Potential Clinical Application of Organs-on-a-Chip in Periodontal Diseases: A Systematic Review of In Vitro Studies
Abstract
1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Selection Criteria
2.3. Question
2.4. Review Process
2.5. Data Collection
2.6. Risk of Bias
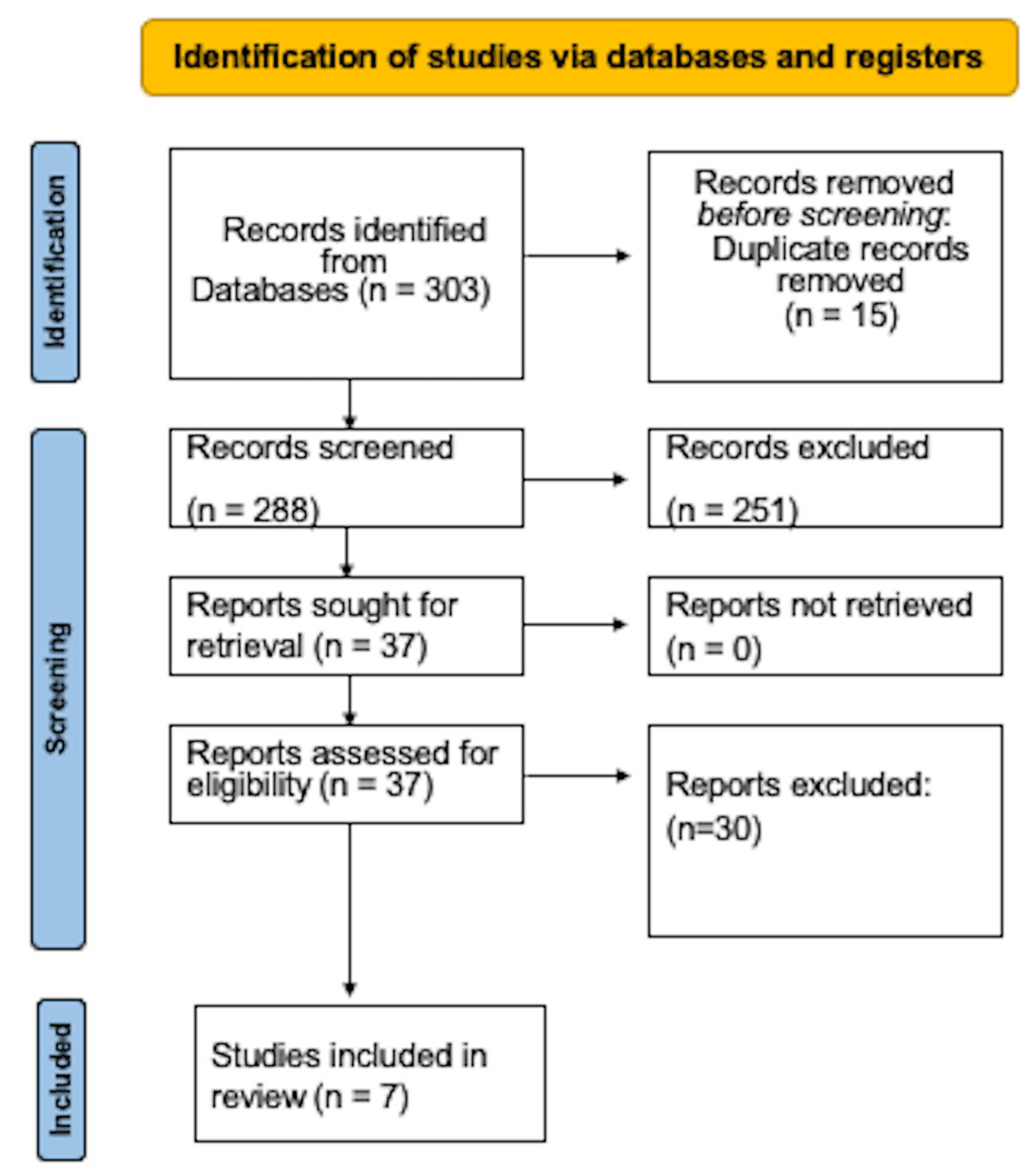
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tonetti, M.S.; Greenwell, H.; Kornman, K.S. Staging and grading of periodontitis: Framework and proposal of a new classification and case definition. J. Periodontol. 2018, 89, S159–S172. [Google Scholar] [CrossRef] [PubMed]
- Kwon, T.; Lamster, I.B.; Levin, L. Current Concepts in the Management of Periodontitis. Int. Dent. J. 2021, 71, 462–476. [Google Scholar] [CrossRef]
- Curtis, M.A.; Diaz, P.I.; Van Dyke, T.E. The role of the microbiota in periodontal disease. Periodontology 2000 2020, 83, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Valm, A.M. The Structure of Dental Plaque Microbial Communities in the Transition from Health to Dental Caries and Periodontal Disease. J. Mol. Biol. 2019, 431, 2957–2969. [Google Scholar] [CrossRef] [PubMed]
- Mombelli, A. Microbial colonization of the periodontal pocket and its significance for periodontal therapy. Periodontology 2000 2018, 76, 85–96. [Google Scholar] [CrossRef]
- Jakubovics, N.S.; Goodman, S.D.; Mashburn-Warren, L.; Stafford, G.P.; Cieplik, F. The dental plaque biofilm matrix. Periodontology 2000 2021, 86, 32–56. [Google Scholar] [CrossRef]
- Takahashi, N.; Sulijaya, B.; Yamada-Hara, M.; Tsuzuno, T.; Tabeta, K.; Yamazaki, K. Gingival epithelial barrier: Regulation by beneficial and harmful microbes. Tissue Barriers 2019, 7, e1651158. [Google Scholar] [CrossRef]
- Stolte, K.N.; Pelz, C.; Yapto, C.V.; Raguse, J.D.; Dommisch, H.; Danker, K. IL-1β strengthens the physical barrier in gingival epithelial cells. Tissue Barriers 2020, 8, 1804249. [Google Scholar] [CrossRef]
- Lee, J.S.; Yilmaz, Ö. Key Elements of Gingival Epithelial Homeostasis upon Bacterial Interaction. J. Dent. Res. 2021, 100, 333–340. [Google Scholar] [CrossRef]
- Lagosz-Cwik, K.B.; Melnykova, M.; Nieboga, E.; Schuster, A.; Bysiek, A.; Dudek, S.; Lipska, W.; Kantorowicz, M.; Tyrakowski, M.; Darczuk, D.; et al. Mapping of DNA methylation-sensitive cellular processes in gingival and periodontal ligament fibroblasts in the context of periodontal tissue homeostasis. Front. Immunol. 2023, 14, 1078031. [Google Scholar] [CrossRef]
- Prado, M.M.; Figueiredo, N.; Pimenta, A.D.L.; Miranda, T.S.; Feres, M.; Figueiredo, L.C.; de Almeida, J.; Bueno-Silva, B. Recent Updates on Microbial Biofilms in Periodontitis: An Analysis of In Vitro Biofilm Models. Periodontitis Adv. Exp. Res. 2022, 1373, 159–174. [Google Scholar]
- Orlandi, M.; Muñoz Aguilera, E.; Marletta, D.; Petrie, A.; Suvan, J.; D’Aiuto, F. Impact of the treatment of periodontitis on systemic health and quality of life: A systematic review. J. Clin. Periodontol. 2022, 49, 314–327. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.C.; Dias-Pereira, A.C.; Branco-de-Almeida, L.S.; Martins, C.C.; Paiva, S.M. Impact of periodontal disease on quality of life: A systematic review. J. Periodontal Res. 2017, 52, 651–665. [Google Scholar] [CrossRef]
- Larvin, H.; Kang, J.; Aggarwal, V.R.; Pavitt, S.; Wu, J. Periodontitis and risk of immune-mediated systemic conditions: A systematic review and meta-analysis. Community Dent. Oral Epidemiol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Luthra, S.; Orlandi, M.; Hussain, S.B.; Leira, Y.; Botelho, J.; Machado, V.; Mendes, J.J.; Marletta, D.; Harden, S.; D’Aiuto, F. Treatment of periodontitis and C-reactive protein: A systematic review and meta-analysis of randomized clinical trials. J. Clin. Periodontol. 2023, 50, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Makkar, H.; Zhou, Y.; Tan, K.S.; Lim, C.T.; Sriram, G. Modeling Crevicular Fluid Flow and Host-Oral Microbiome Interactions in a Gingival Crevice-on-Chip. Adv. Healthc. Mater. 2023, 12, 2202376. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Kou, N.; An, F.; Gao, Z.; Tian, T.; Hui, J.; Chen, C.; Ma, G.; Mao, H.; Liu, H. Analyzing Human Periodontal Soft Tissue Inflammation and Drug Responses In Vitro Using Epithelium-Capillary Interface On-a-Chip. Biosensors 2022, 12, 345. [Google Scholar] [CrossRef]
