A Human Ovarian Tumor & Liver Organ-on-Chip for Simultaneous and More Predictive Toxo-Efficacy Assays
Abstract
1. Introduction
2. Materials and Methods
2.1. Liver and Ovarian Cancer Cell Cultures
2.2. Ovarian Cancer Model
2.3. Static vs. Dynamic In Vitro Model
2.4. Computational Fluid-Dynamic Analyses
2.5. Cell Viability and Pharmacodynamic Evaluation
2.6. Immunofluorescence
2.7. Statistical Analysis
3. Results
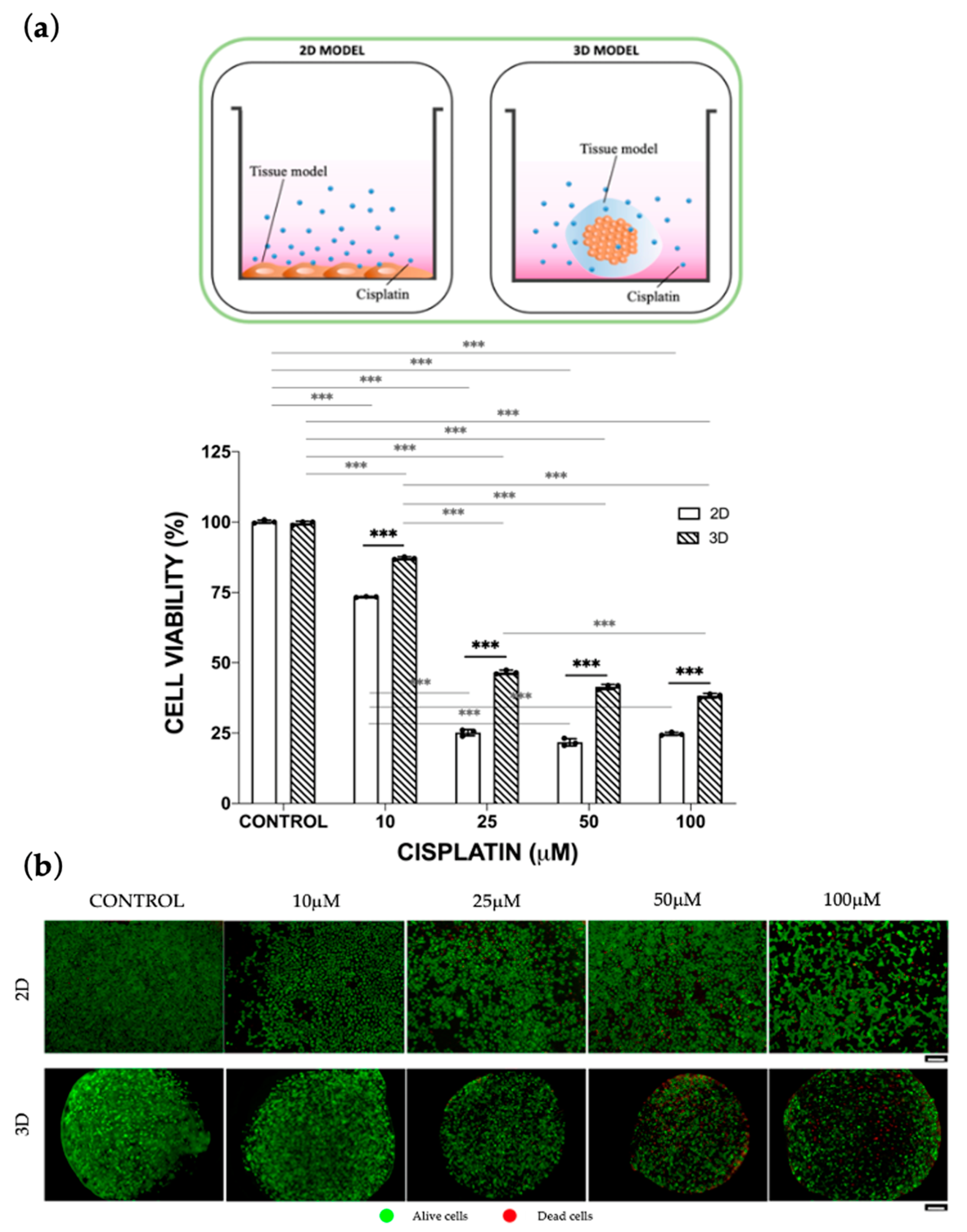
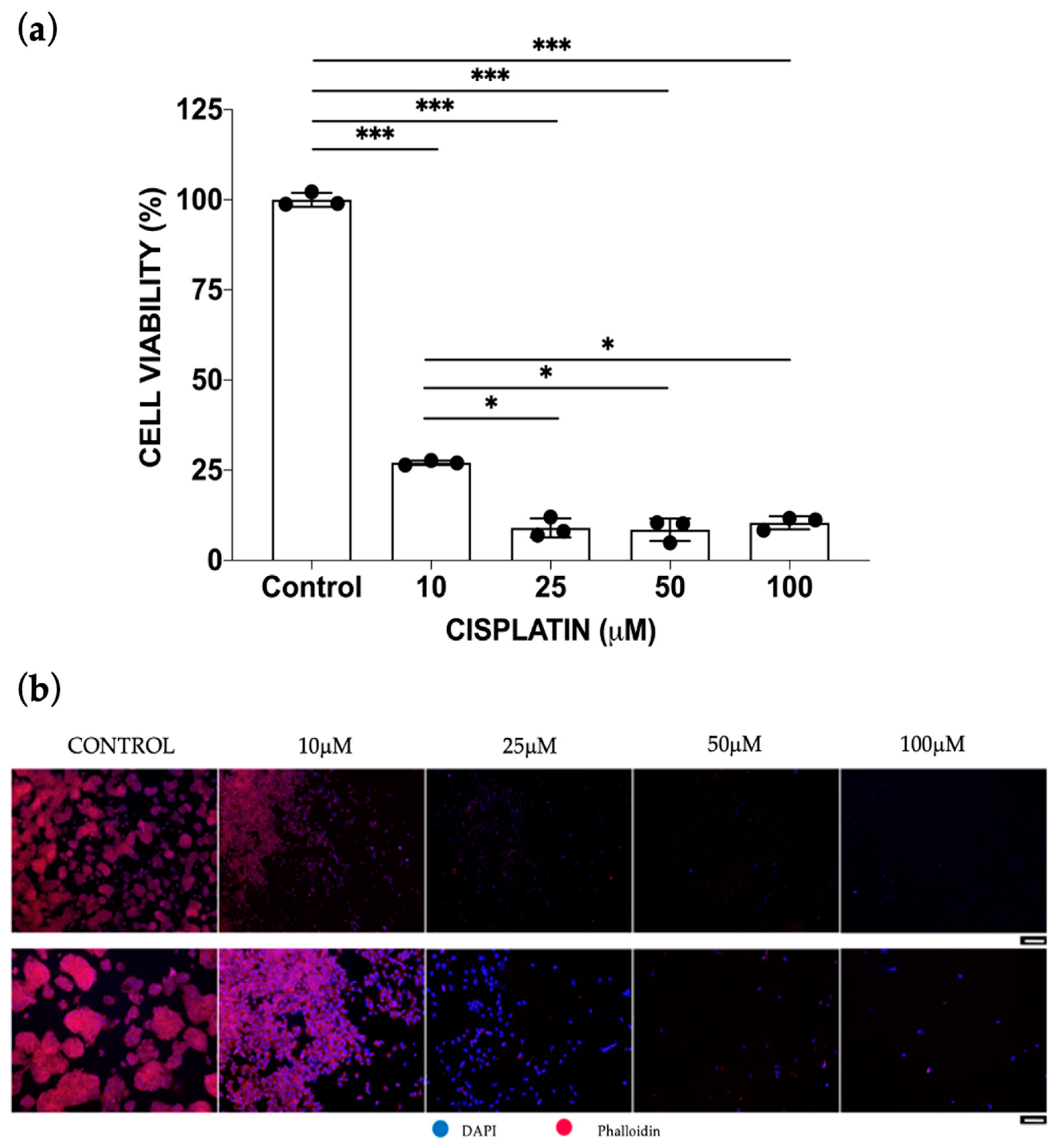
3.1. Static Cisplatin Toxo-Efficacy Evaluation
Mono-Culture vs. Co-Culture Conditions
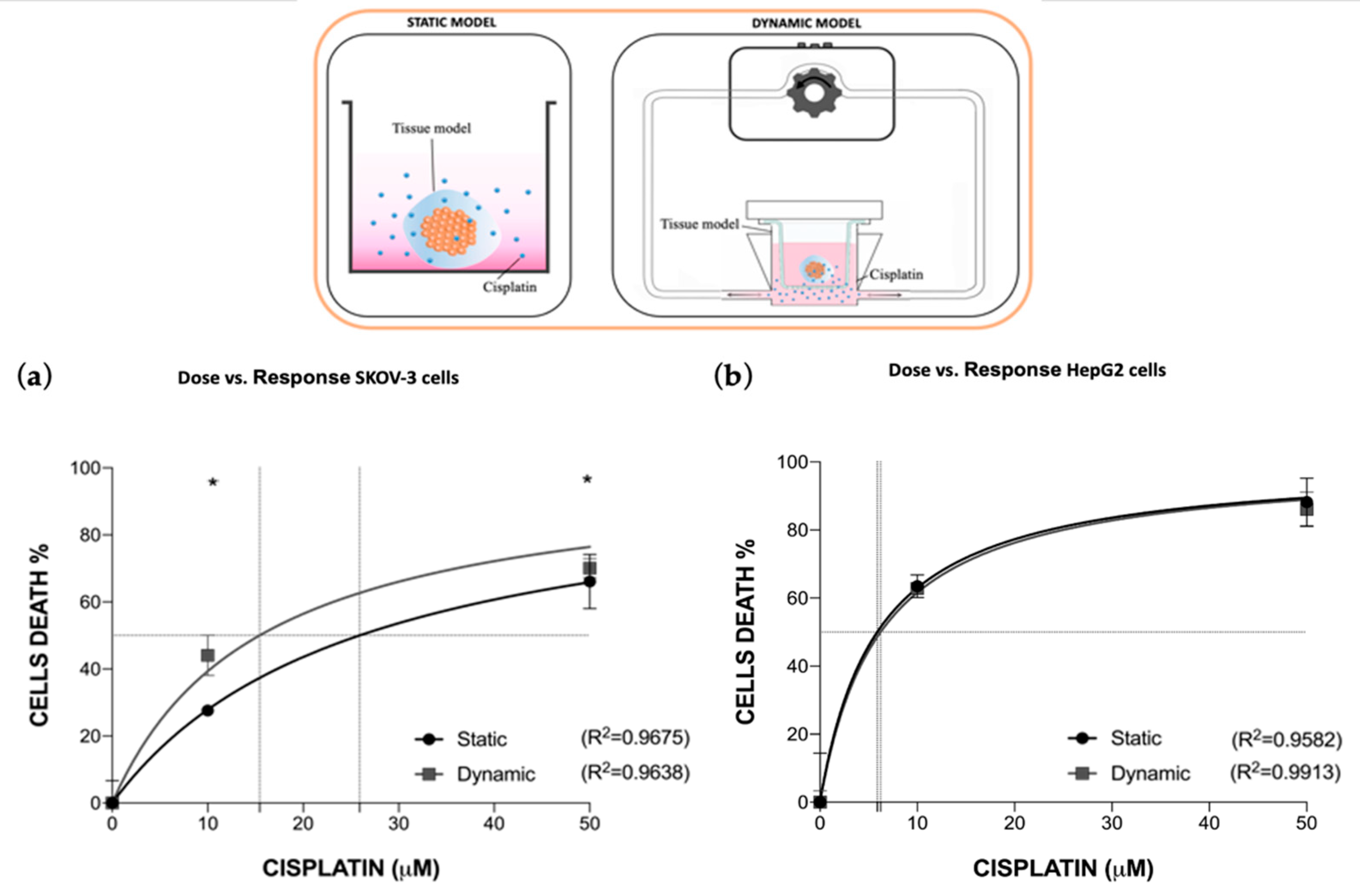
3.2. Dynamic In Vitro Cisplatin Toxo-Efficacy Evaluation
3.2.1. Fluid-Dynamic and Mass Transport Analysis
3.2.2. Cisplatin Toxo-Efficacy Evaluation in Static vs. Dynamic Conditions Models
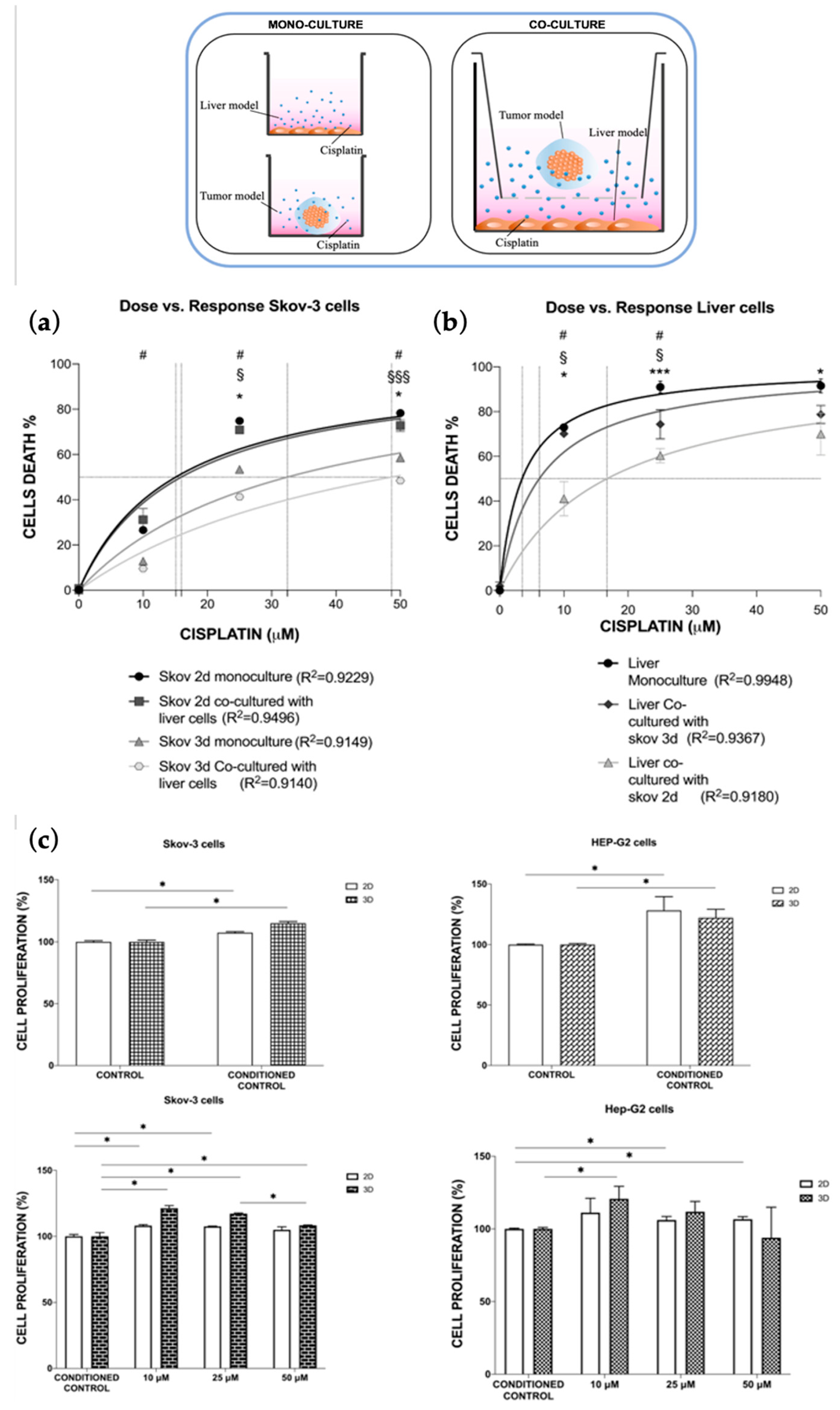
3.3. Multi-Organ-on-Chip Configuration Design and Development
Cisplatin Toxo-Efficacy Evaluation in Single-Organ vs. Multi-Organ Conditions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pound, P.; Ritskes-Hoitinga, M. Is it possible to overcome issues of external validity in preclinical animal research? Why most animal models are bound to fail. J. Transl. Med. 2018, 16, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Marshall, L.J.; Austin, C.P.; Casey, W.; Fitzpatrick, S.C.; Willett, C. Recommendations toward a human pathway-based approach to disease research. Drug Discov. Today 2018, 23, 1824–1832. [Google Scholar] [CrossRef] [PubMed]
- Seyhan, A.A. Lost in translation: The valley of death across preclinical and clinical divide—Identification of problems and overcoming obstacles. Transl. Med. Commun. 2019, 4, 18. [Google Scholar] [CrossRef]
- Jones, R.; Wilsdon, J.R. The Biomedical Bubble: Why UK Research and Innovation Needs a Greater Diversity of Priorities, Politics, Places and People; Nesta: London, UK, 2018. [Google Scholar]
