Hybrid Genome Sequencing and Comparative Analysis of Three Novel Listeria monocytogenes Strains: Insights into Lineage Diversity, Virulence, Antibiotic Resistance, and Defense Systems
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Antibiotic Resistance Testing
2.2. Complete Genome Sequencing
2.3. Bioinformatics Analysis
3. Results and Discussion
3.1. General Characteristics of the Novel L. monocytogenes Complete Genomes
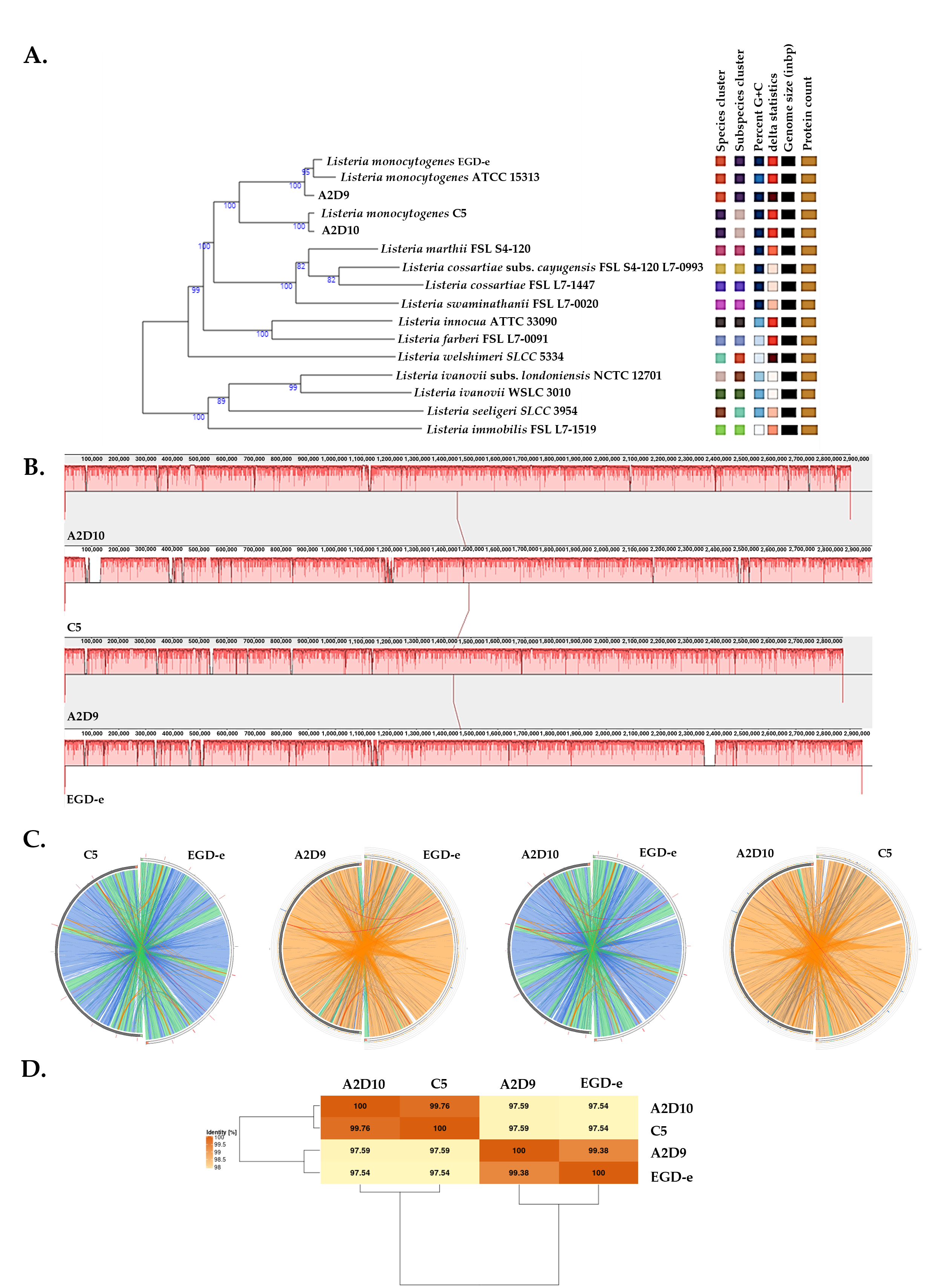
3.2. Phylogenomic and Whole-Genome Comparative Analysis
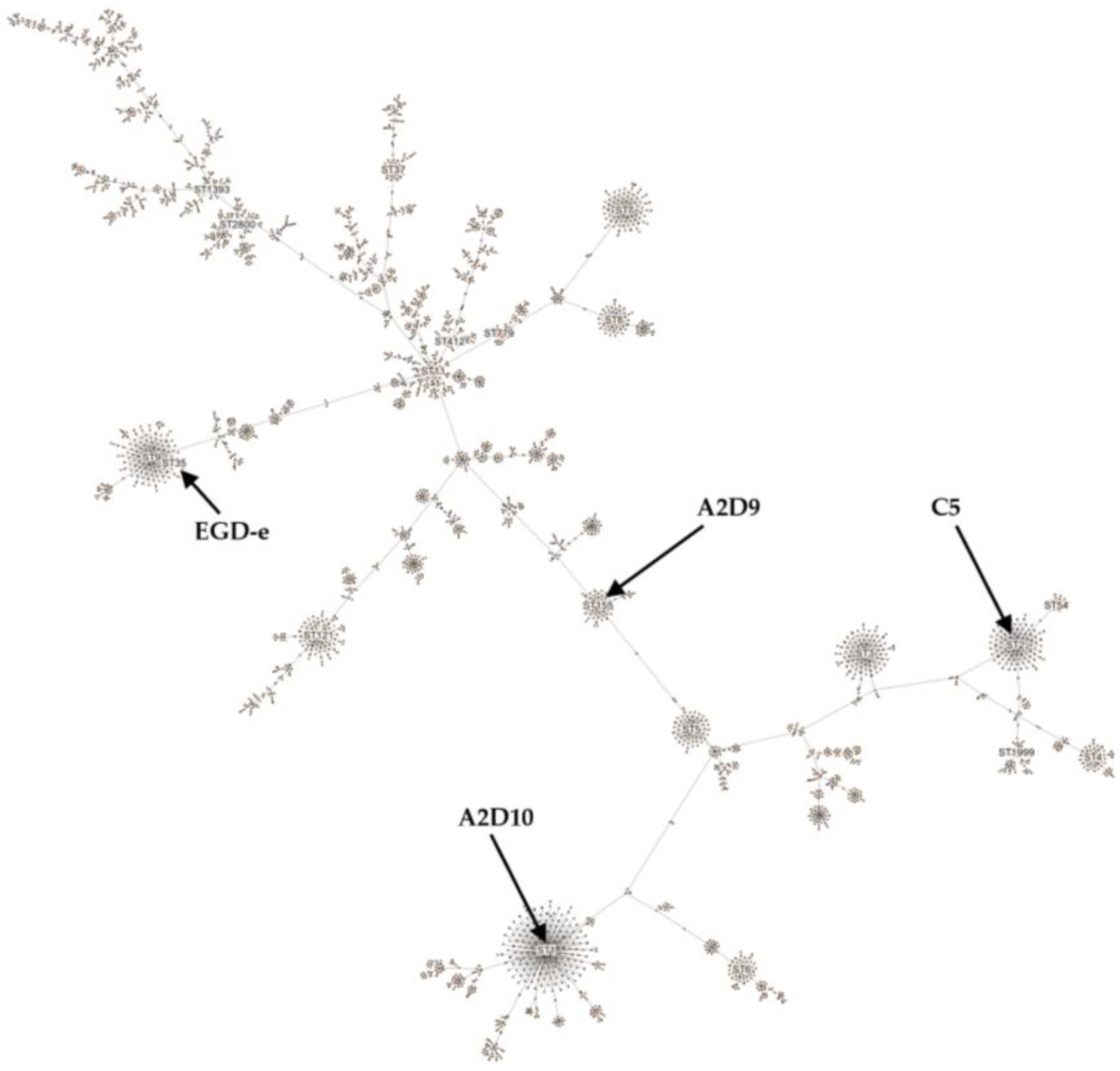
3.3. Typing of L. monocytogenes Strains
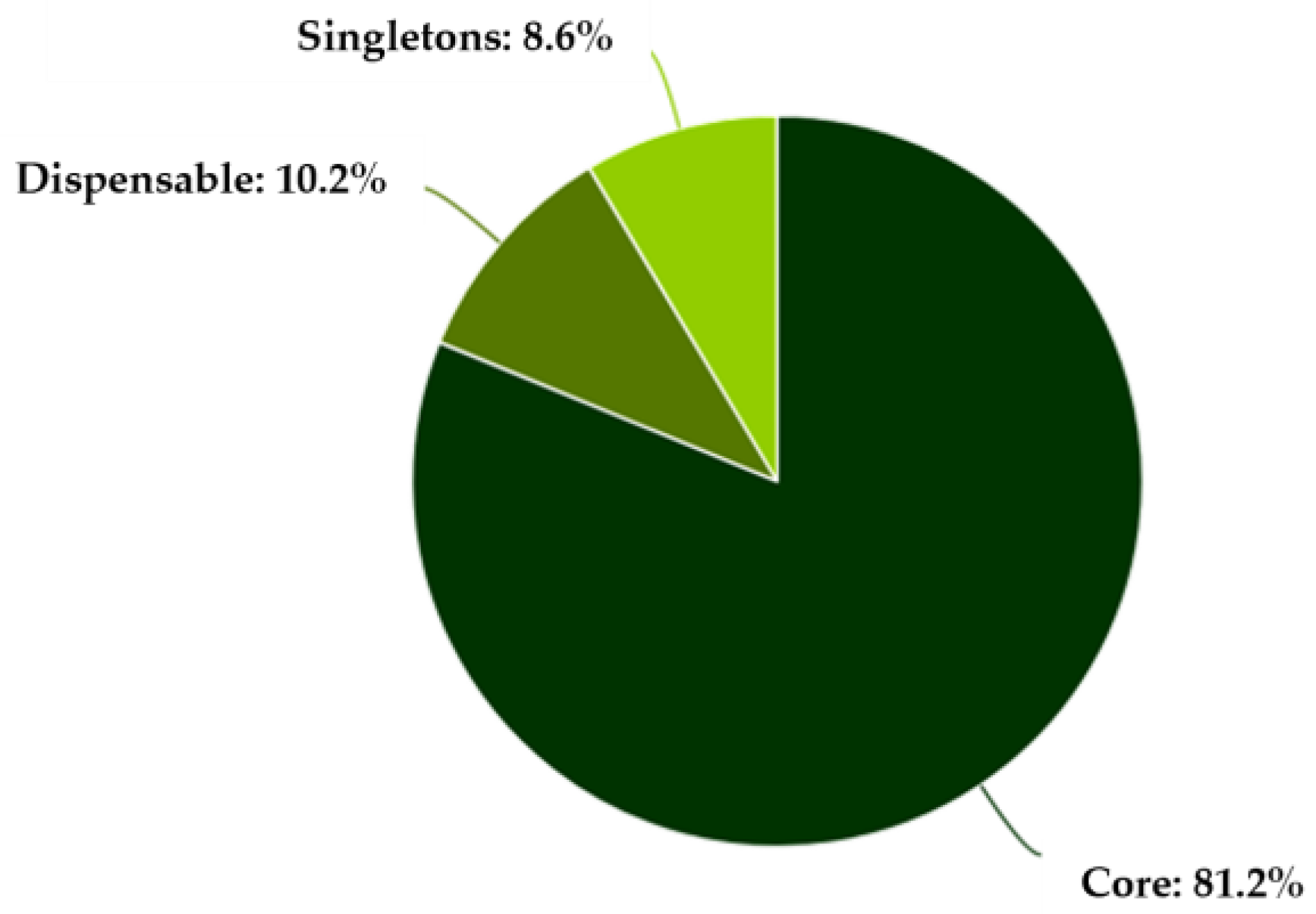
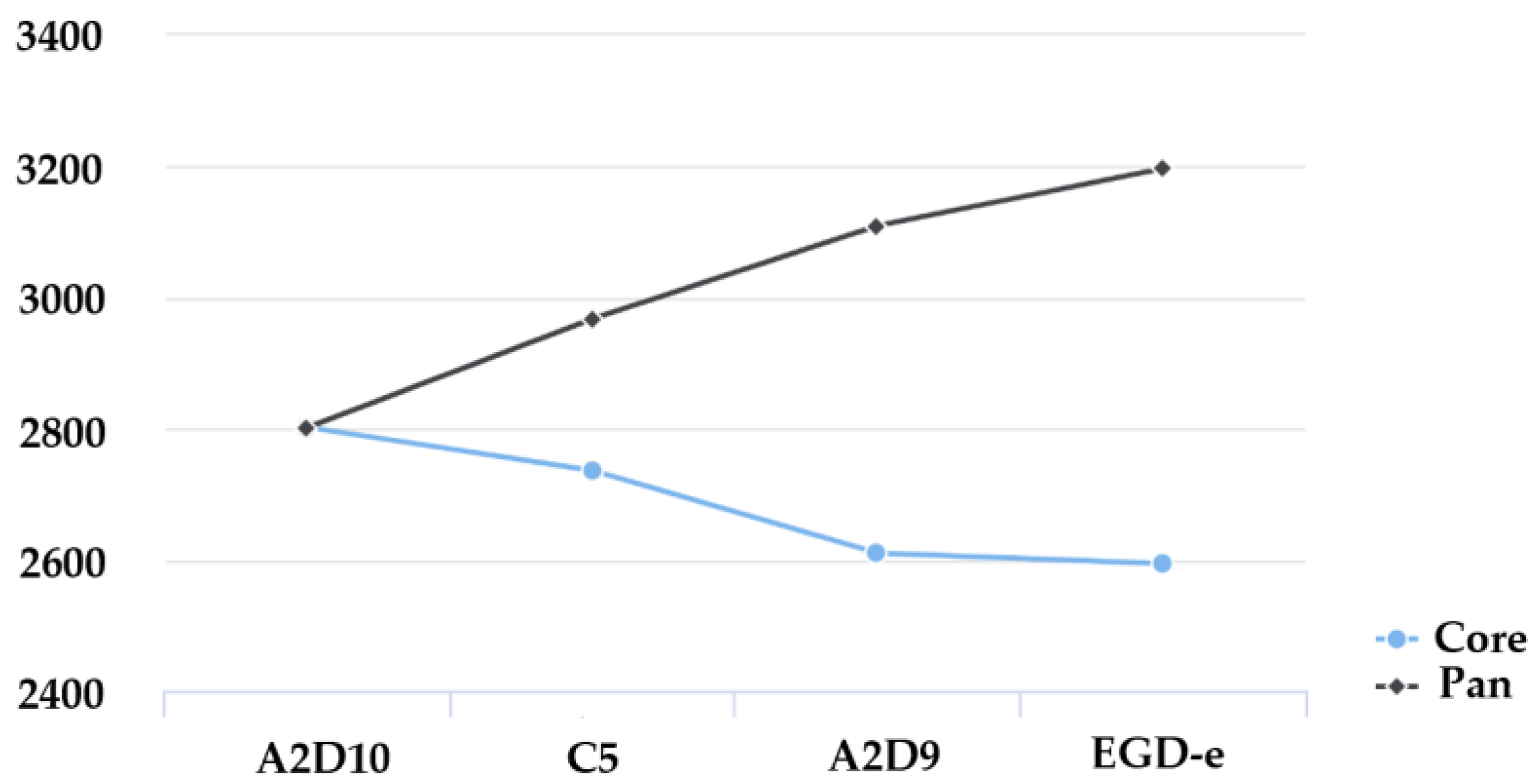
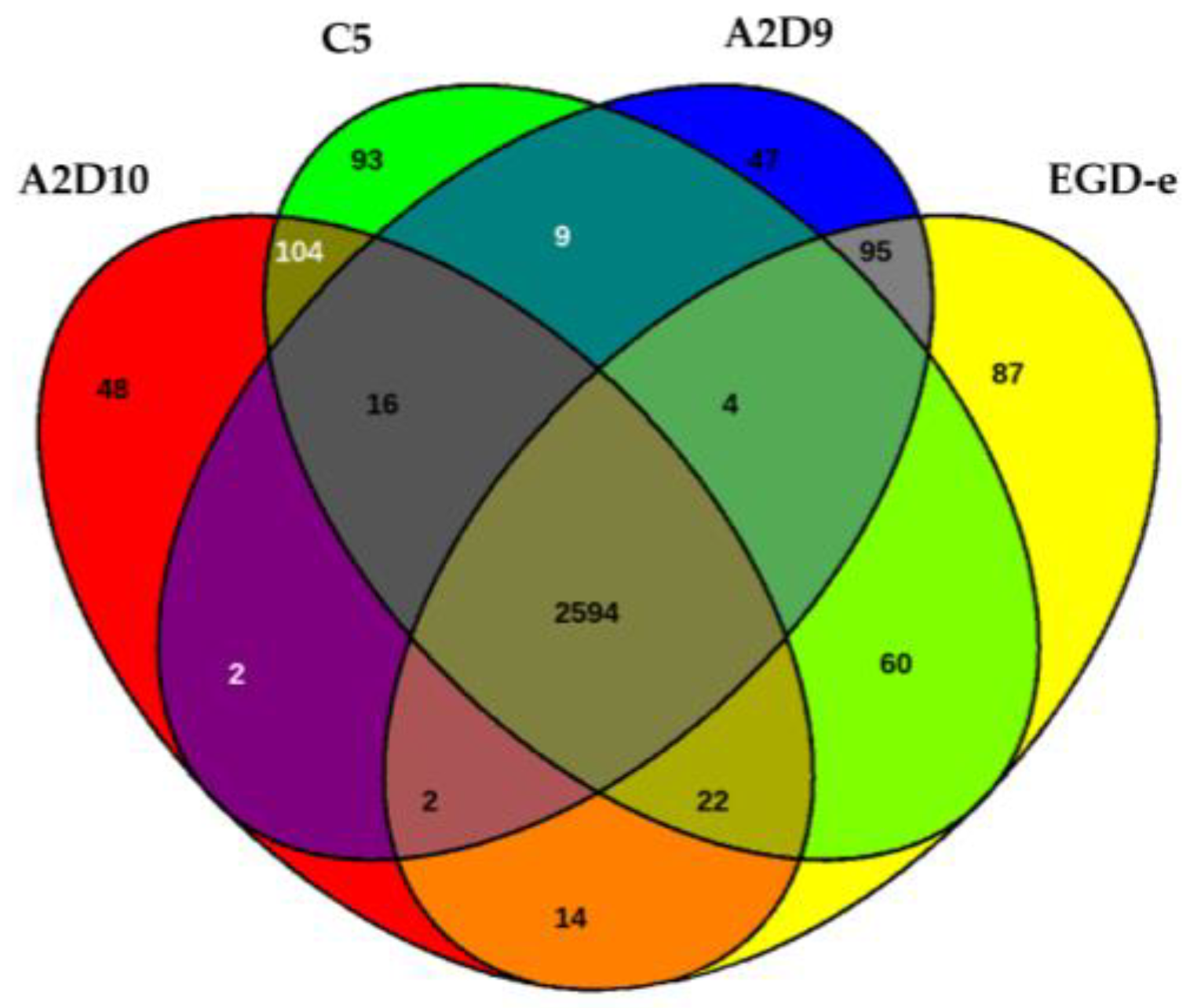
3.4. Pan-Genome and Core-Genome Analysis
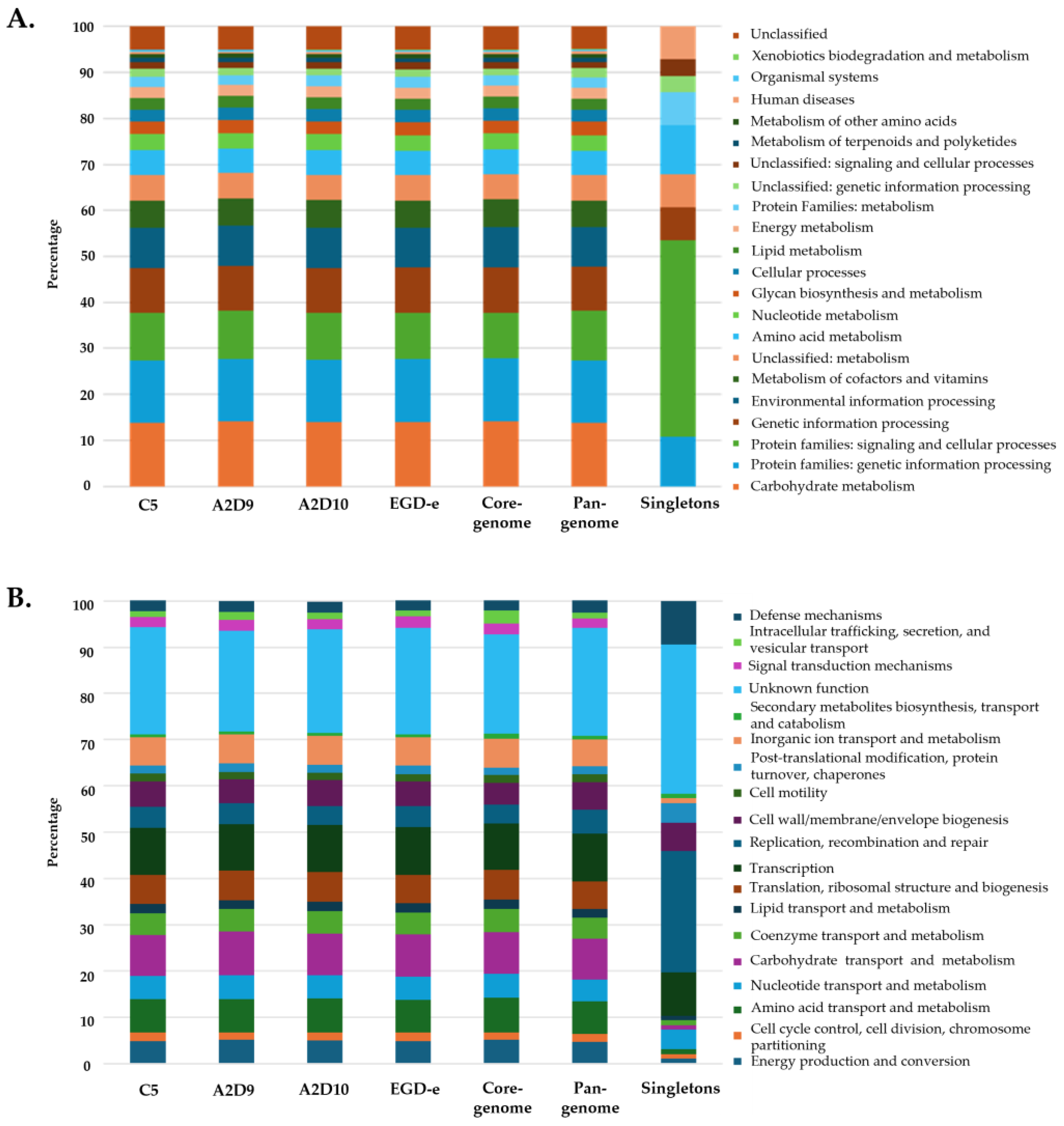
3.5. Functional Annotation
3.6. Virulence Potential
3.7. Antimicrobial Resistance
3.8. Defense Mechanisms and Miscellaneous Traits
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Disson, O.; Charlier, C.; Pérot, P.; Leclercq, A.; Paz, R.N.; Kathariou, S.; Tsai, Y.-H.; Lecuit, M. Listeriosis. Nat. Rev. Dis. Primers 2025, 11, 71. [Google Scholar] [CrossRef] [PubMed]
- Moura, A.; Criscuolo, A.; Pouseele, H.; Maury, M.M.; Leclercq, A.; Tarr, C.; Björkman, J.T.; Dallman, T.; Reimer, A.; Enouf, V.; et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2016, 2, 16185. [Google Scholar] [CrossRef] [PubMed]
