Numerical Approach to a Nonlocal Advection-Reaction-Diffusion Model of Cartilage Pattern Formation
Abstract
1. Introduction
1.1. Mathematical Model
1.2. Numerical Approach
1.3. Outline of Paper
2. Numerical Implementation
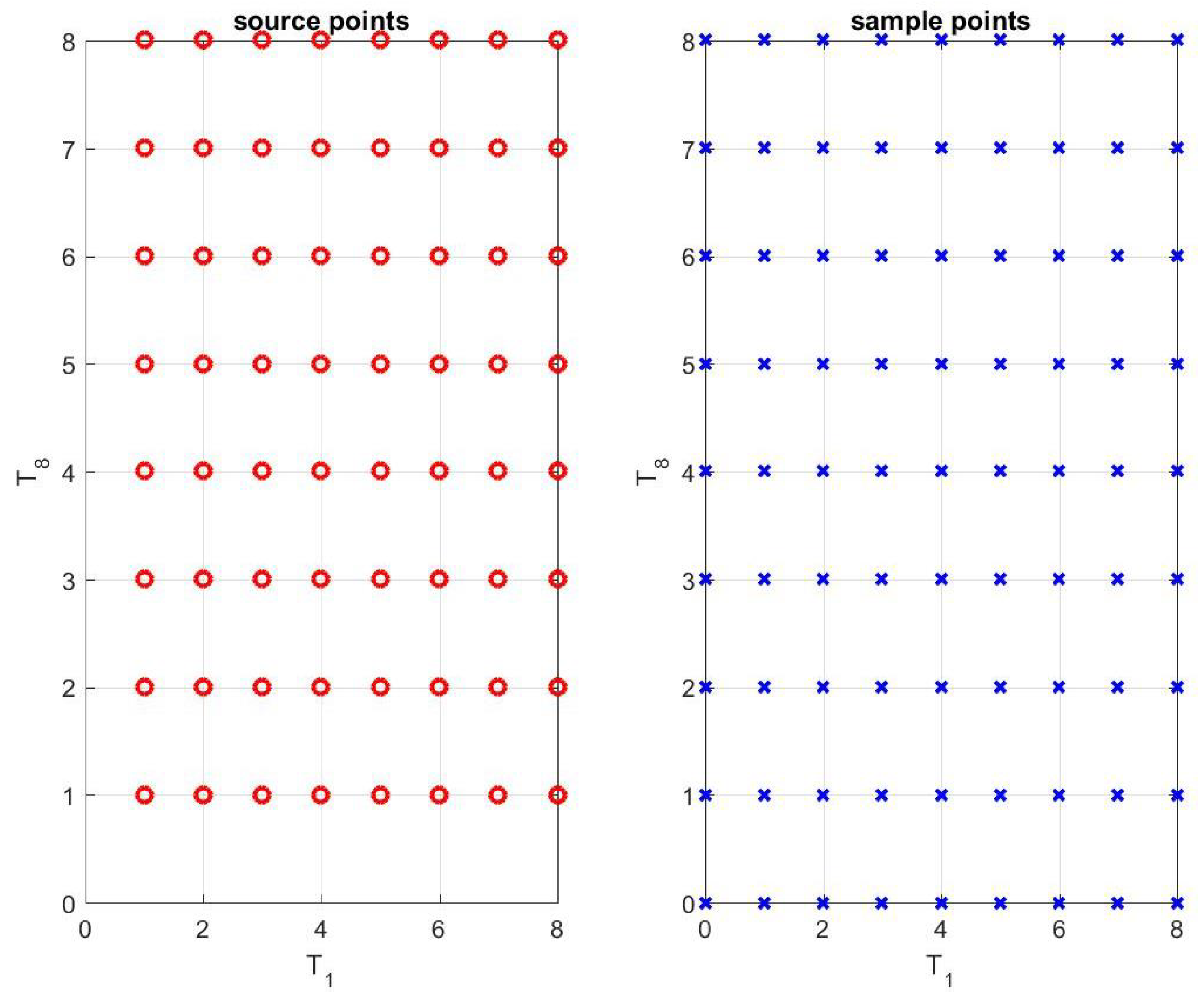
2.1. Radial Basis Function (RBF) Expansion
2.2. Finite Difference WENO5 Discretization
2.3. Time Integration
2.3.1. Method of Integrating Factors
2.3.2. Time Integration Method
2.4. Numerical Algorithm and Complexity
- Let be the number of source points in the -plane. Initialize different R’s over a 2D spatial mesh of grids, associated with the source points.
