; Fong, C.H.H.; Ramanan, S.; Yap, J.L.Y.; Yin, J.N.; Yip, Y.S.; Tan, W.R.; Koh, A.P.F.; Tan, N.S.; et al. Modulated TRPC1 Expression Predicts Sensitivity of Breast Cancer to Doxorubicin and Magnetic Field Therapy: Segue Towards a Precision Medicine Approach. Front. Oncol. 2021, 11, 783803. [Google Scholar] [CrossRef] [PubMed]
- Khizroev, S. Technobiology’s Enabler: The Magnetoelectric Nanoparticle. Cold Spring Harb. Perspect. Med. 2019, 9, a034207. [Google Scholar] [CrossRef]
- Parate, D.; Kadir, N.D.; Celik, C.; Lee, E.H.; Hui, J.H.P.; Franco-Obregon, A.; Yang, Z. Pulsed electromagnetic fields potentiate the paracrine function of mesenchymal stem cells for cartilage regeneration. Stem Cell Res. Ther. 2020, 11, 46. [Google Scholar] [CrossRef]
- Bradlaugh, A.A.; Fedele, G.; Munro, A.L.; Hansen, C.N.; Hares, J.M.; Patel, S.; Kyriacou, C.P.; Jones, A.R.; Rosato, E.; Baines, R.A. Essential elements of radical pair magnetosensitivity in Drosophila. Nature 2023, 615, 111–116. [Google Scholar] [CrossRef]
- Bassetto, M.; Reichl, T.; Kobylkov, D.; Kattnig, D.R.; Winklhofer, M.; Hore, P.J.; Mouritsen, H. No evidence for magnetic field effects on the behaviour of Drosophila. Nature 2023, 620, 595–599. [Google Scholar] [CrossRef]
- Galván, I.; Hassasfar, A.; Adams, B.; Petruccione, F. Isotope effects on radical pair performance in cryptochrome: A new hypothesis for the evolution of animal migration: The quantum biology of migration. BioEssays 2023, 2300152. [Google Scholar] [CrossRef] [PubMed]
- Mouritsen, H. Long-distance navigation and magnetoreception in migratory animals. Nature 2018, 558, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Thoradit, T.; Thongyoo, K.; Kamoltheptawin, K.; Tunprasert, L.; El-Esawi, M.A.; Aguida, B.; Jourdan, N.; Buddhachat, K.; Pooam, M. Cryptochrome and quantum biology: Unraveling the mysteries of plant magnetoreception. Front. Plant Sci. 2023, 14, 1266357. [Google Scholar] [CrossRef] [PubMed]
- Deviers, J.; Cailliez, F.; Lande, A.d.l.; Kattnig, D.R. Anisotropic magnetic field effects in the re-oxidation of cryptochrome in the presence of scavenger radicals. J. Chem. Phys. 2022, 156, 025101. [Google Scholar] [CrossRef] [PubMed]
- Steiner, U.; Ulrich, T. Magnetic field effects in chemical kinetics and related pehnomena. Chem. Rev. 1989, 89, 51–147. [Google Scholar] [CrossRef]
- Hammad, M.; Albaqami, M.; Pooam, M.; Kernevez, E.; Witczak, J.; Ritz, T.; Martino, C.; Ahmad, M. Cryptochrome mediated magnetic sensitivity in Arabidopsis occurs independently of light-induced electron transfer to the flavin. Photochem. Photobiol. Sci. 2020, 19, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Timmel, C.R.; Till, U.; Brocklehurst, B.; McLauchlan, K.A.; Hore, P.J. Effects of weak magnetic fields on free radical recombination reactions. Mol. Phys. 1998, 95, 71–89. [Google Scholar] [CrossRef]
- Hogben, H.J.; Efimova, O.; Wagner-Rundell, N.; Timmel, C.R.; Hore, P.J. Possible involvement of superoxide and dioxygen with cryptochrome in avian magnetoreception: Origin of Zeeman resonances observed by in vivo EPR spectroscopy. Chem. Phys. Lett. 2009, 480, 118–122. [Google Scholar] [CrossRef]
- Player, T.C.; Hore, P.J. Viability of superoxide-containing radical pairs as magnetoreceptors. J. Chem. Phys. 2019, 151, 225101. [Google Scholar] [CrossRef]
