Production of Astaxanthin Using CBFD1/HFBD1 from Adonis aestivalis and the Isopentenol Utilization Pathway in Escherichia coli
Abstract
:1. Introduction
2. Materials and Methods
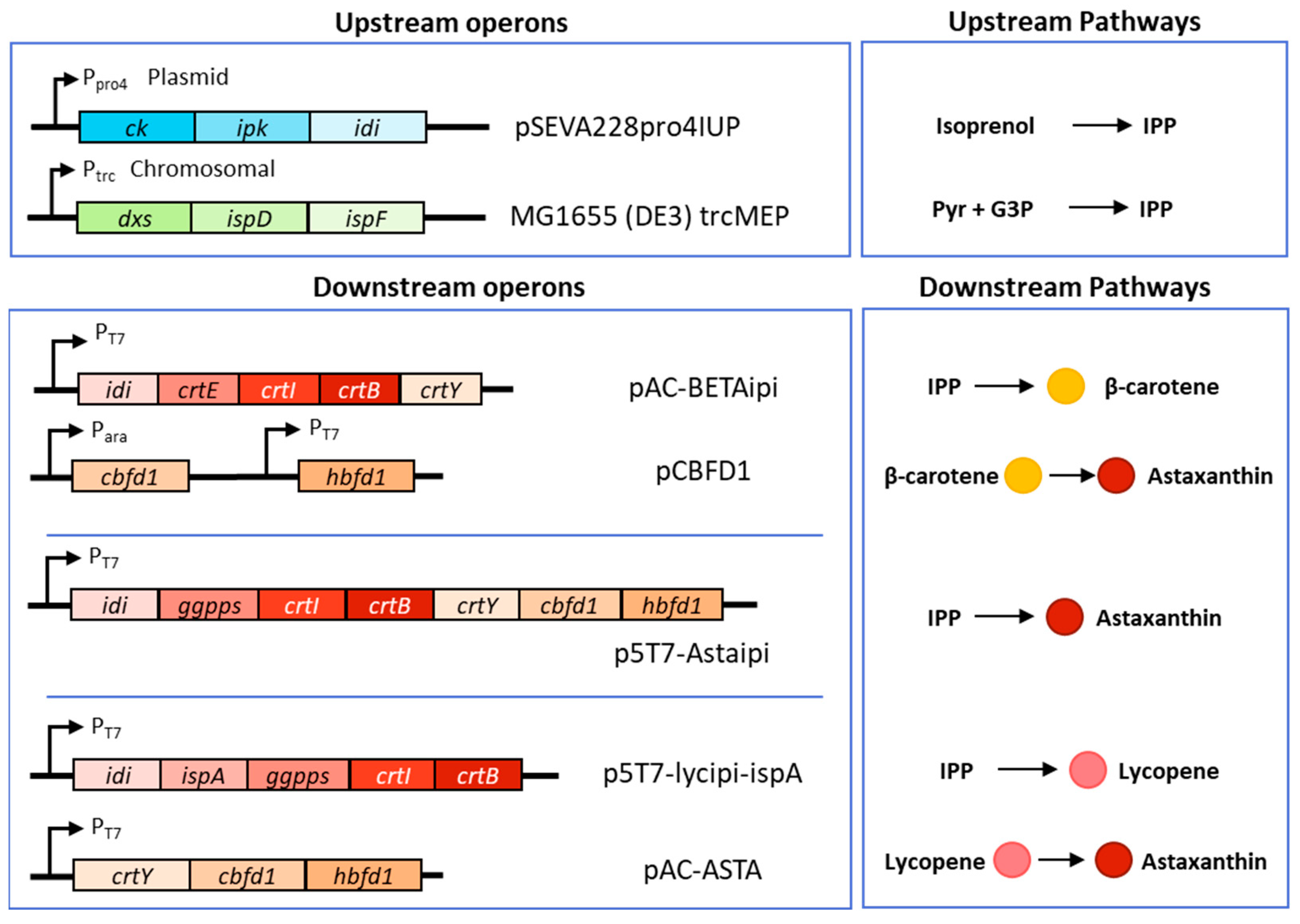
2.1. Strains, Plasmids and Genes
2.2. Cultivation Conditions
2.3. Carotenoid Extraction and UV/Vis Spectroscopy
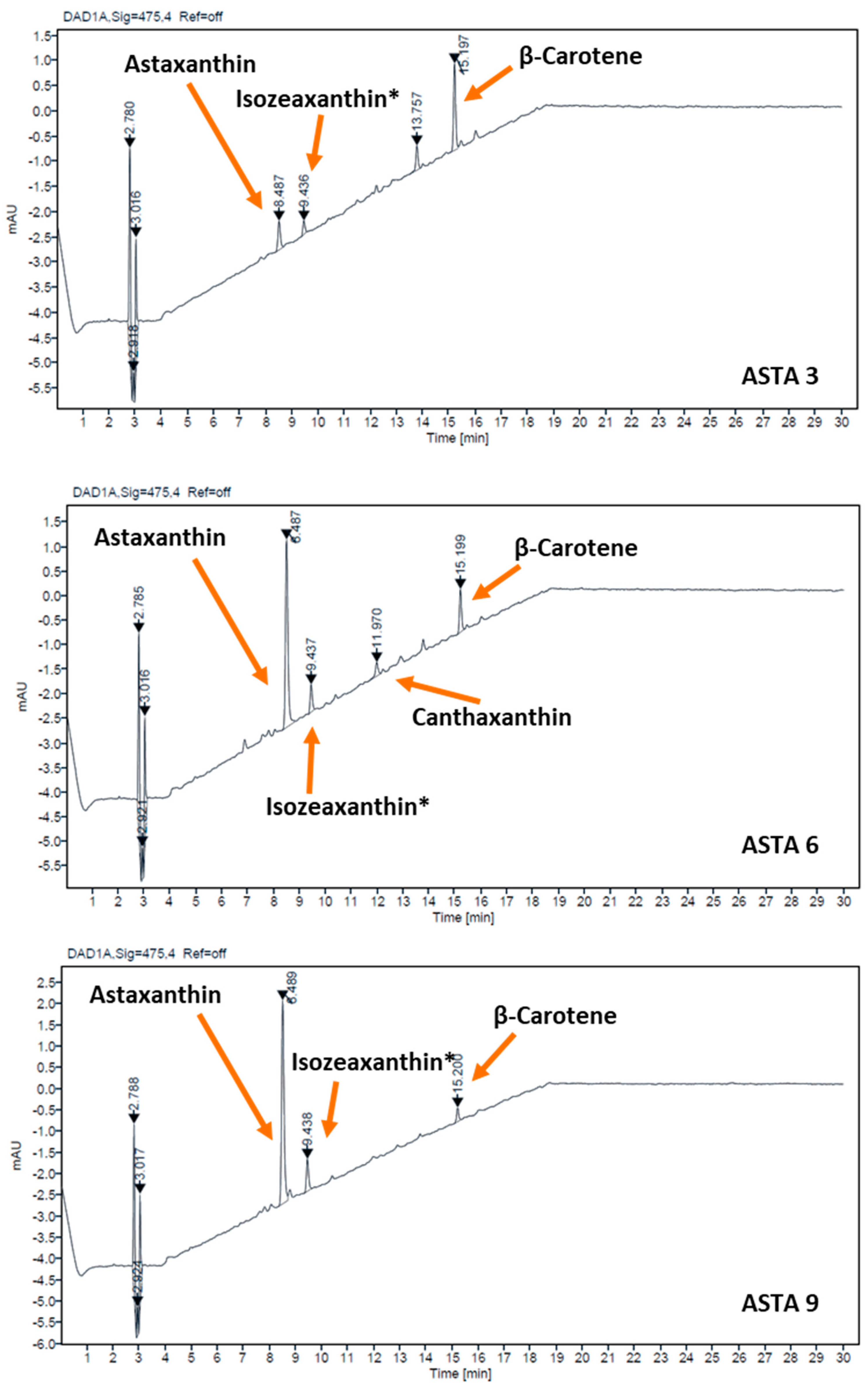
2.4. Carotenoid Characterization
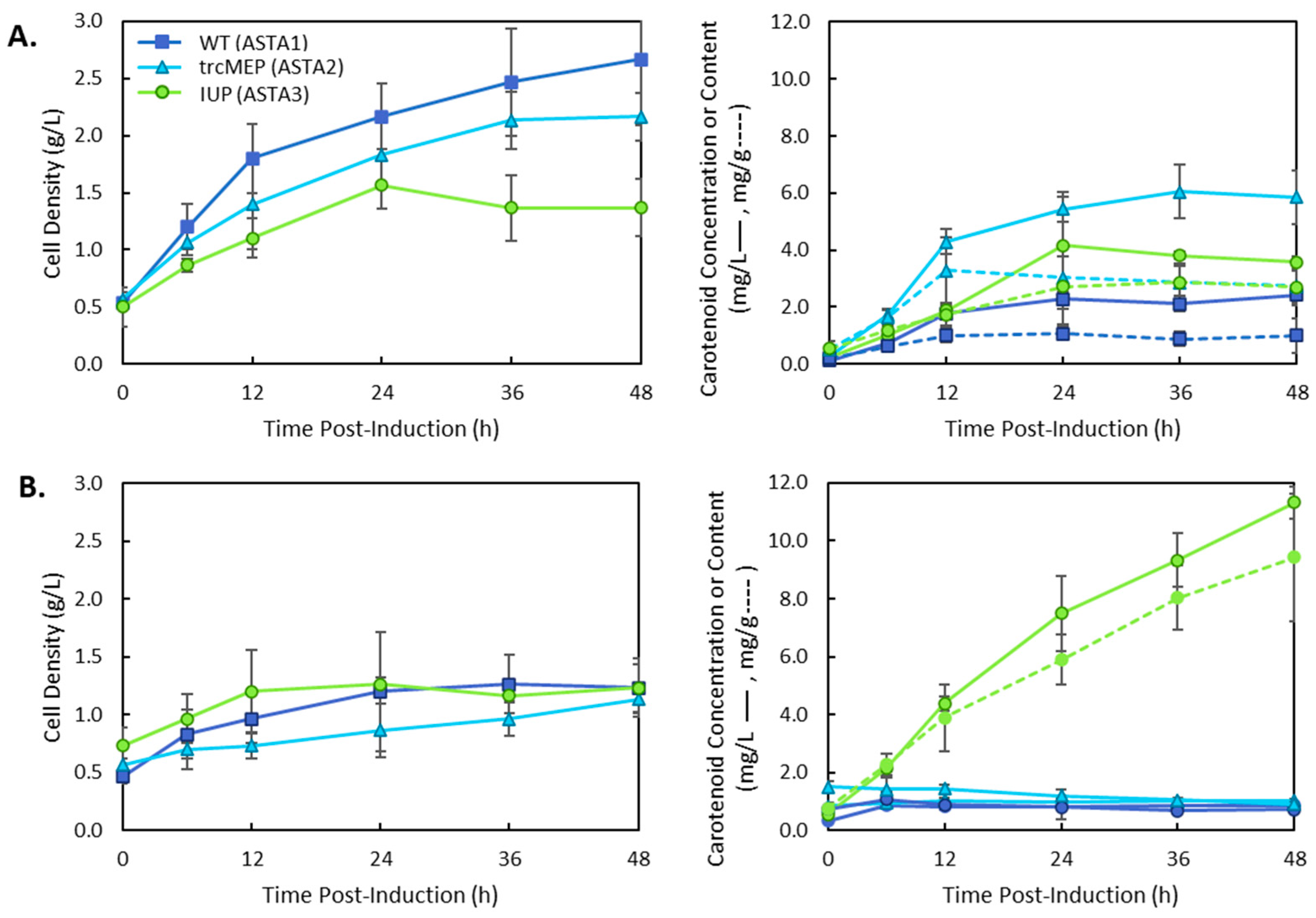
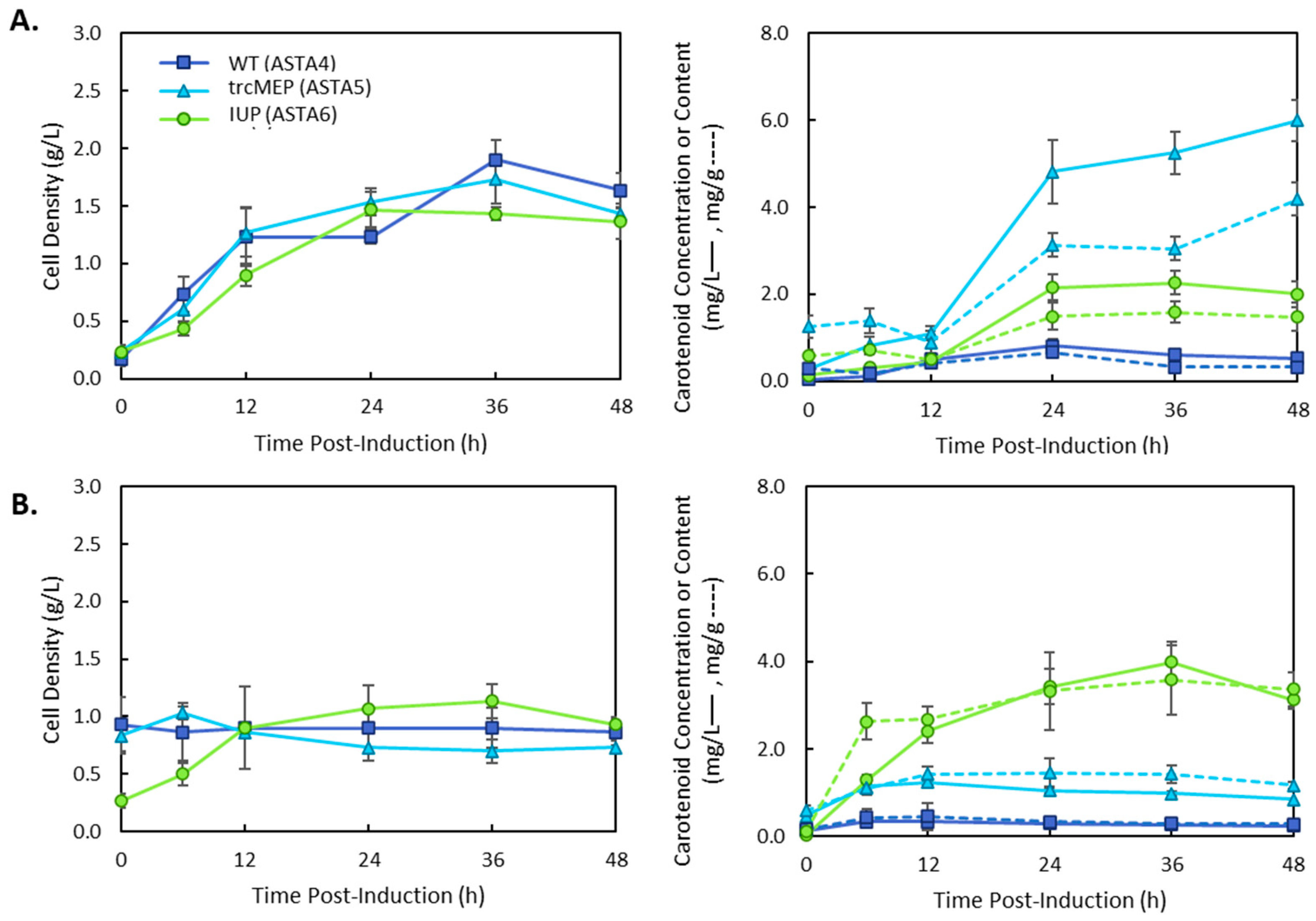
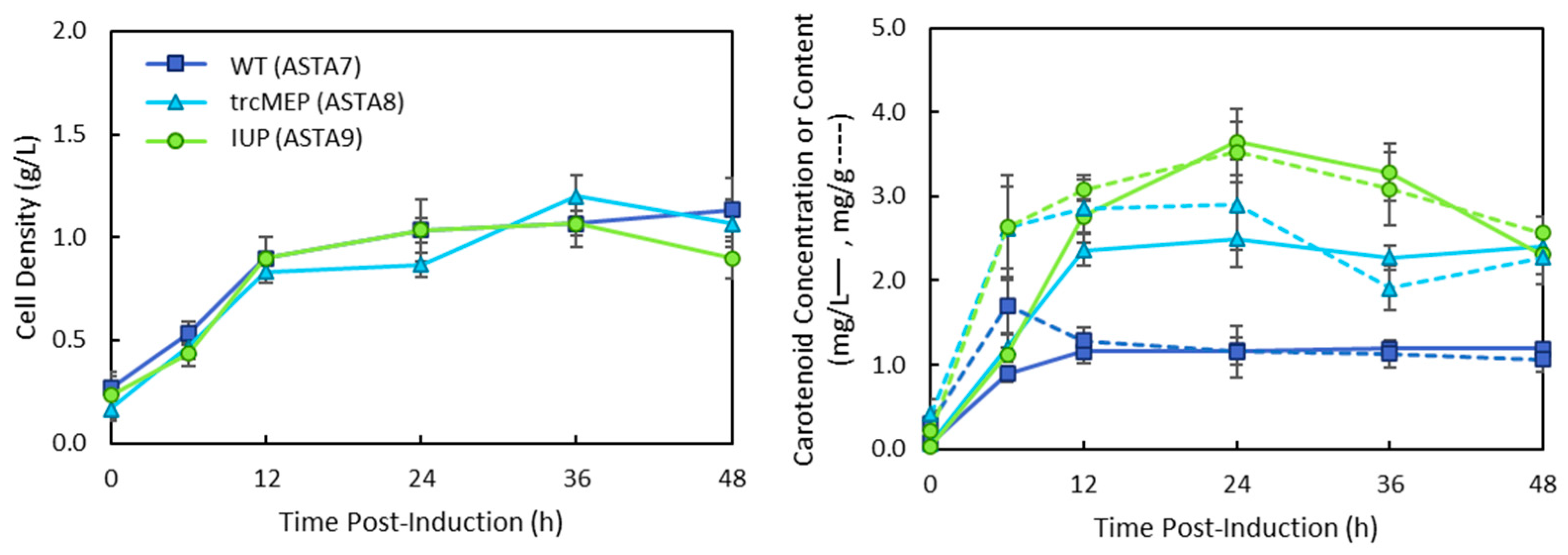
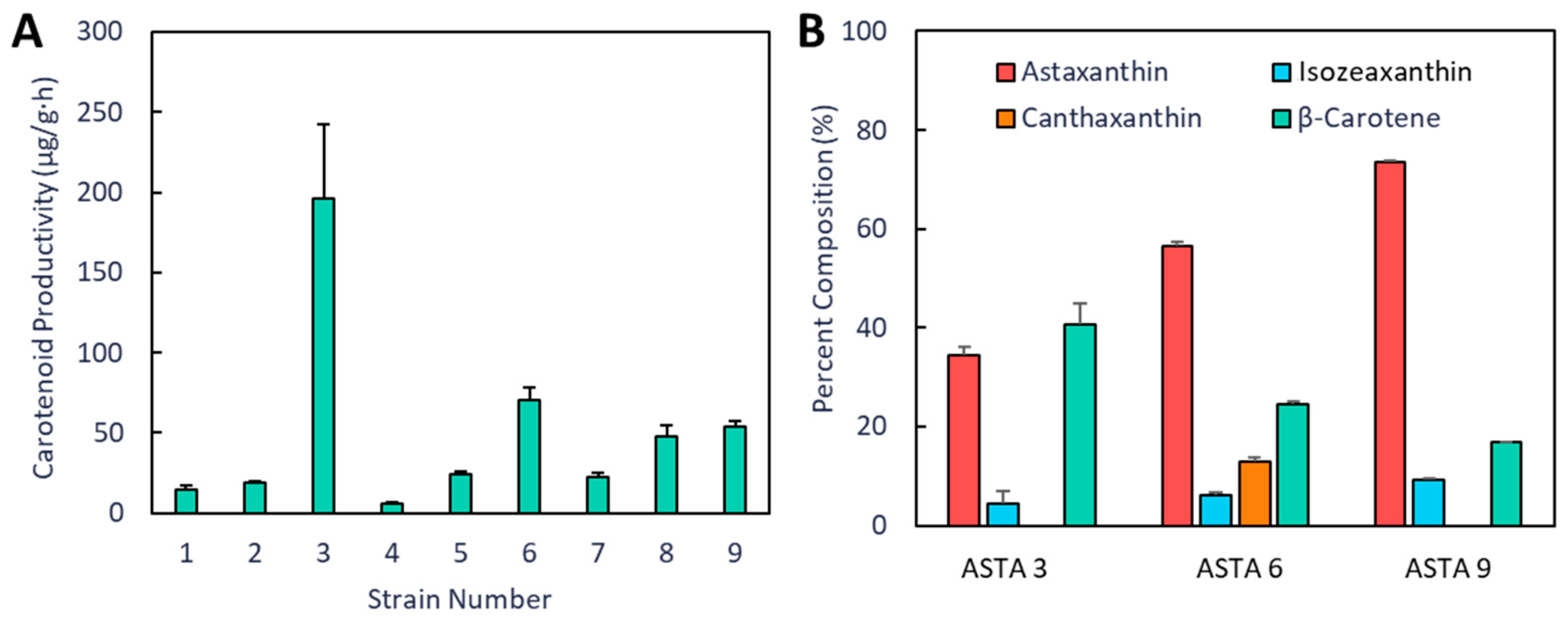
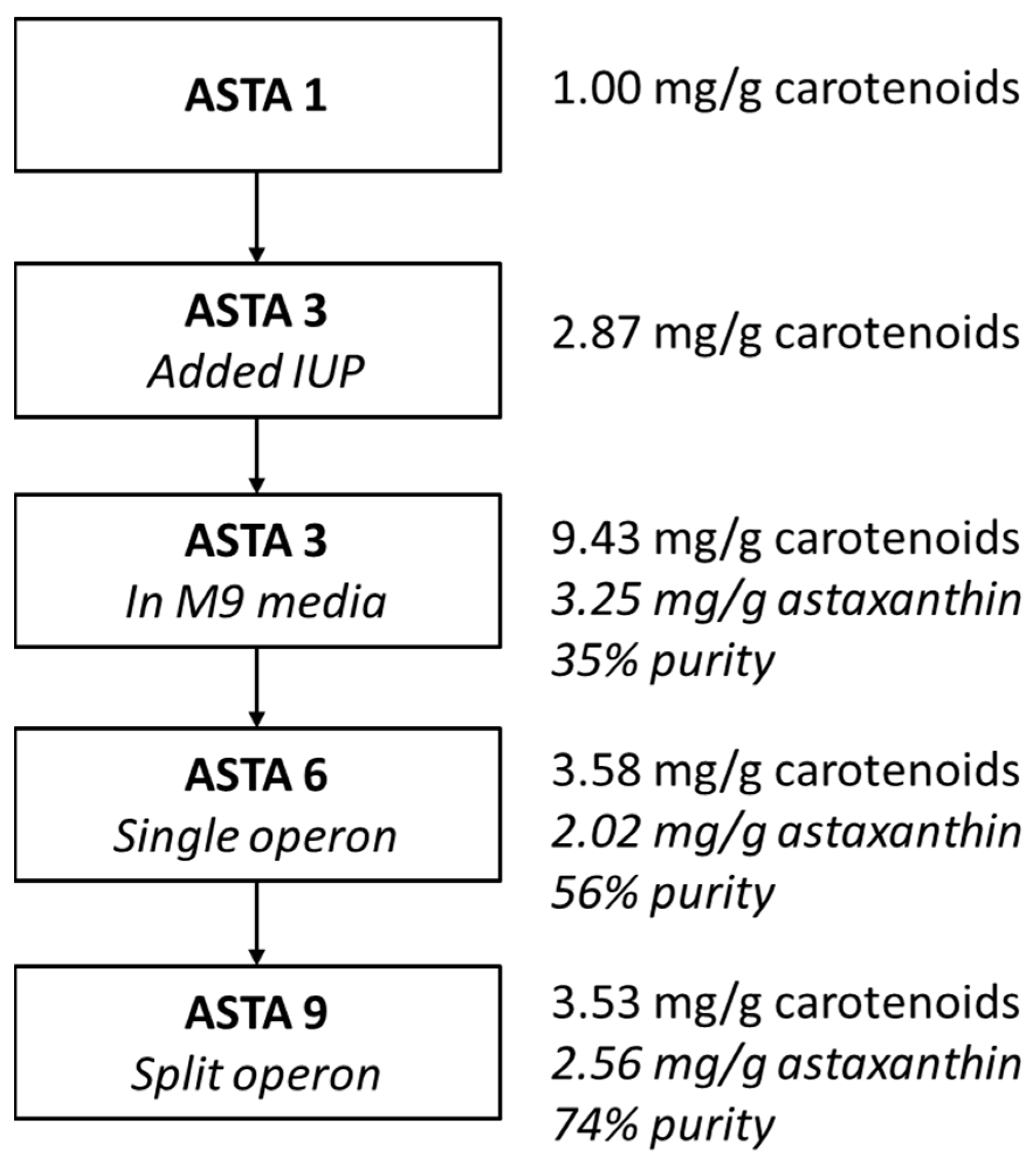
3. Results
4. Discussion
| Isoprenoid Pathway | β-Carotene Biosynthesis Genes | β-Carotene Hydroxylase and Ketolase | Specific Yield | Astaxanthin Purity | Notes | Ref. |
|---|---|---|---|---|---|---|
| Artificial IUP pathway | ||||||
| IUP | crtEBIY—Pantoea agglomerans ggpps—Taxus canadensis | cbfd, hbfd—A. aestivalis | 3.91 mg/g 2.66 mg/g | 34.6% 73.5% |
| This study |
| Endogenous MEP pathway | ||||||
| MEP | crtEBIY—Paracoccus haeundaensis | crtW—P. haeundaensis crtZ—P. haeundaensis | 0.4 mg/g | n.d. | [27] | |
| MEP | crtEBIY—Pantoea agglomerans | crtW—Anabaena variabilis crtZ—S. solfataricus | 0.3 mg/g | 71% |
| [21] |
| MEP | crtEBIY—P. agglomerans | crtW—Nostoc sp. crtZ—P. agglomerans | 1.99 mg/g | >90% |
| [28] |
| MEP | crtEBIY—P. ananatis | crtW—Brevundimonas sp. crtZ—P. ananatis | 7.4 mg/g | 96.6% | [29] | |
| MEP | crtEBIY—P. ananatis | crtW—Brevundimonas sp. crtZ—P. ananatis | 11.92 mg/g | n.d. |
| [30] |
| MEP | crtEBIY—P. ananatis | BKT—Chlamydomonas reinhardtii CHY—H. pluvialis | 4.30 mg/g | n.d. | [24] | |
| MEP | crtEBIY—P. ananatis | crtW—A. aurantiacum crtZ—P. ananatis | 8.3 mg/g | n.d. |
| [31] |
| MEP | crtEBIY—P. ananatis | crtW—Brevundimonas sp. crtZ—Brevundimonas sp. | 0.58 mg/g | ~60% |
| [32] |
| Engineered MEP pathway | ||||||
| MEP + extensive host changes | crtEBIY—P. ananatis | crtW—Brevundimonas sp. crtZ—P. ananatis | ~12 mg/g | n.d. |
| [33] |
| MEP + E. coli idi | Ch(crtEBIY)—P. ananatis | Ch(crtZ)—P. ananatis Ch(crtW148)—N. punctiforme | 1.4 mg/g | >95% | [34] | |
| MEP + E. coli idi | Ggpps—Archaeoglobus fulgidus CrtBIY—A. aurantiacum | crtW—A. aurantiacum crtZ—A. aurantiacum | 1.25 mg/g | n.d. | [35] | |