- Radoshevich, L.; Cossart, P. Listeria monocytogenes: Towards a complete picture of its physiology and pathogenesis. Nat. Rev. Microbiol. 2018, 16, 32–46. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, B.; Gerner-Smidt, P. The epidemiology of human listeriosis. Microbes Infect. 2007, 9, 1236–1243. [Google Scholar] [CrossRef]
- Kuenne, C.; Billion, A.; Mraheil, M.A.; Strittmatter, A.; Daniel, R.; Goesmann, A.; Barbuddhe, S.; Hain, T.; Chakraborty, T. Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genom. 2013, 14, 47. [Google Scholar] [CrossRef]
- Lachtara, B.; Wieczorek, K.; Osek, J. Genetic diversity and relationships of Listeria monocytogenes serogroup IIa isolated in Poland. Microorganisms 2022, 10, 532. [Google Scholar] [CrossRef]
- Liu, D.; Lawrence, M.L.; Wiedmann, M.; Gorski, L.; Mandrell, R.E.; Ainsworth, A.J.; Austin, F.W. Listeria monocytogenes subgroups IIIA, IIIB, and IIIC delineate genetically distinct populations with varied pathogenic potential. J. Clin. Microbiol. 2006, 44, 4229–4233. [Google Scholar] [CrossRef]
- Orsi, R.H.; den Bakker, H.C.; Wiedmann, M. Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 2011, 301, 79–96. [Google Scholar] [CrossRef]