- Sun, D.; Gao, W.; Hu, H.; Zhou, S. Why 90% of clinical drug development fails and how to improve it? Acta Pharm. Sin. B 2022, 12, 3049–3062. [Google Scholar] [CrossRef] [PubMed]
- Mak, I.W.Y.; Evaniew, N.; Ghert, M. Lost in translation: Animal models and clinical trials in cancer treatment. Am. J. Transl. Res. 2014, 6, 114–118. [Google Scholar] [PubMed]
- Harrison, R.K. Phase II and phase III failures: 2013–2015. Nat. Rev. Drug Discov. 2016, 15, 817–818. [Google Scholar] [CrossRef] [PubMed]
- Morse, M.A.; Gwin, W.R.; Mitchell, D.A. Vaccine Therapies for Cancer: Then and Now; Springer International Publishing: Berlin/Heidelberg, Germany, 2021; Volume 16, ISBN 1152302000. [Google Scholar]
- Dowden, H.; Munro, J. Trends in clinical success rates and therapeutic focus. Nat. Rev. Drug Discov. 2019, 18, 495–496. [Google Scholar] [CrossRef] [PubMed]
- Chi, L.H.; Burrows, A.D.; Anderson, R.L. Can preclinical drug development help to predict adverse events in clinical trials? Drug Discov. Today 2022, 27, 257–268. [Google Scholar] [CrossRef]
- Dirnagl, U.; Duda, G.N.; Grainger, D.W.; Reinke, P.; Roubenoff, R. Reproducibility, relevance and reliability as barriers to efficient and credible biomedical technology translation. Adv. Drug Deliv. Rev. 2022, 182, 114118. [Google Scholar] [CrossRef]
- Duval, K.; Grover, H.; Han, L.H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling physiological events in 2D vs. 3D cell culture. Physiology 2017, 32, 266–277. [Google Scholar] [CrossRef]
- Huh, D.; Hamilton, G.A.; Ingber, D.E. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011, 21, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Mermin, N.D. Make mouse studies work. Nature 2014, 507, 7–9. [Google Scholar]