- Sies, H.; Belousov, V.V.; Chandel, N.S.; Davies, M.J.; Jones, D.P.; Mann, G.E.; Murphy, M.P.; Yamamoto, M.; Winterbourn, C. Defining roles of specific reactive oxygen species (ROS) in cell biology and physiology. Nat. Rev. Mol. Cell Biol. 2022, 23, 499–515. [Google Scholar] [CrossRef]
- Rishabh, R.; Zadeh-Haghighi, H.; Salahub, D.; Simon, C. Radical pairs may explain reactive oxygen species-mediated effects of hypomagnetic field on neurogenesis. PLoS Comput. Biol. 2022, 18, e1010198. [Google Scholar] [CrossRef] [PubMed]
- Adams, B.; Petruccione, F. Quantum effects in the brain: A review. AVS Quantum Sci. 2020, 2, 022901. [Google Scholar] [CrossRef]
- Pooam, M.; Jourdan, N.; El Esawi, M.; Sherrard, R.M.; Ahmad, M. HEK293 cell response to static magnetic fields via the radical pair mechanism may explain therapeutic effects of pulsed electromagnetic fields. PLoS ONE 2020, 15, e0243038. [Google Scholar] [CrossRef] [PubMed]
- Bradlaugh, A.; Munro, A.L.; Jones, A.R.; Baines, R.A. Exploiting the Fruitfly, Drosophila melanogaster, to Identify the Molecular Basis of Cryptochrome-Dependent Magnetosensitivity. Quantum Rep. 2021, 3, 127–136. [Google Scholar] [CrossRef]
- Bruice, T.C. Oxygen-Flavin Chemistry. Isr. J. Chem. 1984, 24, 54–61. [Google Scholar] [CrossRef]
- Massey, V. Activation of Molecular-Oxygen by Flavins and Flavoproteins. J. Biol. Chem. 1994, 269, 22459–22462. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Wallace, D.C.; Burke, P.J. Super-Resolution Imaging of Voltages in the Interior of Individual, Vital Mitochondria. ACS Nano 2023. [Google Scholar] [CrossRef] [PubMed]
- Blackiston, D.J.; McLaughlin, K.A.; Levin, M. Bioelectric controls of cell proliferation: Ion channels, membrane voltage and the cell cycle. Cell Cycle 2009, 8, 3527–3536. [Google Scholar] [CrossRef]
- Lautrup, S.; Sinclair, D.A.; Mattson, M.P.; Fang, E.F. NAD+ in Brain Aging and Neurodegenerative Disorders. Cell Metab. 2019, 30, 630–655. [Google Scholar] [CrossRef]
- Diaz-Cuadros, M.; Miettinen, T.P.; Skinner, O.S.; Sheedy, D.; Díaz-García, C.M.; Gapon, S.; Hubaud, A.; Yellen, G.; Manalis, S.R.; Oldham, W.M.; et al. Metabolic regulation of species-specific developmental rates. Nature 2023, 613, 550–557. [Google Scholar] [CrossRef]
- Rhee, S. Fibroblasts in three dimensional matrices: Cell migration and matrix remodeling. Exp. Mol. Med. 2009, 41, 858–865. [Google Scholar] [CrossRef] [PubMed]
- Serafin, A.; Culebras, M.; Collins, M.N. Synthesis and evaluation of alginate, gelatin, and hyaluronic acid hybrid hydrogels for tissue engineering applications. Int. J. Biol. Macromol. 2023, 233, 123438. [Google Scholar] [CrossRef] [PubMed]
- Kharel, P.; Somasekhar, L.; Vecheck, A.; Mitra, K. Self-Contained Three-Dimensional Bioprinter for Applications in Cardiovascular Research. J. Med. Devices 2019, 13, 031010. [Google Scholar] [CrossRef]
- Somasekhar, L.; Huynh, N.D.; Vecheck, A.; Kishore, V.; Bashur, C.A.; Mitra, K. Three-dimensional printing of cell-laden microporous constructs using blended bioinks. J. Biomed. Mater. Res. Part. A 2022, 110, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Kawasumi, M.; Saito, M. Effect of static magnetic field on cell migration. Electr. Eng. Jpn. 2007, 160, 46–52. [Google Scholar] [CrossRef]