| MEP + H. pluvialis idi + E. coli dxs | crtEBI—P. agglomerans LycB—Solanum lycopersicum | crtW—N. sphaeroides crtZ—P. ananatis | 5.8 mg/g | Majority |
| [36] |
| MEP + E. coli idi, ispA | crtE—P. ananatis crtIBY—A. aurantiacum | crtZW—A. aurantiacum | 1.4 mg/g | n.d. | [35] | |
| MEP + E. coli ispDF | crtEBIY—P. ananatis | crtZ—P. ananatis trBKT—Chlamydomonas reinhardtii | 7.12 mg/g | n.d. |
| [12] |
| MEP + E. coli idi, ispA, ispH | crtEYIB—P. haeundaensis | crtW—P. haeundaensis crtZ—P. haeundaensis | 1.2 mg/g | n.d. | [37] | |
| MEP + K. gwangalliensis ispCDEFGH, idi | crtEYIB—P. haeundaensis | crtW—P. haeundaensis crtZ—P. haeundaensis | 1.10 mg/g | ~65% | Jeong 2018 microbial letters | |
| Modified MEP and other pathways | Ch(crtEYIB)—P. agglomerans | crtW—Brevundimonas sp. crtZ—P. agglomerans | 5.88 mg/g | 99% |
| [38] |
| Modified MEP and other pathways | Ch(crtEYIB)—P. agglomerans | Ch(crtW-GlpF) Brevundimonas sp. Ch(crtZ-GlpF) P. agglomerans | ~0.28 mg/g (AX) | n.d. |
| [39] |
| Heterologous MVA pathway | ||||||
| MEP + MVA | Ch(crtEBIY)—P. ananatis Ch (crtY—additional copy)—P. ananatis | crtZ—P. ananatis (2 copies) crtW—P. ananatis (2 copies) crtZ—Paracoccus sp. PC1 (1 copy) | 2.9 mg/g | 65% |
| [14] |
| MEP + MVA | crtEBI—P. ananatis crtY—P. agglomerans | crtW- Brevundimonas sp. crtZ—P. agglomernas | 6.6 mg/g (AX) | n.d. | [40] | |
| MEP + MVA | crtEBI—P. agglomerans crtY—P. ananatis | CrtZ—P. ananatis LMG20103 crtW Brevundimonas sp. SD212 | ~6 mg/g * | ~85% |
| [41] |
| MEP + MVA | Ch(crtEBIY)—P. agglomerans | crtZ—Brevundimonas sp. SD212 crtW—Paracoccus sp. N81106 | 4.67 mg/g | N.d. |
| [42] |
| MEP + MVA | crtEYIB | crtW—P. agglomerans crtZ—P. agglomerans | 6.17 mg/g | 32% |
| [43] |
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Genes | Origin | Accession |
|---|---|---|
| ck | S. cerevisiae, codon-optimized | AAA34499.1 |
| ipk | Arabidopsis thaliana, codon-optimized | AAN12957.1 |
| idi | E. coli | AAD26812.1 |
| ispA | E. coli | |
| ggpps | Taxus canadensis, codon-optimized, truncated first 98 amino acids, methionine added | AAD16018.1 |
| crtE | P. agglomerans | AAA21260.1 |
| crtB | P. agglomerans | AFZ89043.1 |
| crtI | P. agglomerans | AFZ89042.1 |
| crtY | P. agglomerans | AAA64980.1 |
| ipi | P. agglomerans | AAA64978.1 |
| cbfd, hbfd | A. aestivalis | ABK41045.1 |
| hbfd | A. aestivalis | AAV85452.1 |
| Fragment | Template | Assembly | Sequences |