- Lagosz-Cwik, K.B.; Wielento, A.; Lipska, W.; Kantorowicz, M.; Darczuk, D.; Kaczmarzyk, T.; Gibbs, S.; Potempa, J.; Grabiec, A.M. hTERT-immortalized gingival fibroblasts respond to cytokines but fail to mimic primary cell responses to Porphyromonas gingivalis. Sci. Rep. 2021, 11, 10770. [Google Scholar] [CrossRef]
- Stanton, K.A.; McCracken, B.A. An activated-zinc oral rinse reduces pro-inflammatory cytokine secretion and promotes proliferation in Porphyromonas gingivalis LPS-challenged gingival tissues—A pilot study. Clin. Exp. Dent. Res. 2021, 7, 995–1001. [Google Scholar] [CrossRef]
- Dogan, A.A.; Dufva, M. Customized 3D-printed stackable cell culture inserts tailored with bioactive membranes. Sci. Rep. 2022, 12, 3694. [Google Scholar] [CrossRef]
- Alencar-Silva, T.; Díaz-Martín, R.D.; Zonari, A.; Foyt, D.; Guiang, M.; Pogue, R.; Saldanha-Araujo, F.; Dias, S.C.; Franco, O.L.; Carvalho, J.L. The Combination of Synoeca-MP Antimicrobial Peptide with IDR-1018 Stimulates Proliferation, Migration, and the Expression of Pro-Regenerative Genes in Both Human Skin Cell Cultures and 3D Skin Equivalents. Biomolecules. 2023, 13, 804. [Google Scholar] [CrossRef] [PubMed]
- Barua, N.; Huang, L.; Li, C.; Yang, Y.; Luo, M.; Wei, W.I.; Wong, K.T.; Lo, N.W.S.; Kwok, K.O.; Ip, M. Comparative Study of Two-Dimensional (2D) vs. Three-Dimensional (3D) Organotypic Kertatinocyte-Fibroblast Skin Models for Staphylococcus aureus (MRSA) Infection. Int. J. Mol. Sci. 2021, 23, 299. [Google Scholar] [CrossRef] [PubMed]
- Bao, K.; Belibasakis, G.N.; Selevsek, N.; Grossmann, J.; Bostanci, N. Proteomic profiling of host-biofilm interactions in an oral infection model resembling the periodontal pocket. Sci. Rep. 2015, 5, 15999. [Google Scholar] [CrossRef]
- Bostanci, N.; Bao, K.; Wahlander, A.; Grossmann, J.; Thurnheer, T.; Belibasakis, G.N. Secretome of gingival epithelium in response to subgingival biofilms. Mol. Oral Microbiol. 2015, 30, 323–335. [Google Scholar] [CrossRef]
- Sriram, G.; Natu, V.P.; Islam, I.; Fu, X.; Seneviratne, C.J.; Tan, K.S.; Cao, T. Innate Immune Response of Human Embryonic Stem Cell-Derived Fibroblasts and Mesenchymal Stem Cells to Periodontopathogens. Stem Cells Int. 2016, 2016, 8905365. [Google Scholar] [CrossRef] [PubMed]
- Engen, S.A.; Rørvik, G.H.; Schreurs, O.; Blix, I.J.; Schenck, K. The oral commensal Streptococcus mitis activates the aryl hydrocarbon receptor in human oral epithelial cells. Int. J. Oral Sci. 2017, 9, 145–150. [Google Scholar] [CrossRef]
- Zheng, F.; Fu, F.; Cheng, Y.; Wang, C.; Zhao, Y.; Gu, Z. Organ-on-a-Chip Systems: Microengineering to Biomimic Living Systems. Small 2016, 12, 2253–2282. [Google Scholar] [CrossRef] [PubMed]
- Delon, L.C.; Nilghaz, A.; Cheah, E.; Prestidge, C.; Thierry, B. Unlocking the Potential of Organ-on-Chip Models through Pumpless and Tubeless Microfluidics. Adv. Healthc. Mater. 2020, 9, e1901784. [Google Scholar] [CrossRef]
- Shang, L.; Cheng, Y.; Wang, J.; Ding, H.; Rong, F.; Zhao, Y.; Gu, Z. Double emulsions from a capillary array injection microfluidic device. Lab Chip 2014, 14, 3489–3493. [Google Scholar] [CrossRef]
- Jalili-Firoozinezhad, S.; Gazzaniga, F.S.; Calamari, E.L.; Camacho, D.M.; Fadel, C.W.; Bein, A.; Swenor, B.; Nestor, B.; Cronce, M.J.; Tovaglieri, A.; et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 2019, 3, 520–531. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, P.; Luo, R.; Wang, Y.; Li, Z.; Guo, Y.; Yao, Y.; Li, M.; Tao, T.; Chen, W.; et al. Biomimetic Human Disease Model of SARS-CoV-2-Induced Lung Injury and Immune Responses on Organ Chip System. Adv. Sci. 2020, 8, 2002928. [Google Scholar] [CrossRef]