- Van Norman, G.A. Limitations of Animal Studies for Predicting Toxicity in Clinical Trials: Is it Time to Rethink Our Current Approach? JACC Basic Transl. Sci. 2019, 4, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Hoarau-Véchot, J.; Rafii, A.; Touboul, C.; Pasquier, J. Halfway between 2D and animal models: Are 3D cultures the ideal tool to study cancer-microenvironment interactions? Int. J. Mol. Sci. 2018, 19, 181. [Google Scholar] [CrossRef] [PubMed]
- Marrella, A.; Varani, G.; Aiello, M.; Vaccari, I.; Vitale, C.; Mojzisek, M.; Degrassi, C.; Scaglione, S. 3D Fluid-Dynamic Ovarian Cancer Model Resembling Systemic Drug Administration for Efficacy Assay. ALTEX 2021, 38, 82–94. [Google Scholar] [CrossRef]
- Id, A.M.; Fedi, A.; Varani, G.; Vaccari, I.; Id, M.F.; Firpo, G.; Guida, P.; Aceto, N.; Id, S.S. High blood flow shear stress values are associated with circulating tumor cells cluster disaggregation in a multi-channel microfluidic device. PLoS ONE 2021, 16, e0245536. [Google Scholar] [CrossRef]
- Vitale, C.; Fedi, A.; Marrella, A.; Varani, G.; Fato, M.; Scaglione, S. 3D perfusable hydrogel recapitulating the cancer dynamic environment to in vitro investigate metastatic colonization. Polymers 2020, 12, 2467. [Google Scholar] [CrossRef]
- Marzagalli, M.; Pelizzoni, G.; Fedi, A.; Vitale, C.; Fontana, F.; Bruno, S.; Poggi, A.; Dondero, A.; Aiello, M.; Castriconi, R.; et al. A multi-organ-on-chip to recapitulate the infiltration and the cytotoxic activity of circulating NK cells in 3D matrix-based tumor model. Front. Bioeng. Biotechnol. 2022, 10, 1–15. [Google Scholar] [CrossRef]
- Akhtar, A. The Flaws and Human Harms of Animal Experimentation. Camb. Q. Healthc. Ethics 2015, 24, 407–419. [Google Scholar] [CrossRef]
- Junod, S.W.; Beaver, W.T. FDA and Clinical Drug Trials: A Short History. In Quick Guide to Clinical Trials; Davies, M., Kerimani, F., Eds.; BioPlan Associates, Inc.: Washington, DC, USA, 2008; pp. 25–55. [Google Scholar]
- Matthews, R.A.J. Medical progress depends on animal models—Doesn’t it? J. R. Soc. Med. 2008, 101, 95–98. [Google Scholar] [CrossRef]