|---|---|---|---|
| p5T7-lyc_bb | p5T7-lycipi-ggpps | p5T7-Astaipi | F: catgaatcaactcGCAGTACATAACGATGGAAC R: tcactccctgctTTGAACCCAAAAGGGCGG |
| crtY | pAC-BETAipi | p5T7-Astaipi | F: cttttgggttcaaAGCAGGGAGTGAGAGCGTATC R: cctcctgttagccAAAGCCTGCGCCAATCAC |
| cbfd | pCBFD1 | p5T7-Astaipi | F: ggcgcaggctttGGCTAACAGGAGGAATTAAC R: ctttttctttctcataggcgactcctcTCGACGAATTCAGATCTGG |
| hbfd | pCBFD1 | p5T7-Astaipi | F: GGAGTCGCCTATGAGAAAG R: gttatgtactgcGAGTTGATTCATGTAGATGATTGC |
| p5T7-lyc_bb (2) | p5T7-lycipi-ggpps | p5T7-lycipi-ispA | F: ataaaggatcACAGGAGTAGTGATGAATGAAG R: atatctccttTTGAACCCAAAAGGGCGG |
| IspA | p5T7-IspA-ads | p5T7-lycipi-ispA | F: ttgggttcaaAAGGAGATATACCATATGGACTTTC R: ctactcctgtGATCCTTTATTTATTACGCTGGATG |
| beta_bb | pAC-BETAipi | pACT7-CBFD1 | F: aacggcatgaGGCACCAATAACTGCCTTAAAAAAATTAC R: cgggcagtgaCATGAGACGCTGTGCCTTTAG |
| lacI_pT7 | p5T7-lycipi-ggpps | pACT7-CBFD1 | F: gcgtctcatgTCACTGCCCGCTTTCCAG R: ctgttagcccCTACAGGGGAATTGTTATCCG |
| cbfd (2) | pCBFD1 | pACT7-CBFD1 | F: tcccctgtagGGGCTAACAGGAGGAATTAAC R: gatatccaatTCGACGAATTCAGATCTGG |
| T7t | p5T7-lycipi-ggpps | pACT7-CBFD1 | F: aattcgtcgaATTGGATATCGGCCGGCC R: tattggtgccTCATGCCGTTTGTGATGG |
| CBFD_bb | pACT7-CBFD1 | pAC-ASTA | F: tctacatgaaATTGGATATCGGCCGGCC R: actccctgctCCAATCTATTACTCTACTGCTTCATAATG |
| crtY (2) | pAC-BETAipi | pAC-ASTA | F: aatagattggAGCAGGGAGTGAGAGCGTATC R: CACAACGGTTTTTTTCATCCTTTATC |
| hbfd (2) | pCBFD1 | pAC-ASTA | F: ggatgaaaaaaaccgttgtgaaGGAGTCGCCTATGAGAAAG R: gatatccaatTTCATGTAGATGATTGCGTTC |
References
- Ambati, R.R.; Phang, S.-M.; Ravi, S.; Aswathanarayana, R.G. Astaxanthin: Sources, Extraction, Stability, Biological Activities and Its Commercial Applications—A Review. Mar. Drugs 2014, 12, 128–152. [Google Scholar] [CrossRef] [PubMed]
- Panis, G.; Carreon, J.R. Commercial astaxanthin production derived by green alga Haematococcus pluvialis: A microalgae process model and a techno-economic assessment all through production line. Algal Res. 2016, 18, 175–190. [Google Scholar] [CrossRef]
- Lin, Y.-J.; Lin, J.-Y.; Wang, D.-S.; Chen, C.-H.; Chiou, M.-H. Safety assessment of astaxanthin derived from engineered Escherichia coli K-12 using a 13-week repeated dose oral toxicity study and a prenatal developmental toxicity study in rats. Regul. Toxicol. Pharmacol. 2017, 87, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Boussiba, S. Carotenogenesis in the Green Alga Haematococcus Pluvialis: Cellular Physiology and Stress Response. Physiol. Plant. 2000, 108, 111–117. [Google Scholar] [CrossRef]