- Zamudio, R.; Haigh, R.D.; Ralph, J.D.; De Ste Croix, M.; Tasara, T.; Zurfluh, K.; Kwun, M.J.; Millard, A.D.; Bentley, S.D.; Croucher, N.J.; et al. Lineage-specific evolution and gene flow in Listeria monocytogenes are independent of bacteriophages. Environ. Microbiol. 2020, 22, 5058–5072. [Google Scholar] [CrossRef]
- Orsi, R.H.; Wiedmann, M. Characteristics and distribution of Listeria spp., including Listeria species newly described since 2009. Appl. Microbiol. Biotechnol. 2016, 100, 5273–5287. [Google Scholar] [CrossRef]
- De Las Heras, A.; Cain, R.J.; Bielecka, M.K.; Vázquez-Boland, J.A. Regulation of Listeria virulence: PrfA master and commander. Curr. Opin. Microbiol. 2011, 14, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Lecuit, M. Listeria monocytogenes, a model in infection biology. Cell Microbiol. 2020, 22, e13186. [Google Scholar] [CrossRef] [PubMed]
- Luque-Sastre, L.; Arroyo, C.; Fox, E.M.; McMahon, B.J.; Bai, L.; Li, F.; Fanning, S. Antimicrobial resistance in Listeria species. Microbiol. Spectr. 2018, 6, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Scortti, M.; Han, L.; Alvarez, S.; Leclercq, A.; Moura, A.; Lecuit, M.; Vazquez-Boland, J. Epistatic control of intrinsic resistance by virulence genes in Listeria. PLoS Genet. 2018, 14, e1007525. [Google Scholar] [CrossRef]