- Fine, B.; Vunjak-Novakovic, G. Shortcomings of Animal Models and the Rise of Engineered Human Cardiac Tissue. ACS Biomater. Sci. Eng. 2017, 3, 1884–1897. [Google Scholar] [CrossRef] [PubMed]
- Garner, J.P.; Gaskill, B.N.; Weber, E.M.; Ahloy-Dallaire, J.; Pritchett-Corning, K.R. Introducing Therioepistemology: The study of how knowledge is gained from animal research. Lab Anim. 2017, 46, 103–113. [Google Scholar] [CrossRef] [PubMed]
- U.S. FOOD & DRUG. Innovation or Stagnation: Challenge and Opportunity on the Critical Path to New Medical Products. Available online: http//www.fda.gov/ScienceResearch/SpecialTopics/CriticalPathInitiative/default.htm (accessed on 16 March 2004).
- Leenaars, C.H.C.; Kouwenaar, C.; Stafleu, F.R.; Bleich, A.; Ritskes-Hoitinga, M.; De Vries, R.B.M.; Meijboom, F.L.B. Animal to human translation: A systematic scoping review of reported concordance rates. J. Transl. Med. 2019, 17, 223. [Google Scholar] [CrossRef] [PubMed]
- Kamb, A. What’s wrong with our cancer models? Nat. Rev. Drug Discov. 2005, 4, 161–165. [Google Scholar] [CrossRef]
- Rahbari, M.; Rahlfs, S.; Jortzik, E.; Bogeski, I.; Becker, K. Can animal models of disease reliably inform human studies? PLoS ONE 2017, 12, 1–8. [Google Scholar] [CrossRef]
- Ingber, D.E. Human organs-on-chips for disease modelling, drug development and personalized medicine. Nat. Rev. Genet. 2022, 23, 467–491. [Google Scholar] [CrossRef]
- Lee, M.J.; Hatton, B.A.; Villavicencio, E.H.; Khanna, P.C.; Friedman, S.D.; Ditzler, S.; Pullar, B.; Robison, K.; White, K.F.; Tunkey, C.; et al. Hedgehog pathway inhibitor saridegib (IPI-926) increases lifespan in a mouse medulloblastoma model. Proc. Natl. Acad. Sci. USA 2012, 109, 7859–7864. [Google Scholar] [CrossRef]
- Attarwala, H. TGN1412: From discovery to disaster. J. Young Pharm. 2010, 2, 332–336. [Google Scholar] [CrossRef]