- Initialize and over the same 2D spatial mesh of grids.
- Compute exactly the partial derivatives and integrals of radio basis functions to be used later, including
- Perform the following at each time step up to the desired .
- Determine the coefficients ’s in the RBF expansion for all the R’s.
- Compute using WENO5 finite difference discretization.
- Update R explicitly using SEIF3.
- Determine and via linear combinations of the known information from the radio basis functions.
- Update and explicitly using SEIF3.
3. Numerical Results
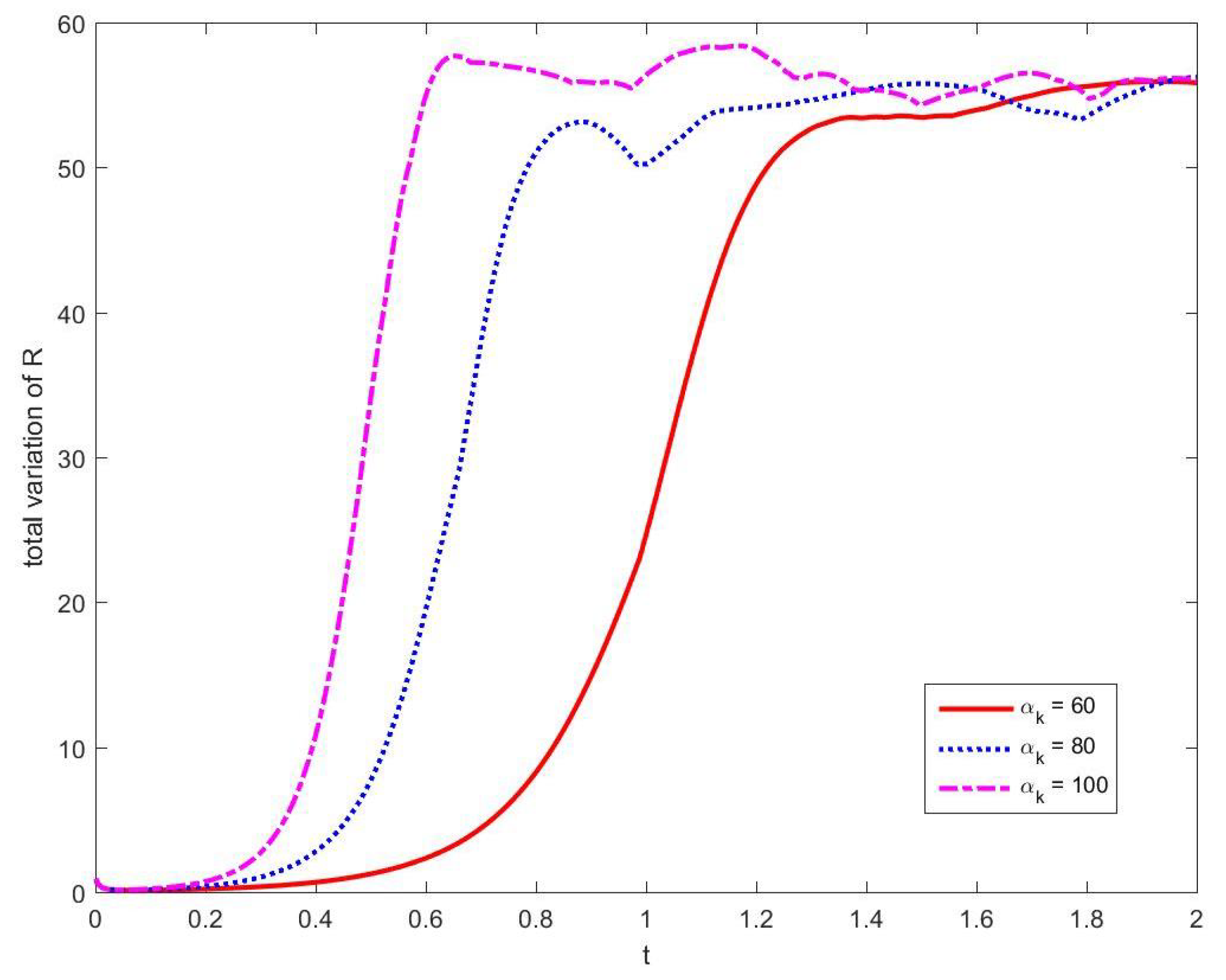
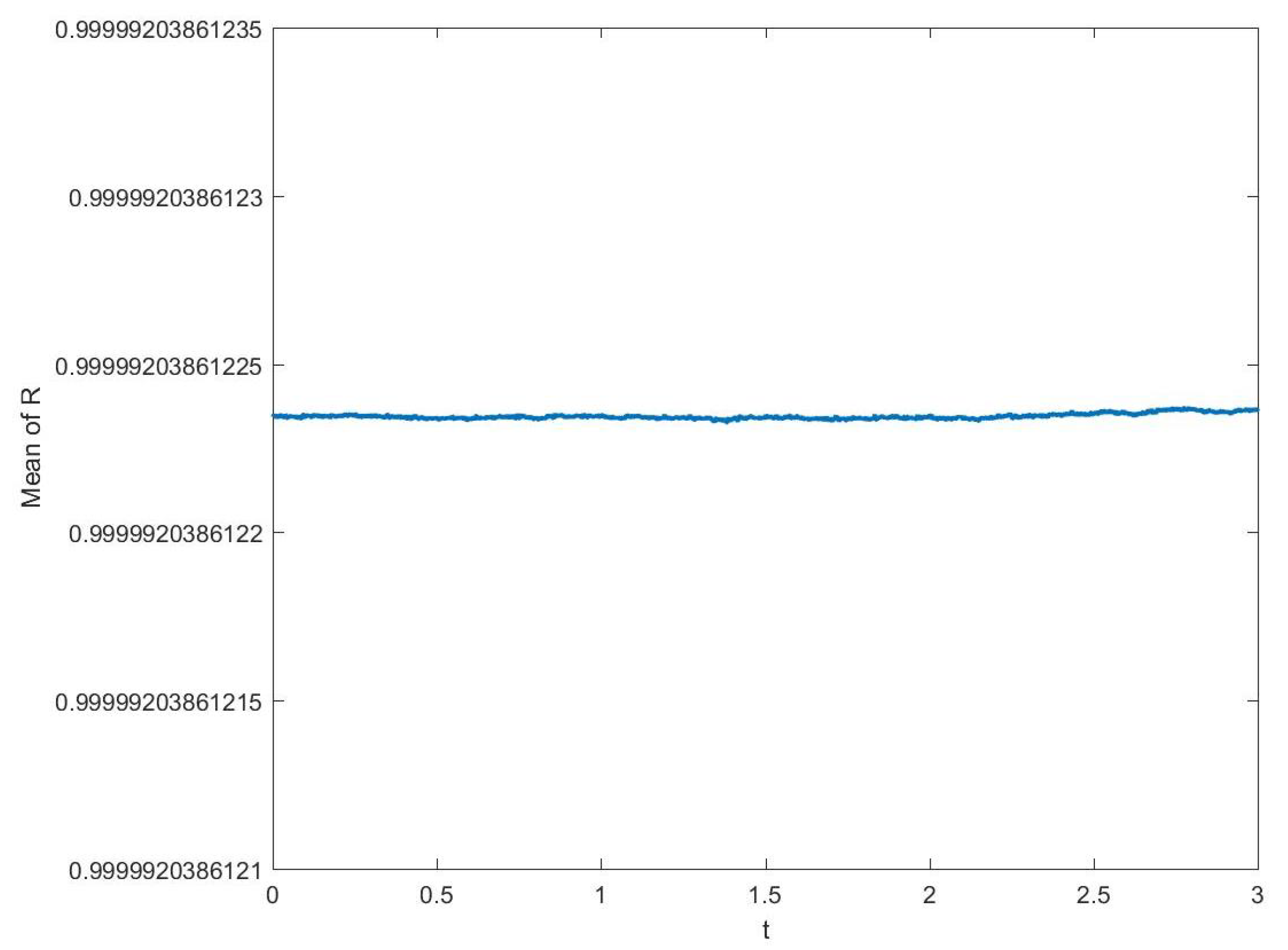
3.1. Convergence and Stability of Numerical Methods
3.1.1. Stability of Time Integration Method
3.1.2. Convergence of RBF Approximation
- A Gaussian function with shape parameter is more accurately approximated by the Gaussian basis functions with shape parameter closest to , as shown in Table 3.
- The Gaussian basis approximation to a flatter Gaussian function achieves higher order of accuracy with matching Gaussian basis functions (i.e., with closest to ), as shown in Table 3.