- Huang, C.; Sanaei, F.; Verdurmen, W.P.R.; Yang, F.; Ji, W.; Walboomers, X.F. The Application of Organs-on-a-Chip in Dental, Oral, and Craniofacial Research. J. Dent. Res. 2023, 102, 364–375. [Google Scholar] [CrossRef] [PubMed]
- Franca, C.M.; Balbinot, G.S.; Cunha, D.; Saboia, V.P.A.; Ferracane, J.; Bertassoni, L.E. In-vitro models of biocompatibility testing for restorative dental materials: From 2D cultures to organs on-a-chip. Acta Biomater. 2022, 150, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Almansoori, A.A.; Kim, B.; Lee, J.H.; Tran, S.D. Tissue Engineering of Oral Mucosa and Salivary Gland: Disease Modeling and Clinical Applications. Micromachines 2020, 11, 1066. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Int. J. Surg. 2021, 88, 105906. [Google Scholar] [CrossRef]
- Ehsani, S.; Mandich, M.A.; El-Bialy, T.H.; Flores-Mir, C. Frictional resistance in self-ligating orthodontic brackets and conventionally ligated brackets. A systematic review. Angle Orthod. 2009, 79, 592–601. [Google Scholar] [CrossRef]
- Jalali, F.; Ellett, F.; Balani, P.; Duncan, M.J.; Dewhirst, F.E.; Borisy, G.G.; Irimia, D. No man’s land: Species-specific formation of exclusion zones bordering Actinomyces graevenitzii microcolonies in nanoliter cultures. Microbiologyopen 2021, 10, e1137. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, M.F.; Leonhardt, D.; Neland, M.L.B.; Schlafer, S. A 3D printed microfluidic flow-cell for microscopy analysis of in situ-grown biofilms. J. Microbiol. Methods 2020, 171, 105876. [Google Scholar] [CrossRef]
- Luo, T.L.; Hayashi, M.; Zsiska, M.; Circello, B.; Eisenberg, M.; Gonzalez-Cabezas, C.; Foxman, B.; Marrs, C.F.; Rickard, A.H. Introducing BAIT (Biofilm Architecture Inference Tool): A software program to evaluate the architecture of oral multi-species biofilms. Microbiology 2019, 165, 527–537. [Google Scholar] [CrossRef]
- Rahimi, C.; Rahimi, B.; Padova, D.; Rooholghodos, S.A.; Bienek, D.R.; Luo, X.; Kaufman, G.; Raub, C.B. Oral mucosa-on-a-chip to assess layer-specific responses to bacteria and dental materials. Biomicrofluidics 2018, 12, 054106. [Google Scholar] [CrossRef]
- Lam, R.H.; Cui, X.; Guo, W.; Thorsen, T. High-throughput dental biofilm growth analysis for multiparametric microenvironmental biochemical conditions using microfluidics. Lab Chip 2016, 16, 1652–1662. [Google Scholar] [CrossRef] [PubMed]
- Azuma, M.M.; Balani, P.; Boisvert, H.; Gil, M.; Egashira, K.; Yamaguchi, T.; Hasturk, H.; Duncan, M.; Kawai, T.; Movila, A. Endogenous acid ceramidase protects epithelial cells from Porphyromonas gingivalis-induced inflammation in vitro. Biochem. Biophys. Res. Commun. 2018, 495, 2383–2389. [Google Scholar] [CrossRef] [PubMed]
- Makkar, H.; Atkuru, S.; Tang, Y.L.; Sethi, T.; Lim, C.T.; Tan, K.S.; Sriram, G. Differential immune responses of 3D gingival and periodontal connective tissue equivalents to microbial colonization. J. Tissue Eng. 2022, 13, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ingendoh-Tsakmakidis, A.; Mikolai, C.; Winkel, A.; Szafrański, S.P.; Falk, C.S.; Rossi, A.; Walles, H.; Stiesch, M. Commensal and pathogenic biofilms differently modulate peri-implant oral mucosa in an organotypic model. Cell. Microbiol. 2019, 21, e13078. [Google Scholar] [CrossRef] [PubMed]
- Lu, E.M.; Hobbs, C.; Ghuman, M.; Hughes, F.J. Development of an in vitro model of the dentogingival junction using 3D organotypic constructs. J. Periodontal Res. 2021, 56, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Lu, E.M.; Hobbs, C.; Dyer, C.; Ghuman, M.; Hughes, F.J. Differential regulation of epithelial growth by gingival and periodontal fibroblasts in vitro. J. Periodontal Res. 2020, 55, 859–867. [Google Scholar] [CrossRef]
- Zou, H.; Zhou, N.; Cheng, X.; Qiu, Y.; Hou, W.; Sun, J. Gingipains are the important virulence factors of Porphyromonas gingivalis downregulating B10 cells. Mol. Oral Microbiol. 2023. [Google Scholar] [CrossRef]
- Neilands, J.; Svensäter, G.; Boisen, G.; Robertsson, C.; Wickström, C.; Davies, J.R. Formation and Analysis of Mono-species and Polymicrobial Oral Biofilms in Flow-Cell Models. Bact. Pathog. Methods Mol. Biol. 2023, 2674, 33–54. [Google Scholar]