- Perel, P.; Roberts, I.; Sena, E.; Wheble, P.; Briscoe, C.; Sandercock, P.; Macleod, M.; Mignini, L.E.; Jayaram, P.; Khan, K.S. Comparison of treatment effects between animal experiments and clinical trials: Systematic review. BMJ 2007, 334, 197–200. [Google Scholar] [CrossRef]
- Hackam, D.G.; Redelmeier, D.A. Translation of research evidence from animals to humans. JAMA 2006, 296, 1727–1732. [Google Scholar] [CrossRef]
- Ahmed, H.M.M.A.M.; Moreira Teixeira, L.S. New Endeavors of (Micro)Tissue Engineering: Cells Tissues Organs on-Chip and Communication Thereof. Cells Tissues Organs 2021, 211, 88–102. [Google Scholar] [CrossRef] [PubMed]
- Shinha, K.; Nihei, W.; Ono, T.; Nakazato, R.; Kimura, H. A pharmacokinetic-pharmacodynamic model based on multi-organ-on-a-chip for drug-drug interaction studies. Biomicrofluidics 2020, 14, 044108. [Google Scholar] [CrossRef] [PubMed]
- Ronaldson-Bouchard, K.; Baldassarri, I.; Tavakol, D.N.; Graney, P.L.; Samaritano, M.; Cimetta, E.; Vunjak-Novakovic, G. Engineering complexity in human tissue models of cancer. Adv. Drug Deliv. Rev. 2022, 184, 114181. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.; Park, D.; Lee, S.; Gumuscu, B.; Jeon, N.L. Engineering Organ-on-a-Chip to Accelerate Translational Research. Micromachines 2022, 13, 1200. [Google Scholar] [CrossRef]
- Leung, C.M.; de Haan, P.; Ronaldson-Bouchard, K.; Kim, G.A.; Ko, J.; Rho, H.S.; Chen, Z.; Habibovic, P.; Jeon, N.L.; Takayama, S.; et al. A guide to the organ-on-a-chip. Nat. Rev. Methods Prim. 2022, 2, 33. [Google Scholar] [CrossRef]
- Esch, E.W.; Bahinski, A.; Huh, D. Organs-on-chips at the frontiers of drug discovery. Nat. Rev. Drug Discov. 2015, 14, 248–260. [Google Scholar] [CrossRef]
- Wikswo, J.P. The relevance and potential roles of microphysiological systems in biology and medicine. Exp. Biol. Med. 2014, 239, 1061–1072. [Google Scholar] [CrossRef]