- Kim, J.W.; Kathariou, S. Temperature-dependent phage resistance of Listeria monocytogenes epidemic clone II. Appl. Environ. Microbiol. 2009, 75, 2433–2438. [Google Scholar] [CrossRef]
- Osuna, B.A.; Karambelkar, S.; Mahendra, C.; Christie, K.A.; Garcia, B.; Davidson, A.R.; Kleinstiver, B.P.; Kilcher, S.; Bondy-Denomy, J. Listeria phages induce Cas9 degradation to protect lysogenic genomes. Cell Host Microbe 2020, 28, 31–40.e39. [Google Scholar] [CrossRef]
- Xu, X.; Gu, P. Overview of phage defense systems in bacteria and their applications. Mol. Sci. 2024, 25, 13316. [Google Scholar] [CrossRef]
- Georjon, H.; Bernheim, A. The highly diverse antiphage defence systems of bacteria. Nat. Rev. Microbiol. 2023, 21, 686–700. [Google Scholar] [CrossRef]
- Bobay, L.M.; Ochman, H. The evolution of bacterial genome architecture. Front. Genet. 2017, 8, 72. [Google Scholar] [CrossRef]
- Gkerekou, M.A.; Kaparakou, E.H.; Tarantilis, P.A.; Skandamis, P.N. Studying the metabolic factors that may impact the growth of co-cultured Listeria monocytogenes strains at low temperature. Food Res. Int. 2023, 171, 113056. [Google Scholar] [CrossRef]
- EUCAST. European Committee on Antimicrobial Susceptibility Testing Disk Diffusion Test Methodology. 2024. Available online: https://www.eucast.org/ast_of_bacteria/disk_diffusion_methodology (accessed on 10 January 2025).