- A Gaussian function centered close to the boundary is less accurately approximated by any Gaussian basis, as shown in Table 4. This effect is most obvious for the flattest Gaussian function (left most column in Table 4) since it does not decay fast enough to smoothly match the homogeneous Dirichlet condition at boundaries and . When the variance of a Gaussian function decreases, it decays fast enough so that the boundary disagreement is negligible compared to the interpolating error caused by the non-flatness of the Gaussian function. This explains why the minimum error does not have to occur exactly at the center of , referring to the right most column in Table 4.
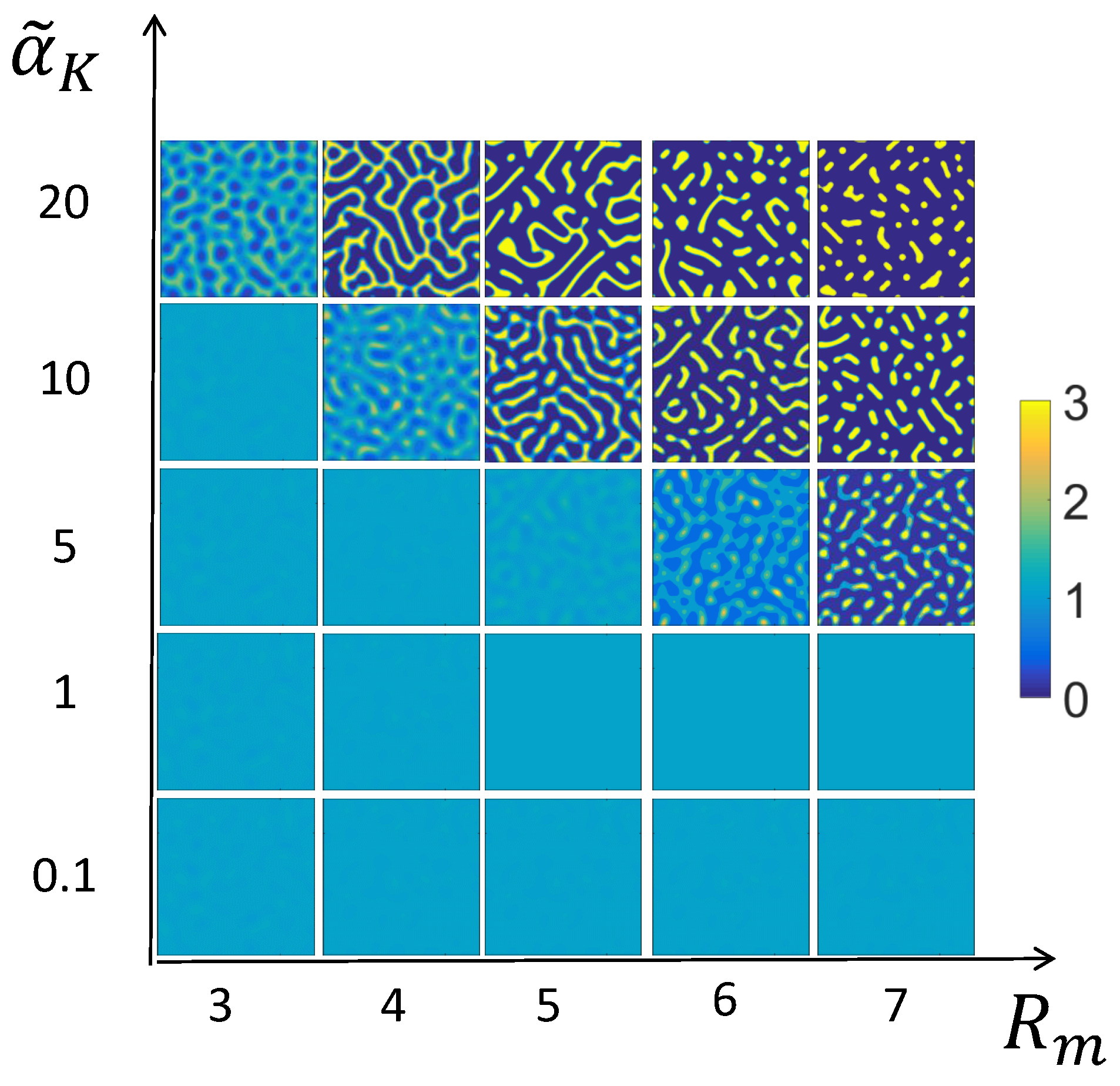
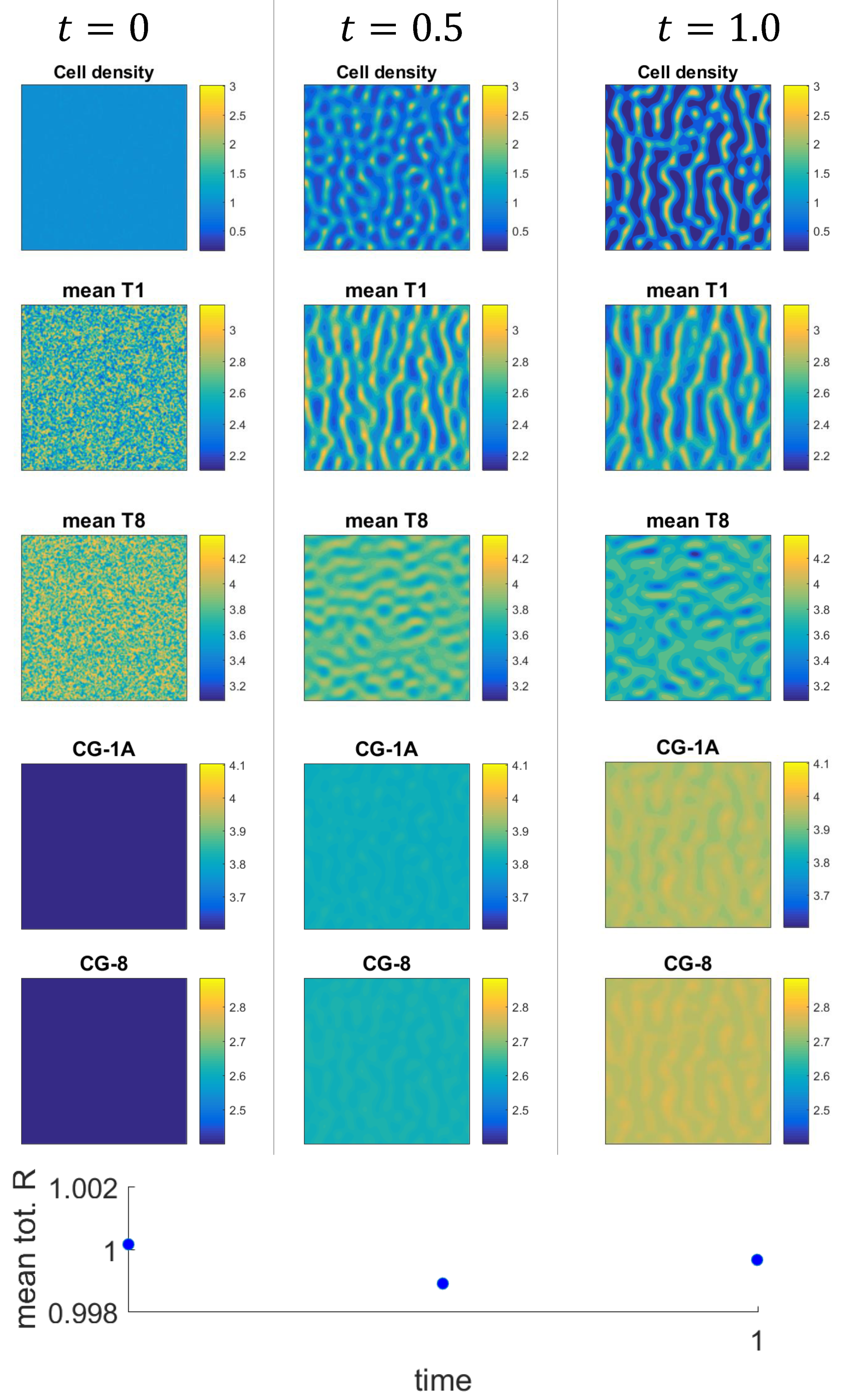
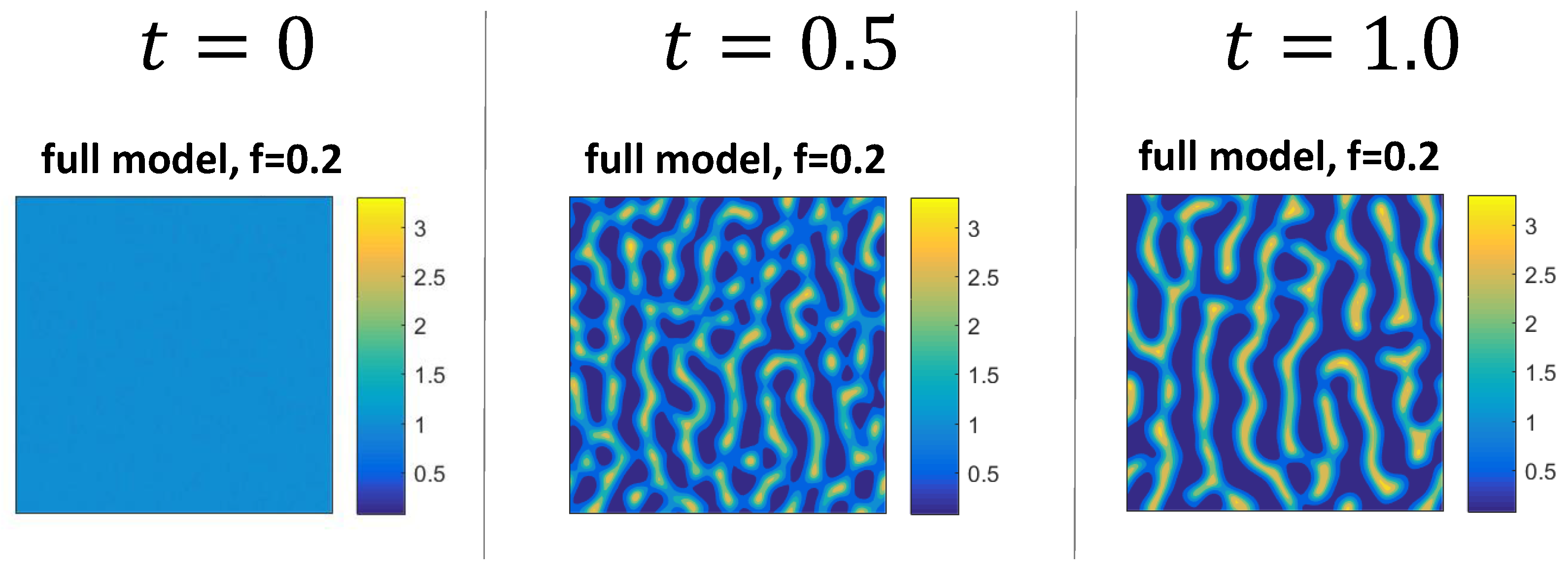