- Jiang, G.; Yang, Z.; Wang, Y.; Yao, M.; Chen, Y.; Xiao, W.; Yuan, Y. Enhanced astaxanthin production in yeast via combined mutagenesis and evolution. Biochem. Eng. J. 2020, 156, 107519. [Google Scholar] [CrossRef]
- Lu, M.; Zhang, Y.; Zhao, C.; Zhou, P.; Yu, L. Analysis and Identification of Astaxanthin and its Carotenoid Precursors from Xanthophyllomyces dendrorhous by High-Performance Liquid Chromatography. Z. Für Naturforschung-Sect. C J. Biosci. 2010, 65, 489–494. [Google Scholar] [CrossRef]
- Kaewpintong, K.; Shotipruk, A.; Powtongsook, S.; Pavasant, P. Photoautotrophic high-density cultivation of vegetative cells of Haematococcus pluvialis in airlift bioreactor. Bioresour. Technol. 2007, 98, 288–295. [Google Scholar] [CrossRef]
- Olaizola, M. Commercial production of astaxanthin from Haematococcus pluvialis using 25,000-liter outdoor photobioreactors. J. Appl. Phycol. 2000, 12, 499–506. [Google Scholar] [CrossRef]
- Shah, M.M.R.; Liang, Y.; Cheng, J.J.; Daroch, M. Astaxanthin-Producing Green Microalga Haematococcus pluvialis: From Single Cell to High Value Commercial Products. Front. Plant Sci. 2016, 7, 531. [Google Scholar] [CrossRef]
- Gao, Z.; Meng, C.; Gao, H.; Zhang, X.; Xu, D.; Su, Y.; Zhao, Y.; Ye, N. Analysis of MRNA Expression Profi Les of Carotenogenesis and Astaxanthin Production of Haematococcus Pluvialis under Exogenous. Biol. Res. 2013, 46, 201–206. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, X.; Too, H.-P. Microbial astaxanthin biosynthesis: Recent achievements, challenges, and commercialization outlook. Appl. Microbiol. Biotechnol. 2020, 104, 5725–5737. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Binkley, R.M.; Kim, W.J.; Lee, M.H.; Lee, S.Y. Metabolic engineering of Escherichia coli for high-level astaxanthin production with high productivity. Metab. Eng. 2018, 49, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Ward, V.C.A.; Chatzivasileiou, A.O.; Stephanopoulos, G. Metabolic engineering of Escherichia coli for the production of isoprenoids. FEMS Microbiol. Lett. 2018, 365, fny079. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Gong, Z.; Tang, J.; Lu, F.; Li, Q.; Zhang, X. Improving astaxanthin production in Escherichia coli by co-utilizing CrtZ enzymes with different substrate preference. Microb. Cell Factories 2022, 21, 71. [Google Scholar] [CrossRef]