- Dama, A.C.; Kim, K.S.; Leyva, D.M.; Lunkes, A.P.; Schmid, N.S.; Jijakli, K.; Jensen, P.A. BacterAI maps microbial metabolism without prior knowledge. Nat. Microbiol. 2023, 8, 1018–1025. [Google Scholar] [CrossRef]
- Mark Welch, J.L.; Rossetti, B.J.; Rieken, C.W.; Dewhirst, F.E.; Borisy, G.G. Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. USA 2016, 113, E791–E800. [Google Scholar] [CrossRef]
- Esteves, G.M.; Pereira, J.A.; Azevedo, N.F.; Azevedo, A.S.; Mendes, L. Friends with Benefits: An Inside Look of Periodontal Microbes’ Interactions Using Fluorescence In Situ Hybridization—Scoping Review. Microorganisms 2021, 9, 1504. [Google Scholar] [CrossRef] [PubMed]
- Kolderman, E.; Bettampadi, D.; Samarian, D.; Dowd, S.E.; Foxman, B.; Jakubovics, N.S.; Rickard, A.H. L-arginine destabilizes oral multi-species biofilm communities developed in human saliva. PLoS ONE 2015, 10, e0121835. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.B.; Klein, O.D. Oral epithelial stem cells in tissue maintenance and disease: The first steps in a long journey. Int. J. Oral Sci. 2013, 5, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Nance, W.C.; Dowd, S.E.; Samarian, D.; Chludzinski, J.; Delli, J.; Battista, J.; Rickard, A.H. A high-throughput microfluidic dental plaque biofilm system to visualize and quantify the effect of antimicrobials. J. Antimicrob. Chemother. 2013, 68, 2550–2560. [Google Scholar] [CrossRef]
- Deutsch, A. An alternate technique of care using silver fluoride followed by stannous fluoride in the management of root caries in aged care. Spec. Care Dent. 2016, 36, 85–92. [Google Scholar] [CrossRef]
- Geisinger, M.L.; Geurs, N.C.; Novy, B.; Otomo-Corgel, J.; Cobb, C.M.; Jacobsen, P.L.; Takesh, T.; Wilder-Smith, P. A randomized double-blind clinical trial evaluating comparative plaque and gingival health associated with commercially available stannous fluoride-containing dentifrices as compared to a sodium fluoride control dentifrice. J. Periodontol. 2023. [Google Scholar] [CrossRef]
- Friesen, L.R.; Goyal, C.R.; Qaqish, J.G.; He, T.; Eusebio, R.; Zsiska, M.; Farmer, T.; Schneiderman, E. Comparative Antiplaque Effect of Two Antimicrobial Dentifrices: Laboratory and Clinical Evaluations. J. Clin. Dent. 2017, 28, B6–B11. [Google Scholar]
- Malone, M.; Goeres, D.M.; Gosbell, I.; Vickery, K.; Jensen, S.; Stoodley, P. Approaches to biofilm-associated infections: The need for standardized and relevant biofilm methods for clinical applications. Expert. Rev. Anti Infect. Ther. 2017, 15, 147–156. [Google Scholar] [CrossRef]
- Duckworth, P.F.; Rowlands, R.S.; Barbour, M.E.; Maddocks, S.E. A novel flow-system to establish experimental biofilms for modelling chronic wound infection and testing the efficacy of wound dressings. Microbiol. Res. 2018, 215, 141–147. [Google Scholar] [CrossRef]
- Hwang, G.; Liu, Y.; Kim, D.; Sun, V.; Aviles-Reyes, A.; Kajfasz, J.K.; Lemos, J.A.; Koo, H. Simultaneous spatiotemporal mapping of in situ pH and bacterial activity within an intact 3D microcolony structure. Sci. Rep. 2016, 6, 32841. [Google Scholar] [CrossRef]
- Schlafer, S.; Dige, I. Ratiometric Imaging of Extracellular pH in Dental Biofilms. J. Vis. Exp. 2016, 109, 53622. [Google Scholar]
- Carvalho, V.; Gonçalves, I.; Lage, T.; Rodrigues, R.O.; Minas, G.; Teixeira, S.F.C.F.; Moita, A.S.; Hori, T.; Kaji, H.; Lima, R.A. 3D Printing Techniques and Their Applications to Organ-on-a-Chip Platforms: A Systematic Review. Sensors 2021, 21, 3304. [Google Scholar] [CrossRef] [PubMed]
- Baptista, L.S.; Porrini, C.; Kronemberger, G.S.; Kelly, D.J.; Perrault, C.M. 3D organ-on-a-chip: The convergence of microphysiological systems and organoids. Front. Cell Dev. Biol. 2022, 10, 1043117. [Google Scholar] [CrossRef] [PubMed]
| Authors, Publication Date | Application of the Organ on a Chip | Materials Used | Chip Design | Cell, Tissues/Microorganisms | Culture | Main Results |