- Mölder, F.; Jablonski, K.P.; Letcher, B.; Hall, M.B.; van Dyken, P.C.; Tomkins-Tinch, C.H.; Sochat, V.; Forster, J.; Vieira, F.G.; Meesters, C.; et al. Sustainable data analysis with Snakemake. F1000Research 2021, 10, 33. [Google Scholar] [CrossRef] [PubMed]
- De Coster, W.; Rademakers, R. NanoPack2: Population-scale evaluation of long-read sequencing data. Bioinformatics 2023, 39, btad311. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Completing bacterial genome assemblies with multiplex MinION sequencing. Microb. Genom. 2017, 3, e000132. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 8 July 2025).
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Lee, J.Y.; Kong, M.; Oh, J.; Lim, J.; Chung, S.H.; Kim, J.M.; Kim, J.-S.; Kim, K.-H.; Yoo, J.-C.; Kwak, W. Comparative evaluation of Nanopore polishing tools for microbial genome assembly and polishing strategies for downstream analysis. Sci. Rep. 2021, 11, 20740. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Hunt, M.; Silva, N.D.; Otto, T.D.; Parkhill, J.; Keane, J.A.; Harris, S.R. Circlator: Automated circularization of genome assemblies using long sequencing reads. Genome Biol. 2015, 16, 294. [Google Scholar] [CrossRef]
- Pellow, D.; Mizrahi, I.; Shamir, R. PlasClass improves plasmid sequence classification. PLoS Comput. Biol. 2020, 16, e1007781. [Google Scholar] [CrossRef]
- Chaumeil, P.A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk v2: Memory friendly classification with the genome taxonomy database. Bioinformatics 2022, 38, 5315–5316. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO update: Novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef] [PubMed]
- Chklovski, A.; Parks, D.H.; Woodcroft, B.J.; Tyson, G.W. CheckM2: A rapid, scalable and accurate tool for assessing microbial genome quality using machine learning. Nat. Methods 2023, 20, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Goker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Darling, A.C.E.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Darzentas, N. Circoletto: Visualizing sequence similarity with Circos. Bioinformatics 2010, 26, 2620–2621. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Felsenstein, J. PHYLIP—Phylogeny Inference Package. 2018. Available online: https://phylipweb.github.io/phylip (accessed on 2 March 2025).
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2015, 32, 929–931. [Google Scholar] [CrossRef]
- Pasteur BIGSdb. BIGSdb. 2024. Available online: http://bigsdb.pasteur.fr/news/ (accessed on 2 October 2025).
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. ΕggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Seemann, T. Abricate. 2014. Available online: https://github.com/tseemann/abricate (accessed on 5 September 2025).
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.M. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Doster, E.; Lakin, S.M.; Dean, C.J.; Wolfe, C.; Young, J.G.; Boucher, C.; Belk, K.E.; Noyes, N.R.; Morley, P.S. MEGARes 2.0: A database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data. Nucleic Acids Res. 2019, 48, D561–D569. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef]
- Tesson, F.; Hervé, A.; Mordret, E.; Touchon, M.; d’Humières, C.; Cury, J.; Bernheim, A. Systematic and quantitative view of the antiviral arsenal of prokaryotes. Nat. Commun. 2022, 13, 2561. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.L. IslandViewer 4: Expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef]
- Qi, Y.; Cao, Q.; Zhao, X.; Tian, C.; Li, T.; Shi, W.; Wei, H.; Song, C.; Xue, H.; Gou, H. Comparative genomic analysis of pathogenic factors of Listeria spp. using whole-genome sequencing. BMC Genom. 2024, 25, 935. [Google Scholar] [CrossRef]
- Lachtara, B.; Osek, J.; Wieczorek, K. Molecular typing of Listeria monocytogenes IVb serogroup isolated from food and food production environments in Poland. Pathogens 2021, 10, 482. [Google Scholar] [CrossRef]
- Briers, Y.; Klumpp, J.; Schuppler, M.; Loessner, M.J. Genome sequence of Listeria monocytogenes Scott A, a clinical isolate from a food-borne listeriosis outbreak. J. Bacteriol. 2011, 193, 4284–4285. [Google Scholar] [CrossRef]
- Liu, D. Identification, subtyping and virulence determination of Listeria monocytogenes, an important foodborne pathogen. J. Med. Microbiol. 2006, 55, 645–659. [Google Scholar] [CrossRef] [PubMed]
- Maijala, R.; Lyytikäinen, O.; Johansson, T.; Autio, T.; Aalto, T.; Haavisto, L.; Honkanen-Buzalski, T. Exposure of Listeria monocytogenes within an epidemic caused by butter in Finland. Int. J. Food Microbiol. 2001, 70, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Rychli, K.; Müller, A.; Zaiser, A.; Schoder, D.; Allerberger, F.; Wagner, M.; Schmitz-Esser, S. Genome sequencing of Listeria monocytogenes “Quargel” listeriosis outbreak strains reveals two different strains with distinct in vitro virulence potential. PLoS ONE 2014, 9, e89964. [Google Scholar] [CrossRef]
- Nelson, K.E.; Fouts, D.E.; Mongodin, E.F.; Ravel, J.; DeBoy, R.T.; Kolonay, J.F.; Rasko, D.A.; Angiuoli, S.V.; Gill, S.R.; Paulsen, I.T.; et al. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 2004, 32, 2386–2395. [Google Scholar] [CrossRef]
- Tan, W.; Wang, G.; Pan, Z.; Yin, Y.; Jiao, X. Complete genome sequence of Listeria monocytogenes NTSN, a serovar 4b and animal source strain. Genome Announc. 2015, 3, e01403-14. [Google Scholar] [CrossRef]
- Kichemazova, N.; Zaytsev, S.; Saltykov, Y.; Larionova, O.; Zaberezhny, A.; Feodorova, V. MLST evidence of two different sequence types of Listeria monocytogenes strains used for commercial veterinary listeriosis vaccines. Zoonoses 2025, 5, 992. [Google Scholar] [CrossRef]
- Salcedo, C.; Arreaza, L.; Alcala, B.; de la Fuente, L.; Vazquez, J.A. Development of a multilocus sequence typing method for analysis of Listeria monocytogenes clones. J. Clin. Microbiol. 2003, 41, 757–762. [Google Scholar] [CrossRef]