- Chatzivasileiou, A.O.; Ward, V.; Edgar, S.M.; Stephanopoulos, G. A Novel Two-step pathway for isoprenoid synthesis. Proc. Natl. Acad. Sci. USA 2018, 116, 506–511. [Google Scholar] [CrossRef]
- Cunningham, F.X., Jr.; Gantt, E. Elucidation of the Pathway to Astaxanthin in the Flowers of Adonis aestivalis. Plant Cell 2011, 23, 3055–3069. [Google Scholar] [CrossRef]
- Ajikumar, P.K.; Xiao, W.-H.; Tyo, K.E.J.; Wang, Y.; Simeon, F.; Leonard, E.; Mucha, O.; Phon, T.H.; Pfeifer, B.; Stephanopoulos, G. Isoprenoid Pathway Optimization for Taxol Precursor Overproduction in Escherichia coli. Science 2010, 330, 70–74. [Google Scholar] [CrossRef]
- Cunningham, F.X.; Gantt, E. A study in scarlet: Enzymes of ketocarotenoid biosynthesis in the flowers of Adonis aestivalis. Plant J. 2005, 41, 478–492. [Google Scholar] [CrossRef]
- Wolfe, R.A.; Fowler, R.G. An Application of Spectrographic Methods to Chemical Concentrations of Trace Elements in Iron and Steel Analysis. J. Opt. Soc. Am. 1945, 35, 86–87. [Google Scholar] [CrossRef]
- Khachik, F.; Beecher, G.R. Separation of Carotenol Fatty Acid Esters by High-Performance Liquid Chromatograph. J. Chromatogr. A 1988, 449, 119–133. [Google Scholar] [CrossRef]
- Scaife, M.A.; Ma, C.A.; Ninlayarn, T.; Wright, P.C.; Armenta, R.E. Comparative Analysis of β-Carotene Hydroxylase Genes for Astaxanthin Biosynthesis. J. Nat. Prod. 2012, 75, 1117–1124. [Google Scholar] [CrossRef] [PubMed]
- Pitera, D.J.; Paddon, C.J.; Newman, J.D.; Keasling, J.D. Balancing a heterologous mevalonate pathway for improved isoprenoid production in Escherichia coli. Metab. Eng. 2007, 9, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.N.; Lee, Y.; Hussein, R. Fundamental relationship between operon organization and gene expression. Proc. Natl. Acad. Sci. USA 2011, 108, 10626–10631. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Huang, J.-C. Assessment of Expression Cassettes and Culture Media for Different Escherichia coli Strains to Produce Astaxanthin. Nat. Prod. Bioprospect. 2018, 8, 397–403. [Google Scholar] [CrossRef]