|---|---|---|---|---|---|---|
| Makkar et al. 2023 [16] | Gingival connective tissue equivalent cultured on a microfluidic substrate called “gingival crevice-on-chip” under dynamic interstitial fluid flow that mimics gingival crevicular fluid. | Polydimethylsiloxane, an optically clear, flexible material, is used to create the microfluidic device. | Parallel chamber | A human gingival crevice-on-chip microfluidic system was created to allow the cultivation of gingival fibroblasts embedded within a 3D extracellular matrix in the presence of interstitial fluid flow that is physiologically appropriate and unidirectional, simulating the fluid flow in gingival crevices, utilizing live commensal bacteria, such as Streptococcus oralis, live pathogenic bacteria, such as Fusobacterium nucleatum, and a Toll-like receptor-2 (TLR-2) agonist. A model of the healthy and inflamed states of the connective tissue wall of the gingival crevice was developed utilizing the gingival crevice-on-chip. | To recreate the healthy state of the gingival crevice, which is defined by the colonization of commensal bacteria, long-term bacterial co-culture and colonization investigations were carried out. The crevicular tube was coated with human saliva overnight and then filled with oral commensal bacteria, Streptococcus oralis, to recreate the microbial colonization. | It is possible to replicate microbial colonization, the development of biofilm-like structures at the tissue-microbiome interface, long-term co-culture, and bacterial clearance because of simulating gingival crevicular fluid flow utilizing oral symbiont (Streptococcus oralis) on a chip. Furthermore, on-chip exposure of the gingival connective tissue analog to the periodontal pathogen Fusobacterium nucleatum or the Toll-like receptor-2 (TLR-2) agonist has the capacity to simulate early gingival inflammation. The protective impact of gingival crevicular fluid flow is demonstrated by the generation of simulated gingival crevicular fluid flow toward the bacterial front, which reduces the secretion of inflammatory mediators in contrast to direct exposure. |
| Jin et al. 2022 [17] | The in vivo gingival epithelial barrier with an immunological micro-environment was closely mimicked by the development of a microfluidic epithelium–capillary barrier with a thin culture membrane. | Negative photoresist and polydimethylsiloxane were used to create the upper and lower layers of the microfluidic device in accordance with conventional soft lithography and microfabrication techniques. | Parallel chamber | Surgery was used to remove one to two mm of healthy gingival tissue from patients between the ages of 18 and 30. The lamina propria and epithelium were divided. The tissue fragments and cell pellets were then gathered and re-suspended in keratinocyte growth media. In the endothelial basal medium, human vascular endothelial cells were grown. | The epithelial and endothelial cell suspensions were seeded on the upper and lower sides of the porous membrane in the device, respectively, to establish a bilayer epithelial capillary on the chip. The sterilized chip’s chambers were filled with liquid and left in a culture medium for a whole night before being inoculated with cells. Human vascular endothelial cells were then added to the chip’s lower chamber after it had been turned over so that it could face upwards. The chip was turned over once more so that its upper chamber faced upward, and human gingival epithelial cells were injected into it after resting for two hours to allow cells to attach. | This periodontal soft tissue device was demonstrated to be capable of simulating the inflammatory process induced by lipopolysaccharides or tumor necrosis factor-alpha in major periodontal soft tissue cell lines, while assessing multiple biomarkers of each periodontal soft tissue cell line, to comprehend the intercellular communication between one another. This in vitro epithelium–capillary interface microarray system may be used as a platform to study how drugs affect the health and function of periodontal soft tissues. |