- Zhang, Y.; Yan, J.; Xu, H.; Yang, Y.; Li, W.; Wu, H.; Liu, C. Extremely low frequency electromagnetic fields promote mesenchymal stem cell migration by increasing intracellular Ca2+ and activating the FAK/Rho GTPases signaling pathways in vitro. Stem Cell Res. Ther. 2018, 9, 143. [Google Scholar] [CrossRef]
- Ebrahimdamavandi, S.; Mobasheri, H. Application of a static magnetic field as a complementary aid to healing in an in vitro wound model. J. Wound Care 2019, 28, 40–52. [Google Scholar] [CrossRef]
- Solbu, A.A.; Caballero, D.; Damigos, S.; Kundu, S.C.; Reis, R.L.; Halaas, Ø.; Chahal, A.S.; Strand, B.L. Assessing cell migration in hydrogels: An overview of relevant materials and methods. Mater. Today Bio 2023, 18, 100537. [Google Scholar] [CrossRef]
- Rosenfeldt, H.; Grinnell, F. Fibroblast Quiescence and the Disruption of ERK Signaling in Mechanically Unloaded Collagen Matrices*. J. Biol. Chem. 2000, 275, 3088–3092. [Google Scholar] [CrossRef]
- Tschumperlin, D.J. Fibroblasts and the Ground They Walk On. Physiology 2013, 28, 380–390. [Google Scholar] [CrossRef]
- Bott, K.; Upton, Z.; Schrobback, K.; Ehrbar, M.; Hubbell, J.A.; Lutolf, M.P.; Rizzi, S.C. The effect of matrix characteristics on fibroblast proliferation in 3D gels. Biomaterials 2010, 31, 8454–8464. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, T. Optically detected magnetic resonance microscopy. Oyo Buturi 2020, 89, 196–202. [Google Scholar] [CrossRef]
- Habibalahi, A.; Moghari, M.D.; Campbell, J.M.; Anwer, A.G.; Mahbub, S.B.; Gosnell, M.; Saad, S.; Pollock, C.; Goldys, E.M. Non-invasive real-time imaging of reactive oxygen species (ROS) using auto-fluorescence multispectral imaging technique: A novel tool for redox biology. Redox Biol. 2020, 34, 101561. [Google Scholar] [CrossRef] [PubMed]
- Stephen, A.; Boppart, S.Y.; Li, L.; Chen, J.; Tu, H. Simultaneous label-free autofluorescence-multiharmonic microscopy and beyond. APL Photonics 2019, 4, 100901. [Google Scholar]
- Murphy, M.P.; Bayir, H.; Belousov, V.; Chang, C.J.; Davies, K.J.A.; Davies, M.J.; Dick, T.P.; Finkel, T.; Forman, H.J.; Janssen-Heininger, Y.; et al. Guidelines for measuring reactive oxygen species and oxidative damage in cells and in vivo. Nat. Metab. 2022, 4, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Ng, K.K.; Hu, J.J.; Ye, S.; Yang, D. Small-Molecule-Based Fluorescent Sensors for Selective Detection of Reactive Oxygen Species in Biological Systems. Annu. Rev. Biochem. 2019, 88, 605–633. [Google Scholar] [CrossRef]
- Shen, C.L.; Lou, Q.; Zang, J.H.; Liu, K.K.; Qu, S.N.; Dong, L.; Shan, C.X. Near-Infrared Chemiluminescent Carbon Nanodots and Their Application in Reactive Oxygen Species Bioimaging. Adv. Sci. 2020, 7, 1903525. [Google Scholar] [CrossRef]
- Duanghathaipornsuk, S.; Farrell, E.J.; Alba-Rubio, A.C.; Zelenay, P.; Kim, D.S. Detection Technologies for Reactive Oxygen Species: Fluorescence and Electrochemical Methods and Their Applications. Biosensors 2021, 11, 30. [Google Scholar] [CrossRef]
- Qian, Y.; Celiker, O.T.; Wang, Z.; Guner-Ataman, B.; Boyden, E.S. Temporally multiplexed imaging of dynamic signaling networks in living cells. Cell 2023, 186, 5656–5672.E21. [Google Scholar] [CrossRef]
- Semashko, V.V.; Pudovkin, M.S.; Cefalas, A.-C.; Zelenikhin, P.V.; Gavriil, V.E.; Nizamutdinov, A.S.; Kollia, Z.; Ferraro, A.; Sarantopoulou, E. Tiny Rare-Earth Fluoride Nanoparticles Activate Tumour Cell Growth via Electrical Polar Interactions. Nanoscale Res. Lett. 2018, 13, 370. [Google Scholar] [CrossRef]