- Terol, G.L.; Gallego-Jara, J.; Martínez, R.A.S.; Vivancos, A.M.; Díaz, M.C.; de Diego Puente, T. Impact of the Expression System on Recombinant Protein Production in Escherichia coli BL21. Front. Microbiol. 2021, 12, 682001. [Google Scholar] [CrossRef]
- Davis, J.H.; Rubin, A.J.; Sauer, R.T. Design, construction and characterization of a set of insulated bacterial promoters. Nucleic Acids Res. 2011, 39, 1131–1141. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, Y.T. Cloning and characterization of the astaxanthin biosynthesis gene cluster from the marine bacterium Paracoccus haeundaensis. Gene 2006, 370, 86–95. [Google Scholar] [CrossRef]
- Scaife, M.A.; Burja, A.M.; Wright, P.C. Characterization of cyanobacterial β-carotene ketolase and hydroxylase genes in Escherichia coli, and their application for astaxanthin biosynthesis. Biotechnol. Bioeng. 2009, 103, 944–955. [Google Scholar] [CrossRef]
- Lu, Q.; Bu, Y.-F.; Liu, J.-Z. Metabolic Engineering of Escherichia coli for Producing Astaxanthin as the Predominant Carotenoid. Mar. Drugs 2017, 15, 296. [Google Scholar] [CrossRef]
- Lu, Q.; Liu, J.-Z. Enhanced Astaxanthin Production in Escherichia coli via Morphology and Oxidative Stress Engineering. J. Agric. Food Chem. 2019, 67, 11703–11709. [Google Scholar] [CrossRef]
- Chou, Y.-L.; Ko, C.-Y.; Yen, C.-C.; Chen, L.-F.O.; Shaw, J.-F. Multiple promoters driving the expression of astaxanthin biosynthesis genes can enhance free-form astaxanthin production. J. Microbiol. Methods 2019, 160, 20–28. [Google Scholar] [CrossRef]
- Nogueira, M.; Enfissi, E.M.; Welsch, R.; Beyer, P.; Zurbriggen, M.D.; Fraser, P.D. Construction of a fusion enzyme for astaxanthin formation and its characterisation in microbial and plant hosts: A new tool for engineering ketocarotenoids. Metab. Eng. 2019, 52, 243–252. [Google Scholar] [CrossRef]
- Lu, Q.; Zhou, X.-L.; Liu, J.-Z. Adaptive laboratory evolution and shuffling of Escherichia coli to enhance its tolerance and production of astaxanthin. Biotechnol. Biofuels Bioprod. 2022, 15, 17. [Google Scholar] [CrossRef]
- Lemuth, K.; Steuer, K.; Albermann, C. Engineering of a plasmid-free Escherichia coli strain for improved in vivo biosynthesis of astaxanthin. Microb. Cell Factories 2011, 10, 29. [Google Scholar] [CrossRef]
- Wang, C.-W.; Oh, M.-K.; Liao, J.C. Engineered isoprenoid pathway enhances astaxanthin production in Escherichia coli. Biotechnol. Bioeng. 1999, 62, 235–241. [Google Scholar] [CrossRef]