| Jalali et al. 2021 [36] | The dynamic interactions between Streptococcus species, Staphylococcus aureus, and Actinomyces species were evaluated, and a microfluidic-based co-culture system and time-lapse imaging were used. | On four-inch wafers, devices were created using conventional soft-lithography methods. Through a photolithography mask, the photoresist was spin-coated onto a silicon wafer and subjected to ultraviolet light. In a 10:1 mixture with a cross-linking agent, polydimethylsiloxane was applied to wafers. | Multiarray | Three species of Actinomycetaceae were co-loaded in combination with four species of Streptococcus—one strain of Streptococcus salivarius, three strains of Streptococcus mitis, two strains of Streptococcus oralis, and one strain of Streptococcus cristatus—to study the co-culture characteristics of Actinomycetaceae and Streptococcal species in the microfluidic chambers. | On Chacolate II agar, Actinomyces graevenitzii was cultivated at 37 °C in an anaerobic incubator. Overnight incubation of Streptococcus cristatus was performed at 37 °C with shaking. Hemocytometer measurements of bacterial suspension concentrations were used to alter the final bacterial concentration, which was then diluted in Iscove’s Modified Dulbecco’s Medium with 20% fetal bovine serum. Infusion agar plates made from the brain and heart were frequently used to produce bacterial cultures. It was used in nanoliter co-cultures and microfluidic devices. | When Streptococcus cristatus or Streptococcus salivarius were co-cultured in nanoliter containers with Actinomyces graevenitzii, it was discovered that these bacteria were specifically excluded from the media around the microcolonies of A. graevenitzii. This community structure did not exist in co-cultures containing other Actinomycetaceae species such as Streptococcus odontolyticus or Actinomyces naeslundii or with Streptococcus mitis, Streptococcus oralis or isolates of those species. Additionally, compartments containing both A. graevenitzii and Staphylococcus aureus drew fewer neutrophils than compartments having an equivalent amount of either species alone, indicating a potential survival advantage when immune responses are combined. |
| Kristensen et al. 2020 [37] | It was shown how to create a brand-new, adaptable flow cell for microscopy that can handle samples of various geometries and offers shear-controlled flow. The flow cell is created using specialized 3D software, which makes it simple to alter the geometry to fit the given sample. | A flow cell with a customizable shape was created for microscopy examination of samples of in situ-grown biofilms in shear-controlled flow. The flow cells were made as single-piece disposable models, printed in three dimensions from resin, and sealed with a coverslip after the biofilm sample was inserted. | One chamber | The flow cell was used to monitor pH variations in various microenvironments within intraorally developed biofilms from one participant on glass slabs. | On the days of biofilm collection, the participant submitted paraffin-stimulated saliva samples to obtain a realistic flow medium. The saliva samples were cleaned by centrifugation (5 min, 1150 g), filtered through sterile gauze, and then utilized instantly. The biofilms were exposed to a flow rate of 5 mm/min, which corresponds to stimulated saliva flow in the oral cavity, after 30 min of static incubation. | pH decreased in the biofilms under static conditions, with noticeable variations between individual biofilms as well as between various microscopic fields of view inside a single biofilm. The pH of the biofilms’ top layer tended to be lower than that of their bottom layer. The pH of all biofilms increased to neutral or slightly alkaline values when saliva flow was induced, and the vertical gradients were reversed, with the bottom of the biofilms being more acidic than the top. The significance of flow for the investigation of pH in dental biofilms was established. |