- Mafuna, T.; Matle, I.; Magwedere, K.; Pierneef, R.E.; Reva, O.N. Whole genome-based characterization of Listeria monocytogenes isolates recovered from the food chain in South Africa. Front. Microbiol. 2021, 12, 669287. [Google Scholar] [CrossRef]
- Nielsen, E.M.; Björkman, J.T.; Kiil, K.; Grant, K.; Dallman, T.; Painset, A.; Amar, C.; Roussel, S.; Guillier, L.; Félix, B.; et al. Closing gaps for performing a risk assessment on Listeria monocytogenes in ready-to-eat (RTE) foods: Activity 3, the comparison of isolates from different compartments along the food chain, and from humans using whole genome sequencing (WGS) analysis. EFSA Support. Publ. 2017, 14, 1151E. [Google Scholar] [CrossRef]
- Rychli, K.; Stessl, B.; Szakmary-Brändle, K.; Strauß, A.; Wagner, M.; Schoder, D. Listeria monocytogenes isolated from illegally imported food products into the European Union harbor different virulence factor variants. Genes 2018, 9, 428. [Google Scholar] [CrossRef]
- Toledo, V.; den Bakker, H.C.; Hormazábal, J.C.; González-Rocha, G.; Bello-Toledo, H.; Toro, M.; Moreno-Switt, A.I. Genomic diversity of Listeria monocytogenes isolated from clinical and non-clinical samples in Chile. Genes 2018, 9, 396. [Google Scholar] [CrossRef] [PubMed]
- Cabal, A.; Pietzka, A.; Huhulescu, S.; Allerberger, F.; Ruppitsch, W.; Schmid, D. Isolate-based surveillance of Listeria monocytogenes by whole genome sequencing in Austria. Front. Microbiol. 2019, 10, 2282. [Google Scholar] [CrossRef] [PubMed]
- Wagner, E.; Zaiser, A.; Leitner, R.; Quijada, N.M.; Pracser, N.; Pietzka, A.; Ruppitsch, W.; Schmitz-Esser, S.; Wagner, M.; Rychli, K. Virulence characterization and comparative genomics of Listeria monocytogenes sequence type 155 strains. BMC Genom. 2020, 21, 847. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Dong, Q.; Liu, Y.; Liu, H.; Zhang, H.; Wang, X. Systematic review of Listeria monocytogenes from food and clinical samples in Chinese mainland from 2010 to 2019. Food Qual. Saf. 2022, 6, fyac021. [Google Scholar] [CrossRef]
- Dreyer, M.; Aguilar-Bultet, L.; Rupp, S.; Guldimann, C.; Stephan, R.; Schock, A.; Otter, A.; Schupbach, G.; Brisse, S.; Lecuit, M.; et al. Listeria monocytogenes sequence type 1 is predominant in ruminant rhombencephalitis. Sci. Rep. 2016, 6, 36419. [Google Scholar] [CrossRef]
- Silva, A.; Silva, V.; Gomes, J.P.; Coelho, A.; Batista, R.; Saraiva, C.; Esteves, A.; Martins, Â.; Contente, D.; Diaz-Formoso, L.; et al. Listeria monocytogenes from food products and food associated environments: Antimicrobial resistance, genetic clustering and biofilm insights. Antibiotics 2024, 13, 447. [Google Scholar] [CrossRef]
- Wieczorek, K.; Bomba, A.; Osek, J. Whole-genome sequencing-based characterization of Listeria monocytogenes from fish and fish production environments in Poland. Mol. Sci. 2020, 21, 9419. [Google Scholar] [CrossRef]
- Ragon, M.; Wirth, T.; Hollandt, F.; Lavenir, R.; Lecuit, M.; Le Monnier, A.; Brisse, S. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 2008, 4, e1000146. [Google Scholar] [CrossRef]
- Li, H.; Wang, P.; Lan, R.; Luo, L.; Cao, X.; Wang, Y.; Wang, Y.; Li, H.; Zhang, L.; Ji, S.; et al. Risk factors and level of Listeria monocytogenes contamination of raw pork in retail markets in China. Front. Microbiol. 2018, 9, 1090. [Google Scholar] [CrossRef]
- Glomski, I.J.; Gedde, M.M.; Tsang, A.W.; Swanson, J.A.; Portnoy, D.A. The Listeria monocytogenes hemolysin has an acidic pH optimum to compartmentalize activity and prevent damage to infected host cells. J. Cell Biol. 2002, 156, 1029–1038. [Google Scholar] [CrossRef]
- Mitchell, G.; Ge, L.; Huang, Q.; Chen, C.; Kianian, S.; Roberts, M.F.; Schekman, R.; Portnoy, D.A. Avoidance of autophagy mediated by PlcA or ActA is required for Listeria monocytogenes growth in macrophages. Infect. Immun. 2015, 83, 2175–2184. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Gao, B.; Cai, H.; Qin, X.; Xia, X.; Dong, Q.; Hirata, T.; Li, Z. Comparative analysis of virulence in Listeria monocytogenes: Insights from genomic variations and in vitro cell-based studies. Int. J. Food Microbiol. 2025, 435, 111188. [Google Scholar] [CrossRef] [PubMed]
- Wiktorczyk-Kapischke, N.; Skowron, K.; Wałecka-Zacharska, E. Genomic and pathogenicity islands of Listeria monocytogenes—Overview of selected aspects. Front. Mol. Biosci. 2023, 10, 1161486. [Google Scholar] [CrossRef] [PubMed]
- Pedroni, L.; Ghidini, S.; Vazquez, J.; Luque, F.J.; Dellafiora, L. Modeling the anti-adhesive role of punicalagin against Listeria monocytogenes from the analysis of the interaction between internalin a and e-cadherin. Int. J. Mol. Sci. 2025, 26, 7327. [Google Scholar] [CrossRef]
- Cabanes, D.; Sousa, S.; Cebriá, A.; Lecuit, M.; García-del Portillo, F.; Cossart, P. Gp96 is a receptor for a novel Listeria monocytogenes virulence factor, Vip, a surface protein. Embo J. 2005, 24, 2827–2838. [Google Scholar] [CrossRef]
- Camejo, A.; Carvalho, F.; Reis, O.; Leitao, E.; Sousa, S.; Cabanes, D. The arsenal of virulence factors deployed by Listeria monocytogenes to promote its cell infection cycle. Virulence 2011, 2, 379–394. [Google Scholar] [CrossRef]
- Lebreton, A.; Lakisic, G.; Job, V.; Fritsch, L.; Tham, T.N.; Camejo, A.; Matteï, P.-J.; Regnault, B.; Nahori, M.-A.; Cabanes, D.; et al. A Bacterial protein targets the BAHD1 chromatin complex to stimulate type III interferon response. Science 2011, 331, 1319–1321. [Google Scholar] [CrossRef]
- Agbavor, C.; Zimnicka, A.; Kumar, A.; George, J.L.; Torres, M.; Prehna, G.; Alonzo, F.; Durrant, J.D.; Freitag, N.E.; Cahoon, L.A. The chaperone PrsA2 regulates the secretion, stability, and folding of listeriolysin O during Listeria monocytogenes infection. mBio 2024, 15, e00743-24. [Google Scholar] [CrossRef]