- Ori, H.; Duque, M.; Frank Hayward, R.; Scheibner, C.; Tian, H.; Ortiz, G.; Vitelli, V.; Cohen, A.E. Observation of topological action potentials in engineered tissues. Nat. Phys. 2023, 19, 290–296. [Google Scholar] [CrossRef]
- Brieger, K.; Schiavone, S.; Miller, F.J.; Krause, K.H. Reactive oxygen species: From health to disease. Swiss Med. Wkly. 2012, 142, w13659. [Google Scholar] [CrossRef] [PubMed]
- Calvillo, L.; Redaelli, V.; Ludwig, N.; Qaswal, A.B.; Ghidoni, A.; Faini, A.; Rosa, D.; Lombardi, C.; Pengo, M.; Bossolasco, P.; et al. Quantum Biology Research Meets Pathophysiology and Therapeutic Mechanisms: A Biomedical Perspective. Quantum Rep. 2022, 4, 148–172. [Google Scholar] [CrossRef]
- Huang, W.J.; Zhang, X.; Chen, W.W. Role of oxidative stress in Alzheimer’s disease. Biomed. Rep. 2016, 4, 519–522. [Google Scholar] [CrossRef]
- Angelova, P.R.; Abramov, A.Y. Role of mitochondrial ROS in the brain: From physiology to neurodegeneration. FEBS Lett. 2018, 592, 692–702. [Google Scholar] [CrossRef] [PubMed]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The Hallmarks of Aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]
- Bigarella, C.L.; Liang, R.; Ghaffari, S. Stem cells and the impact of ROS signaling. Development 2014, 141, 4206–4218. [Google Scholar] [CrossRef] [PubMed]
- Marycz, K.; Kornicka, K.; Röcken, M. Static Magnetic Field (SMF) as a Regulator of Stem Cell Fate—New Perspectives in Regenerative Medicine Arising from an Underestimated Tool. Stem Cell Rev. Rep. 2018, 14, 785–792. [Google Scholar] [CrossRef]
- Stoddard-Bennett, T.; Reijo Pera, R. Treatment of Parkinson’s Disease through Personalized Medicine and Induced Pluripotent Stem Cells. Cells 2019, 8, 26. [Google Scholar] [CrossRef]
- Traber, J.; Wild, T.; Marotz, J.; Berli, M.C.; Franco-Obregón, A. Concurrent Optical- and Magnetic-Stimulation-Induced Changes on Wound Healing Parameters, Analyzed by Hyperspectral Imaging: An Exploratory Case Series. Bioengineering 2023, 10, 750. [Google Scholar] [CrossRef]

| Sample | R50 (µm) | Number of Cells | Density (Cell/µm2) |
|---|---|---|---|
| Control Day 0 | 259 | 263 | 6.2 × 10−4 |
| Control Day 1 | 251 | 300 | 7.6 × 10−4 |
| Control Day 7 | 233 | 159 | 4.7 × 10−4 |
| RF Day 0 | 218 | 177 | 5.9 × 10−4 |
| RF Day 1 | 158 | 195 | 12.5 × 10−4 |
| RF Day 7 | 161 | 385 | 23.5 × 10−4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vecheck, A.M.; McNamee, C.M.; Reijo Pera, R.; Usselman, R.J. Magnetic Field Intervention Enhances Cellular Migration Rates in Biological Scaffolds. Bioengineering 2024, 11, 9. https://doi.org/10.3390/bioengineering11010009
Vecheck AM, McNamee CM, Reijo Pera R, Usselman RJ. Magnetic Field Intervention Enhances Cellular Migration Rates in Biological Scaffolds. Bioengineering. 2024; 11(1):9. https://doi.org/10.3390/bioengineering11010009
Chicago/Turabian StyleVecheck, Amy M., Cameron M. McNamee, Renee Reijo Pera, and Robert J. Usselman. 2024. "Magnetic Field Intervention Enhances Cellular Migration Rates in Biological Scaffolds" Bioengineering 11, no. 1: 9. https://doi.org/10.3390/bioengineering11010009
APA StyleVecheck, A. M., McNamee, C. M., Reijo Pera, R., & Usselman, R. J. (2024). Magnetic Field Intervention Enhances Cellular Migration Rates in Biological Scaffolds. Bioengineering, 11(1), 9. https://doi.org/10.3390/bioengineering11010009