- Low, L.A.; Mummery, C.; Berridge, B.R.; Austin, C.P.; Tagle, D.A. Organs-on-chips: Into the next decade. Nat. Rev. Drug Discov. 2021, 20, 345–361. [Google Scholar] [CrossRef]
- Picollet-D’hahan, N.; Zuchowska, A.; Lemeunier, I.; Le Gac, S. Multiorgan-on-a-Chip: A Systemic Approach To Model and Decipher Inter-Organ Communication. Trends Biotechnol. 2021, 39, 788–810. [Google Scholar] [CrossRef]
- Saiding, Q.; Ma, J.; Ke, C.; Cui, W. From “organs on a chip” to “patient on a chip”. Innovation 2022, 3, 100282. [Google Scholar] [CrossRef]
- van Berlo, D.; van de Steeg, E.; Amirabadi, H.E.; Masereeuw, R. The potential of multi-organ-on-chip models for assessment of drug disposition as alternative to animal testing. Curr. Opin. Toxicol. 2021, 27, 8–17. [Google Scholar] [CrossRef]
- McAleer, C.W.; Long, C.J.; Elbrecht, D.; Sasserath, T.; Bridges, L.R.; Rumsey, J.W.; Martin, C.; Schnepper, M.; Wang, Y.; Schuler, F.; et al. Multi-organ system for the evaluation of efficacy and off-target toxicity of anticancer therapeutics. Sci. Transl. Med. 2019, 11, eaav1386. [Google Scholar] [CrossRef] [PubMed]
- Zuchowska, A.; Skorupska, S. Multi-organ-on-chip approach in cancer research. Organs A-Chip 2022, 4, 100014. [Google Scholar] [CrossRef]
- Vitale, C.; Marzagalli, M.; Scaglione, S.; Dondero, A.; Bottino, C.; Castriconi, R. Tumor Microenvironment and Hydrogel-Based 3D Cancer Models for In Vitro Testing Immunotherapies. Cancers 2022, 14, 1013. [Google Scholar] [CrossRef] [PubMed]
- Ronaldson-Bouchard, K.; Teles, D.; Yeager, K.; Tavakol, D.N.; Zhao, Y.; Chramiec, A.; Tagore, S.; Summers, M.; Stylianos, S.; Tamargo, M.; et al. A Multi-Organ Chip with Matured Tissue Niches Linked by Vascular Flow. Nat. Biomed. Eng. 2022, 6, 351–371. [Google Scholar] [CrossRef] [PubMed]