- Burkholder, K.M.; Bhunia, A.K. Listeria monocytogenes uses Listeria adhesion protein (LAP) to promote bacterial transepithelial translocation and induces expression of LAP receptor Hsp60. Infect. Immun. 2010, 78, 5062–5073. [Google Scholar] [CrossRef]
- Ingeborg, H.; Klein, D.K.; Lehner, A.; Bubert, A.; Branda Ernst, B.; Wagner, M. Detection and quantification of the iap gene of Listeria monocytogenes and Listeria innocua by a new real-time quantitative PCR assay. Res. Microbiol. 2001, 152, 37–46. [Google Scholar] [CrossRef]
- Quereda, J.J.; Morón-García, A.; Palacios-Gorba, C.; Dessaux, C.; García-Del Portillo, F.; Pucciarelli, M.G.; Ortega, A.D. Pathogenicity and virulence of Listeria monocytogenes: A trip from environmental to medical microbiology. Virulence 2021, 12, 2509–2545. [Google Scholar] [CrossRef]
- Sabet, C.; Toledo-Arana, A.; Personnic, N.; Lecuit, M.; Dubrac, S.; Poupel, O.; Gouin, E.; Nahori, M.A.; Cossart, P.; Bierne, H. The Listeria monocytogenes virulence factor InlJ is specifically expressed in vivo and behaves as an adhesin. Infect. Immun. 2008, 76, 1368–1378. [Google Scholar] [CrossRef] [PubMed]
- Parra-Flores, J.; Daza-Prieto, B.; Chavarria, P.; Troncoso, M.; Stoger, A.; Figueroa, G.; Mancilla-Rojano, J.; Cruz-Cordova, A.; Martinovic, A.; Ruppitsch, W. From traditional typing to genomic precision: Whole-genome sequencing of Listeria monocytogenes isolated from refrigerated foods in Chile. Foods 2025, 14, 290. [Google Scholar] [CrossRef] [PubMed]
- Pyz-Lukasik, R.; Paszkiewicz, W.; Kielbus, M.; Ziomek, M.; Gondek, M.; Domaradzki, P.; Michalak, K.; Pietras-Ozga, D. Genetic diversity and potential virulence of Listeria monocytogenes isolates originating from polish artisanal cheeses. Foods 2022, 11, 2805. [Google Scholar] [CrossRef] [PubMed]
- Fern Tan, M.; Siow, C.C.; Dutta, A.; Mutha, N.V.; Wee, W.Y.; Heydari, H.; Tan, S.Y.; Ang, M.Y.; Wong, G.J.; Choo, S.W. Development of ListeriaBase and comparative analysis of Listeria monocytogenes. BMC Genom. 2015, 16, 755. [Google Scholar] [CrossRef]
- Milohanic, E.; Jonquières, R.; Cossart, P.; Berche, P.; Gaillard, J. The autolysin Ami contributes to the adhesion of Listeria monocytogenes to eukaryotic cells via its cell wall anchor. Mol. Microbiol. 2001, 39, 1212–1224. [Google Scholar] [CrossRef]
- Lee, S. Bacteriocins of Listeria monocytogenes and their potential as a virulence factor. Toxins 2020, 12, 103. [Google Scholar] [CrossRef]
- Meza-Torres, J.; Lelek, M.; Quereda, J.J.; Sachse, M.; Manina, G.; Ershov, D.; Tinevez, J.Y.; Radoshevich, L.; Maudet, C.; Chaze, T.; et al. Listeriolysin S: A bacteriocin from Listeria monocytogenes that induces membrane permeabilization in a contact-dependent manner. Proc. Natl. Acad. Sci. USA 2021, 118, e2108155118. [Google Scholar] [CrossRef]
- Spanjaard, L.; Vandenbroucke-Grauls, C.M. Activity of daptomycin against Listeria monocytogenes isolates from cerebrospinal fluid. Antimicrob. Agents Chemother. 2008, 52, 1850–1851. [Google Scholar] [CrossRef]
- Charpentier, E.; Courvalin, P. Antibiotic resistance in Listeria spp. Antimicrob. Agents Chemother. 1999, 43, 2103–2108. [Google Scholar] [CrossRef]
- Grayo, S.; Join-Lambert, O.; Desroches, M.C.; Le Monnier, A. Comparison of the in vitro efficacies of moxifloxacin and amoxicillin against Listeria monocytogenes. Antimicrob. Agents Chemother. 2008, 52, 1697–1702. [Google Scholar] [CrossRef] [PubMed]
- Conter, M.; Paludi, D.; Zanardi, E.; Ghidini, S.; Vergara, A.; Ianieri, A. Characterization of antimicrobial resistance of foodborne Listeria monocytogenes. Int. J. Food Microbiol. 2009, 128, 497–500. [Google Scholar] [CrossRef] [PubMed]
- Manqele, A.; Adesiyun, A.; Mafuna, T.; Pierneef, R.; Moerane, R.; Gcebe, N. Virulence potential and antimicrobial resistance of Listeria monocytogenes isolates obtained from beef and beef-based products deciphered using whole-genome sequencing. Microorganisms 2024, 12, 1166. [Google Scholar] [CrossRef]
- Ernst, C.M.; Peschel, A. MprF-mediated daptomycin resistance. Int. J. Med. Microbiol. 2019, 309, 359–363. [Google Scholar] [CrossRef]
- Krawczyk-Balska, A.; Markiewicz, Z. The intrinsic cephalosporin resistome of Listeria monocytogenes in the context of stress response, gene regulation, pathogenesis and therapeutics. J. Appl. Microbiol. 2016, 120, 251–265. [Google Scholar] [CrossRef]
- Lungu, B.; Ricke, S.C.; Johnson, M.G. Growth, survival, proliferation and pathogenesis of Listeria monocytogenes under low oxygen or anaerobic conditions: A review. Anaerobe 2009, 15, 7–17. [Google Scholar] [CrossRef]
- Oliveira, P.H.; Touchon, M.; Rocha, E.P.C. The interplay of restriction-modification systems with mobile genetic elements and their prokaryotic hosts. Nucleic Acids Res. 2014, 42, 10618–10631. [Google Scholar] [CrossRef]
- Shen, B.W.; Xu, D.; Chan, S.H.; Zheng, Y.; Zhu, Z.; Xu, S.Y.; Stoddard, B.L. Characterization and crystal structure of the type IIG restriction endonuclease RM.BpuSI. Nucleic Acids Res. 2011, 39, 8223–8236. [Google Scholar] [CrossRef]
- Tock, M.R.; Dryden, D.T.F. The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 2005, 8, 466–472. [Google Scholar] [CrossRef]
- Gao, L.; Altae-Tran, H.; Böhning, F.; Makarova, K.S.; Segel, M.; Schmid-Burgk, J.L.; Koob, J.; Wolf, Y.I.; Koonin, E.V.; Zhang, F. Diverse enzymatic activities mediate antiviral immunity in prokaryotes. Science 2020, 369, 1077–1084. [Google Scholar] [CrossRef]
- Millman, A.; Bernheim, A.; Stokar-Avihail, A.; Fedorenko, T.; Voichek, M.; Leavitt, A.; Oppenheimer-Shaanan, Y.; Sorek, R. Bacterial retrons function in anti-phage defense. Cell 2020, 183, 1551–1561.e12. [Google Scholar] [CrossRef]
- Davidov, E.; Kaufmann, G. RloC: A wobble nucleotide-excising and zinc-responsive bacterial tRNase. Mol. Microbiol. 2008, 69, 1560–1574. [Google Scholar] [CrossRef]