- Zelcbuch, L.; Antonovsky, N.; Bar-Even, A.; Levin-Karp, A.; Barenholz, U.; Dayagi, M.; Liebermeister, W.; Flamholz, A.; Noor, E.; Amram, S.; et al. Spanning high-dimensional expression space using ribosome-binding site combinatorics. Nucleic Acids Res. 2013, 41, e98. [Google Scholar] [CrossRef]
- Lee, J.-H.; Seo, Y.-B.; Kim, Y.-T. Enhanced Production of Astaxanthin by Metabolic Engineered Isoprenoid Pathway in Escherichia coli. J. Life Sci. 2008, 18, 1764–1770. [Google Scholar] [CrossRef]
- Li, D.; Li, Y.; Xu, J.-Y.; Li, Q.-Y.; Tang, J.-L.; Jia, S.-R.; Bi, C.-H.; Dai, Z.-B.; Zhu, X.-N.; Zhang, X.-L. Engineering CrtW and CrtZ for improving biosynthesis of astaxanthin in Escherichia coli. Chin. J. Nat. Med. 2020, 18, 666–676. [Google Scholar] [CrossRef]
- Ye, L.; Zhu, X.; Wu, T.; Wang, W.; Zhao, D.; Bi, C.; Zhang, X. Optimizing the localization of astaxanthin enzymes for improved productivity. Biotechnol. Biofuels 2018, 11, 278. [Google Scholar] [CrossRef]
- Ma, T.; Zhou, Y.; Li, X.; Zhu, F.; Cheng, Y.; Liu, Y.; Deng, Z.; Liu, T. Genome mining of astaxanthin biosynthetic genes from Sphingomonas sp. ATCC 55669 for heterologous overproduction in Escherichia coli. Biotechnol. J. 2016, 11, 228–237. [Google Scholar] [CrossRef]
- Zhang, C.; Seow, V.Y.; Chen, X.; Too, H.-P. Multidimensional heuristic process for high-yield production of astaxanthin and fragrance molecules in Escherichia coli. Nat. Commun. 2018, 9, 1858. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Yan, P.; Liu, X.; Wang, Z.; Tang, Y.-J.; Chen, T.; Zhao, X. Combinatorial expression of different β-carotene hydroxylases and ketolases in Escherichia coli for increased astaxanthin production. J. Ind. Microbiol. Biotechnol. 2019, 46, 1505–1516. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Wang, H.; Tang, J.; Bi, C.; Li, Q.; Zhang, X. Coordinated Expression of Astaxanthin Biosynthesis Genes for Improved Astaxanthin Production in Escherichia coli. J. Agric. Food Chem. 2020, 68, 14917–14927. [Google Scholar] [CrossRef] [PubMed]

| Plasmids | Description (ori, Antibiotic Marker, Operon) | Reference |
|---|---|---|
| pAC-BETAipi | p15A ori, CmR, Pendogenous (crtE, ipi, crtY, crtI, crtB) | [18] |
| pCBFD1 | pBR322 ori, ApR, PBAD (cbfd), Plac (hbfd) | [16] |
| pSEVA228-pro4IUP | RK2 ori, KnR, Ppro4 (ck, ipk, idi) | [15] |