- Panteix, G.; Beaujard, A.; Garbit, F.; Chaduiron-Faye, C.; Guillaumont, M.; Gilly, F.; Baltassat, P.; Bressolle, F. Population pharmacokinetics of cisplatin in patients with advanced ovarian cancer during intraperitoneal hyperthermia chemotherapy. Anticancer. Res. 2002, 22, 1329–1336. [Google Scholar]
- Cavo, M.; Fato, M.; Peñuela, L.; Beltrame, F.; Raiteri, R.; Scaglione, S. Microenvironment complexity and matrix stiffness regulate breast cancer cell activity in a 3D in vitro model. Sci. Rep. 2016, 6, 35367. [Google Scholar] [CrossRef]
- Moradi, E.; Jalili-firoozinezhad, S.; Solati-hashjin, M. Microfluidic organ-on-a-chip models of human liver tissue. Acta Biomater. 2020, 116, 67–83. [Google Scholar] [CrossRef]
- Guo, L.; Dial, S.; Shi, L.; Branham, W.; Liu, J.; Fang, J.; Green, B.; Deng, H.; Kaput, J.; Ning, B. Similarities and Differences in the Expression of Drug-Metabolizing Enzymes between Human Hepatic Cell Lines and Primary Human Hepatocytes. Drug Metab. Dispos. 2011, 39, 528–538. [Google Scholar] [CrossRef]
- Discovery, D.; Spring, K.; Bryant, K.; Shackel, N.A. In Vitro Models of the Liver: Disease Modeling, Drug Discovery and Clinical Applications. Hepatocell. Carcinoma 2019, 3, 47–67. [Google Scholar]
- Zeilinger, K.; Freyer, N.; Damm, G.; Seehofer, D.; Kno, F. Cell sources for in vitro human liver cell culture models. Exp. Biol. Med. 2016, 241, 1684–1698. [Google Scholar] [CrossRef] [PubMed]
- Soldatow, V.Y.; Lecluyse, E.L.; Rusyn, I. In vitro models for liver toxicity testing. Toxicol. Res. 2013, 2, 23–39. [Google Scholar] [CrossRef] [PubMed]
- Marrella, A.; Buratti, P.; Markus, J.; Firpo, G.; Pesenti, M.; Landry, T.; Ayehunie, S.; Scaglione, S.; Kandarova, H.; Aiello, M. In vitro demonstration of intestinal absorption mechanisms of different sugars using 3D organotypic tissues in a fluidic device. ALTEX-Altern. Anim. Exp. 2020, 37, 255–264. [Google Scholar] [CrossRef]
- Sugarbaker, P.H. Optimizing regional chemotherapy for epithelial ovarian cancer. J. Obstet. Gynaecol. Res. 2022, 48, 1306–1317. [Google Scholar] [CrossRef] [PubMed]