- Aframian, N.; Eldar, A. Abortive infection antiphage defense systems: Separating mechanism and phenotype. Trends Microbiol. 2023, 31, 1003–1012. [Google Scholar] [CrossRef]
- Mestre, M.R.; Gao, L.A.; Shah, S.A.; López-Beltrán, A.; González-Delgado, A.; Martínez-Abarca, F.; Iranzo, J.; Redrejo-Rodríguez, M.; Zhang, F.; Toro, N. UG/Abi: A highly diverse family of prokaryotic reverse transcriptases associated with defense functions. Nucleic Acids Res. 2022, 50, 6084–6101. [Google Scholar] [CrossRef]
- Dy, R.L.; Przybilski, R.; Semeijn, K.; Salmond, G.P.; Fineran, P.C. A widespread bacteriophage abortive infection system functions through a Type IV toxin-antitoxin mechanism. Nucleic Acids Res. 2014, 42, 4590–4605. [Google Scholar] [CrossRef]
| Strain | Completeness (%) | Contamination (%) | GC Content (%) | Size (bp) |
|---|---|---|---|---|
| C5 | 99.99 | 0.61 | 38 | 2,985,720 |
| A2D9 | 99.99 | 0.81 | 38 | 2,873,619 |
| A2D10 | 99.98 | 0.69 | 38 | 2,902,195 |
| Strain | Number of Genes | Number of CDS | Number of tRNAs Genes | Number of rRNAs |
|---|---|---|---|---|
| C5 | 2971 | 2903 | 67 | 18 |
| A2D9 | 2837 | 2769 | 67 | 18 |
| A2D10 | 2870 | 2802 | 67 | 18 |
| Gene | Function | C5 | A2D9 | A2D10 | EGD-e |
|---|---|---|---|---|---|
| prfA | Listeriolysin positive regulatory protein | + | + | + | + |
| plcA | Phosphatidylinositol-specific phospholipase c | + | + | + | + |
| plcB | Phospholipase C | + | + | + | + |
| hly | Listeriolysin O precursor | + | + | + | + |
| mpl | Zinc metalloproteinase precursor | + | + | + | + |
| actA | Actin-assembly inducing protein precursor | + | + | + | + |
| clpC | Endopeptidase Clp ATP-binding chain C | + | + | + | + |
| vip | surface aghesin Vip | + | + | + | + |
| inlA | Internalin A | + | + | + | + |
| inlB | Internalin B | + | + | + | + |
| inlC | Internalin C | + | + | + | + |
| inlF | Internalin F | + | + | + | + |
| inlJ | Internalin J | - | + | - | + |
| inlK | Internalin K | + | + | + | + |
| pdgA | Peptidoglycan N-deacetylase | - | + | - | + |
| lntA | Listeria nuclear targeted protein A | + | + | + | + |
| iap/cwhA | P60 extracellular protein invasion associated protein Iap | + | + | + | + |
| hpt | Hexose phosphate transport protein | + | + | + | + |
| iplA1 | Lipoate protein ligase | + | + | + | + |
| clpE | ATP-dependent protease | + | + | + | + |
| aut | Autolysin | - | + | - | + |
| llsA | Listeriolysin S | - | - | + | - |
| llsB | SagB family dehydrogenase LlsB | - | - | + | - |
| llsD | Streptolysin-assossiated protein LlsD | - | - | + | - |
| llsG | ABC transporter ATP-binding protein LlsG | - | - | + | - |
| llsH | ABC transporter permease protein LlsH | - | - | + | - |
| llsP | CAAX amino terminal protease LlsP | - | - | + | - |
| llsX | Protein LlsX | - | - | + | - |
| llsY | Protein LlsY | - | - | + | - |
| oatA | Peptidoglycan O-acetyltransferase | + | + | + | + |
| lap | Listeria adhesion protein | + | + | + | + |
| lapB | Listeria adhesion protein LapB | + | + | + | + |
| fbpA | Fibronectin-binding protein | + | + | + | + |
| lspA | Signal peptidase II | + | + | + | + |
| lpeA | Lipoprotein promoting cell invasion | + | + | + | + |
| bsh | Bile salt hydrolase | + | + | + | + |
| prsA2 | Post translocation chaperone PrsA2 | + | + | + | + |
| clpP | ATP-dependent Clp protease proteolytic subunit | + | + | + | + |
| gtcA | Wall teichoic acid glycosylation protein GtcA | + | + | + | + |
| ami | Autolysin amidase adhesin | - | + | - | + |
| Gene | Function | Resistance | C5 | A2D9 | A2D10 | EGD-e |
|---|---|---|---|---|---|---|
| lin | Lin protein | Lincosamides | + | + | + | + |
| fosX | FosX enzyme | Fosfomycin | + | + | + | + |
| norB | NorB multidrug efflux pump | Fluoroquinolones | + | + | + | + |
| mprF | MprF integral membrane protein | Cationic peptides | + | + | + | + |
| mdrL | MdrL efflux pump | Multidrug resistance | + | + | + | + |
| Strain | Defense Systems | Prophage Regions | Genomic Islands |
|---|---|---|---|
| C5 | R-M type I, R-M type II, R-M type IIG | 3 | 4 |
| A2D9 | CRISPR-Cas type-IB, IetAS, retron I-C | 2 | 4 |
| A2D10 | R-M type II, RloC, DRT2, Abi2 | 2 | 2 |
| EGD-e | R-M type II, AbiH | 2 | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Pemaj, V.; Slavko, A.; Konandreas, K.; Pavlidis, D.E.; Ioannidis, A.; Panousopoulos, K.; Xydia, N.; Antonopoulou, V.; Papadelli, M.; Drosinos, E.H.; et al. Hybrid Genome Sequencing and Comparative Analysis of Three Novel Listeria monocytogenes Strains: Insights into Lineage Diversity, Virulence, Antibiotic Resistance, and Defense Systems. Foods 2026, 15, 88. https://doi.org/10.3390/foods15010088
Pemaj V, Slavko A, Konandreas K, Pavlidis DE, Ioannidis A, Panousopoulos K, Xydia N, Antonopoulou V, Papadelli M, Drosinos EH, et al. Hybrid Genome Sequencing and Comparative Analysis of Three Novel Listeria monocytogenes Strains: Insights into Lineage Diversity, Virulence, Antibiotic Resistance, and Defense Systems. Foods. 2026; 15(1):88. https://doi.org/10.3390/foods15010088
Chicago/Turabian StylePemaj, Violeta, Aleksandra Slavko, Konstantinos Konandreas, Dimitrios E. Pavlidis, Anastasios Ioannidis, Konstantinos Panousopoulos, Nikoletta Xydia, Vassiliki Antonopoulou, Marina Papadelli, Eleftherios H. Drosinos, and et al. 2026. "Hybrid Genome Sequencing and Comparative Analysis of Three Novel Listeria monocytogenes Strains: Insights into Lineage Diversity, Virulence, Antibiotic Resistance, and Defense Systems" Foods 15, no. 1: 88. https://doi.org/10.3390/foods15010088
APA StylePemaj, V., Slavko, A., Konandreas, K., Pavlidis, D. E., Ioannidis, A., Panousopoulos, K., Xydia, N., Antonopoulou, V., Papadelli, M., Drosinos, E. H., Skandamis, P. N., Magin, S., & Papadimitriou, K. (2026). Hybrid Genome Sequencing and Comparative Analysis of Three Novel Listeria monocytogenes Strains: Insights into Lineage Diversity, Virulence, Antibiotic Resistance, and Defense Systems. Foods, 15(1), 88. https://doi.org/10.3390/foods15010088