| Luo et al. 2019 [38] | A microfluidic biofilm system-based image analysis tool called Biofilm Architecture Inference for quantifying the architecture of oral multi-species biofilms after anti-biofilm interventions was tested. Architectures of treated and untreated biofilms were contrasted, as well as those treated with sodium gluconate (placebo), stannous fluoride, and water (negative control). | Biofilm Architecture Inference Tool, a piece of software that quickly analyzes the structure of biofilms photographed using a confocal laser scanning microscope, was introduced. The program combines plug-and-play automation with a graphical user interface to give users a simple way to examine the architecture of biofilms. | One chamber | At least five healthy, non-smoking individuals’ saliva was collected in batches. Saliva samples were pooled. Both cell-free saliva and cell-containing saliva (the inoculum and growth media, respectively) were prepared. | On 24-well Bioflux plates, a 1 mL sample of saliva-containing cells was added to the outlet wells. The surface of the plate was then treated for 20 min at room temperature. Each treatment well-received additions of various stannous fluoride treatment or placebo treatment concentrations. To imitate higher shear force brought on by brushing or swishing, the treatment schedule was set at 2.0 dynes/cm2 for 2 min. Imaging was performed at five points evenly spaced across the viewing port to record the growth of the biofilm. | Biofilm biovolume, the total number of objects, surface area, fluffiness, connectedness, convex hull porosity, and viability were all calculated using the Biofilm Architecture Inference Tool. Image analysis revealed that 3439 and 10,000 p.p.m. dramatically changed the architecture of the oral biofilm. Treatment with stannous fluoride reduced biovolume, surface area, object count, and connectedness while increasing fluffiness (p < 0.01). The ability of the Biofilm Architecture Inference Tool to evaluate changes in biofilm architecture and identify potential antimicrobial and anti-biofilm effects of candidate drugs was demonstrated. |
| Rahimi et al. 2018 [39] | Mucosal remodeling and the reactions of epithelial and subepithelial layers to stressors frequently seen in the oral environment were assessed. This study set out to create a co-cultured microfluidic mucosal model on a chip. | Eight sets of microchannels were present on each chip. There are six polydimethylsiloxane posts and seven square pores between each set’s three channels. Custom microfluidic device molds were made utilizing conventional photolithography, a spin coater, and an exposure-masking system on a 4 in. silicon wafer. | Parallel chamber | For the mucosa-on-a-chip, two immortalized human cell lines were grown. In polystyrene tissue culture flasks covered in type I collagen, the keratinocyte cell line and human gingival fibroblast cell line were kept alive. | Keratinocytes and fibroblasts were cultured in Prigrow III and IV medium, respectively. A steady co-culture construct was created when human gingival keratinocyte and fibroblast cell lines were added to a 3-channel microfluidic device. | The creation of an oral mucosa-on-a-chip resulted from the formation of a submucosal layer of fibroblasts inserted in collagen in a central conduit, followed by trans-channel planting of keratinocytes into pores between polydimethylsiloxane posts. This structure enabled convenient and accurate tracking of keratinocytes and fibroblasts using conventional microscopy, with posts permitting for dual attachment of keratinocytes to apical and basal surfaces, as appears in the junctional epithelium. After 2-hydroxylethyl methacrylate exposure, cell viability decreased, and transepithelial electrical resistance declined following Streptococcus mutans culture from the luminal channel. These findings establish a microfluidic culture-on-a-chip methodology to test the oral mucosa hypothesis. |