| p5T7-lycipi-ggpps | pSC101 ori, SpR, PT7lacUV (ggpps, ipi, crtI, crtB) | [15] |
| p5T7-IspA-ads | pSC101 ori, SpR, PT7lacUV (ispA, ads) | [15] |
| p5T7-lycipi-ispA | pSC101 ori, SpR, PT7lacUV (ggpps, ispA, ipi, crtI, crtB) | This study |
| p5T7-Astaipi | pSC101 ori, SpR, PT7lacUV (ggpps, crtY, cbfd, hbfd, ipi, crtI, crtB) | This study |
| pAC-ASTA | p15A ori, CmR, PT7lacUV (cbfd, crtY, hbfd) | This study |
| Host/Strain | Genotype, Plasmids | Reference |
| MG1655(DE3) | ΔendA ΔrecA (λ DE3) | [17] |
| MG1655(DE3)-trcMEP | ΔendA ΔrecA (λ DE3) Ptrc dxs-idi-ispDF | [17] |
| NEB-5α | fhuA2 Δ(argF-lacZ)U169 phoA glnV44 Φ80 Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 | NEB |
| ASTA 1 | MG1655(DE3), pAC-BETAipi, pCBFD1 | This study |
| ASTA 2 | MG1655(DE3)-trcMEP, pAC-BETAipi, pCBFD1 | This study |
| ASTA 3 | MG1655(DE3), pSEVA228-pro4IUP, pAC-BETAipi, pCBFD1 | This study |
| ASTA 4 | MG1655(DE3), p5T7-Astaipi | This study |
| ASTA 5 | MG1655(DE3)-trcMEP, p5T7-Astaipi | This study |
| ASTA 6 | MG1655(DE3), pSEVA228-pro4IUP, p5T7-Astaipi | This study |
| ASTA 7 | MG1655(DE3), p5T7-lycipi-ispA, pAC-ASTA | This study |
| ASTA 8 | MG1655(DE3)-trcMEP, p5T7-lycipi-ispA, pAC-ASTA | This study |
| ASTA 9 | MG1655(DE3), pSEVA228-pro4IUP, p5T7-lycipi-ispA, pAC-ASTA | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roth, J.H.; Ward, V.C.A. Production of Astaxanthin Using CBFD1/HFBD1 from Adonis aestivalis and the Isopentenol Utilization Pathway in Escherichia coli. Bioengineering 2023, 10, 1033. https://doi.org/10.3390/bioengineering10091033
Roth JH, Ward VCA. Production of Astaxanthin Using CBFD1/HFBD1 from Adonis aestivalis and the Isopentenol Utilization Pathway in Escherichia coli. Bioengineering. 2023; 10(9):1033. https://doi.org/10.3390/bioengineering10091033
Chicago/Turabian StyleRoth, Jared H., and Valerie C. A. Ward. 2023. "Production of Astaxanthin Using CBFD1/HFBD1 from Adonis aestivalis and the Isopentenol Utilization Pathway in Escherichia coli" Bioengineering 10, no. 9: 1033. https://doi.org/10.3390/bioengineering10091033
APA StyleRoth, J. H., & Ward, V. C. A. (2023). Production of Astaxanthin Using CBFD1/HFBD1 from Adonis aestivalis and the Isopentenol Utilization Pathway in Escherichia coli. Bioengineering, 10(9), 1033. https://doi.org/10.3390/bioengineering10091033