- Atkins, C.D.; Piccart, M.J. Randomized intergroup trial of cisplatin-paclitaxel versus cisplatin-cyclophosphamide in women with advanced epithelial ovarian cancer: Three-year results [4] (multiple letters). J. Natl. Cancer Inst. 2000, 92, 1446–1447. [Google Scholar] [CrossRef]
- Xu, J.; Gewirtz, D.A. Is Autophagy Always a Barrier to Cisplatin Therapy? Biomolecules 2022, 12, 463. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Cui, M.; Liu, K. Therapeutic strategies to overcome cisplatin resistance in ovarian cancer. Eur. J. Med. Chem. 2022, 232, 114205. [Google Scholar] [CrossRef]
- Kaye, S.B.; Lewis, C.R.; Paul, J.; Soukop, M.; Rankin, E.M.; Cassidy, J.; Davis, J.A.; Reed, N.S.; MacLean, A.; Kennedy, J.H. Randomised study of two doses of cisplatin with cyclophosphamide in epithelial ovarian cancer. Lancet 1992, 340, 329–333. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fedi, A.; Vitale, C.; Fato, M.; Scaglione, S. A Human Ovarian Tumor & Liver Organ-on-Chip for Simultaneous and More Predictive Toxo-Efficacy Assays. Bioengineering 2023, 10, 270. https://doi.org/10.3390/bioengineering10020270
Fedi A, Vitale C, Fato M, Scaglione S. A Human Ovarian Tumor & Liver Organ-on-Chip for Simultaneous and More Predictive Toxo-Efficacy Assays. Bioengineering. 2023; 10(2):270. https://doi.org/10.3390/bioengineering10020270
Chicago/Turabian StyleFedi, Arianna, Chiara Vitale, Marco Fato, and Silvia Scaglione. 2023. "A Human Ovarian Tumor & Liver Organ-on-Chip for Simultaneous and More Predictive Toxo-Efficacy Assays" Bioengineering 10, no. 2: 270. https://doi.org/10.3390/bioengineering10020270
APA StyleFedi, A., Vitale, C., Fato, M., & Scaglione, S. (2023). A Human Ovarian Tumor & Liver Organ-on-Chip for Simultaneous and More Predictive Toxo-Efficacy Assays. Bioengineering, 10(2), 270. https://doi.org/10.3390/bioengineering10020270