| Lam et al. 2016 [40] | A high-throughput microfluidic “artificial teeth” device was described that offered controls of various microenvironmental elements (such as nutrients, growth factors, dissolved gases, and seeded cell populations) for quantitative traits of long-term dental bacterial growth and biofilm development. | From top to bottom, the microfluidic device was constructed using a multilayer soft lithography of polydimethylsiloxane for the microchannel layers: gas control channels, oxygenation channels, water jackets, flow channels, and culture chambers. | Multiarray | The biofilm samples were taken from a human oral cavity. Based on their prevalence and significance in the formation of the dental biofilm, the target species were selected to be Sprepptococci, Fusobacterium nucleatum, Actinomyces naeslundii, and Porphyromonas gingivalis. | Dental bacteria samples were parallel cultured under four daily dissolved oxygen cycles to show the impact of the dissolved oxygen profiles on the bacterial species contents in the dental biofilm. A healthy pH buffer and the nutritional equivalent of saliva is basal medium mucin. A basal medium recipe was used to create it. A solution containing 5 g/L trypticase peptone, 10 g/L proteose peptone, 5 g/L yeast extract, 2.5 g/L potassium chloride, 5 mg/L hemin, 1 mg/L menadione, 2.5 g/L gastric mucin, 1 mmol/L urea, and 1 mmol/L L-arginine was used to create a basal medium mucin. | As the dental biofilms developed, the device for artificial teeth was able to evaluate the biofilm’s form, colonization density, and spatial arrangement of bacterial species. By measuring the cluster thickness, cell viability distribution, and percentage of the major dental bacteria (Streptococci, Fusobacterium nucleatum, Actinomyces naeslundii, and Porphyromonas gingivalis), this device may acquire quantitative biofilm characteristics. Moreover, the multiplexed platform’s scalable architecture allowed for the monitoring of several culture samples that were exposed to various environmental conditions, which reduced the considerable experimental efforts required in traditional laboratory settings. |
| Study | Design Objective Sample Baseline Characteristics Co-Interventions | Measures Measurement Method Blinding Examiner Blinding Statistician Described Reliability. Level of Agreement | Analysis Statistical Appropriate Analysis Co-Interventions Subgroup Analysis Statistical Significance Confidence Intervals | Clinical Significance | Total |
|---|---|---|---|---|---|
| Makkar et al. 2023 [16] | 4 | 3 | 4 | 1 | 12 |
| Jin et al. 2022 [17] | 4 | 3 | 4 | 1 | 12 |
| Jalali et al. 2021 [36] | 4 | 3 | 5 | 1 | 13 |
| Kristensen et al. 2020 [37] | 4 | 3 | 4 | 1 | 12 |
| Luo et al. 2019 [38] | 4 | 3 | 4 | 1 | 12 |
| Rahimi et al. 2018 [39] | 4 | 3 | 4 | 1 | 12 |
| Lam et al. 2016 [40] | 4 | 3 | 4 | 1 | 12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ardila, C.M.; Jiménez-Arbeláez, G.A.; Vivares-Builes, A.M. Potential Clinical Application of Organs-on-a-Chip in Periodontal Diseases: A Systematic Review of In Vitro Studies. Dent. J. 2023, 11, 158. https://doi.org/10.3390/dj11070158
Ardila CM, Jiménez-Arbeláez GA, Vivares-Builes AM. Potential Clinical Application of Organs-on-a-Chip in Periodontal Diseases: A Systematic Review of In Vitro Studies. Dentistry Journal. 2023; 11(7):158. https://doi.org/10.3390/dj11070158
Chicago/Turabian StyleArdila, Carlos M., Gustavo A. Jiménez-Arbeláez, and Annie Marcela Vivares-Builes. 2023. "Potential Clinical Application of Organs-on-a-Chip in Periodontal Diseases: A Systematic Review of In Vitro Studies" Dentistry Journal 11, no. 7: 158. https://doi.org/10.3390/dj11070158
APA StyleArdila, C. M., Jiménez-Arbeláez, G. A., & Vivares-Builes, A. M. (2023). Potential Clinical Application of Organs-on-a-Chip in Periodontal Diseases: A Systematic Review of In Vitro Studies. Dentistry Journal, 11(7), 158. https://doi.org/10.3390/dj11070158