3.2. Numerical Examples for the Full System
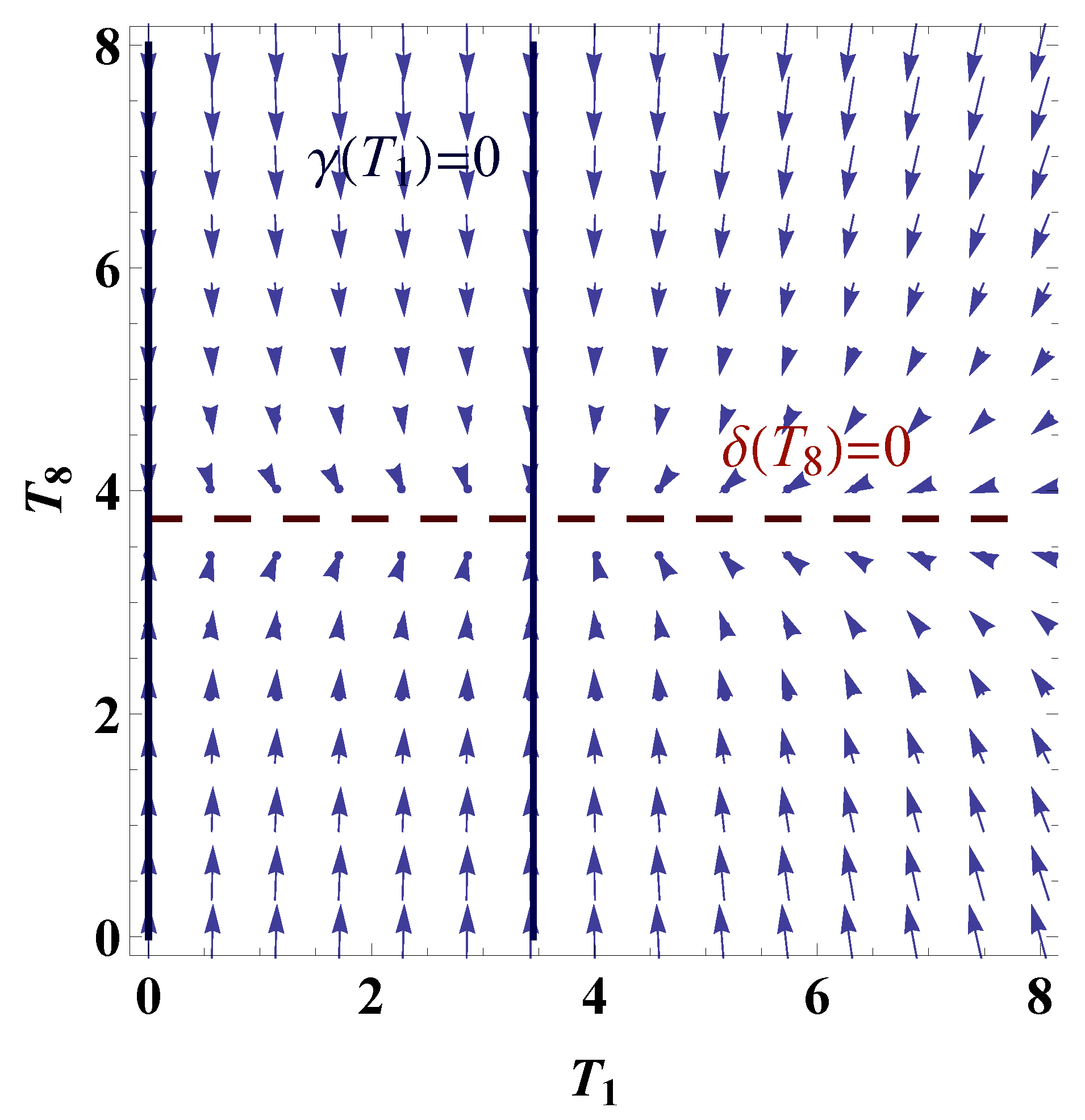
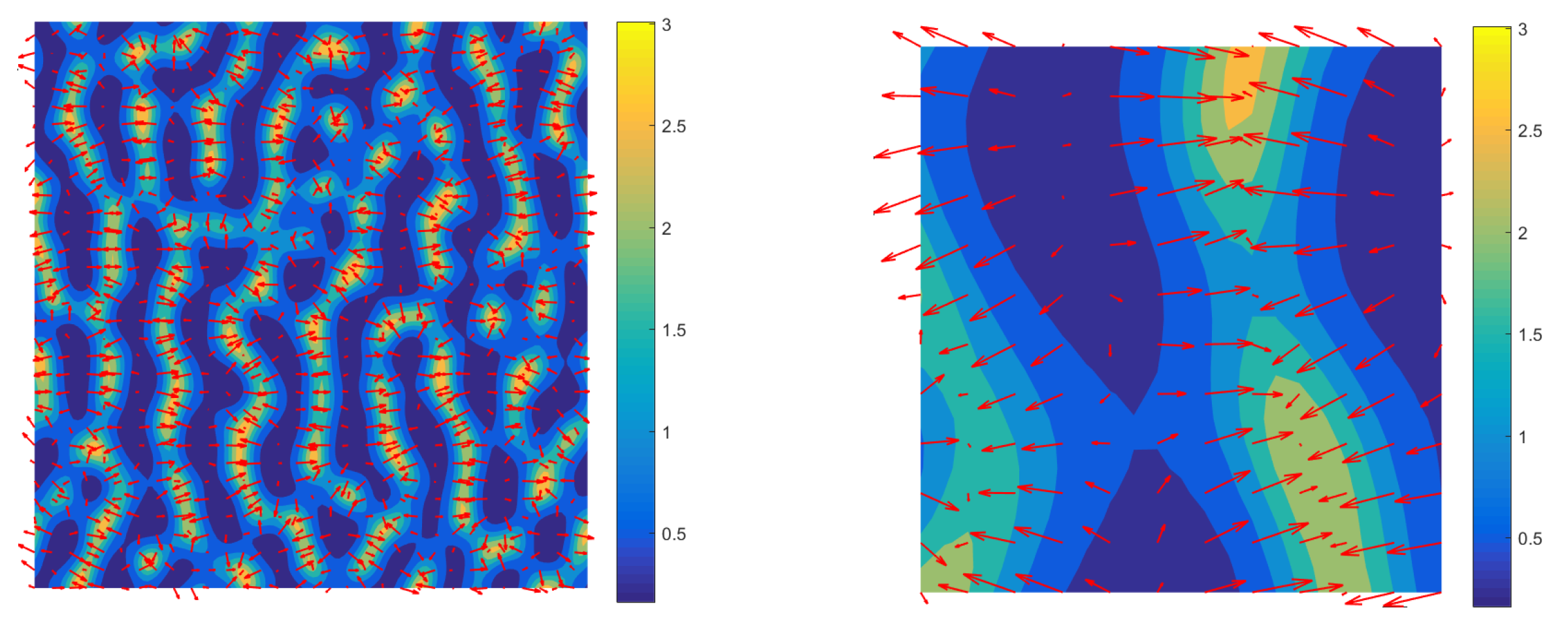
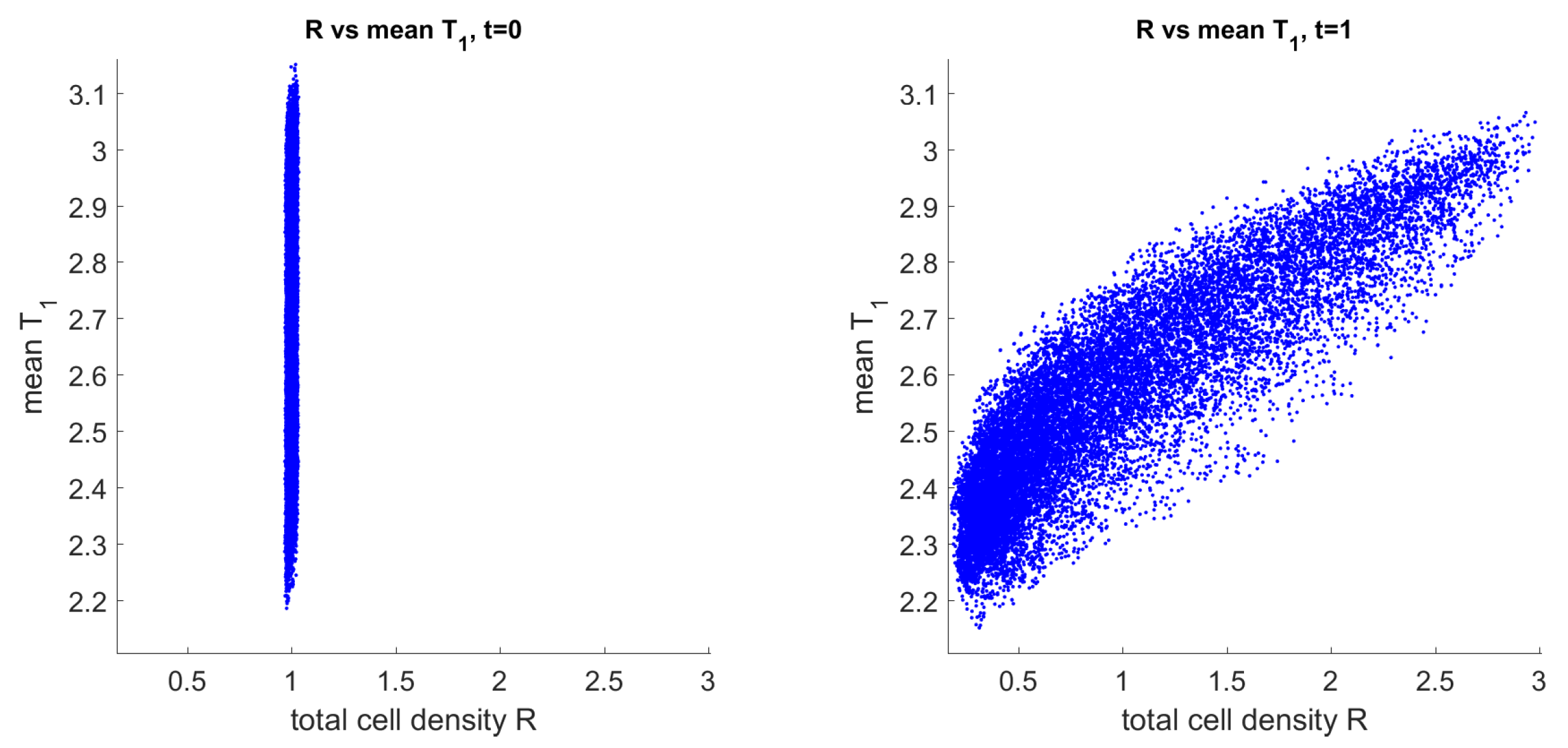
3.3. Mechanism of Pattern Formation
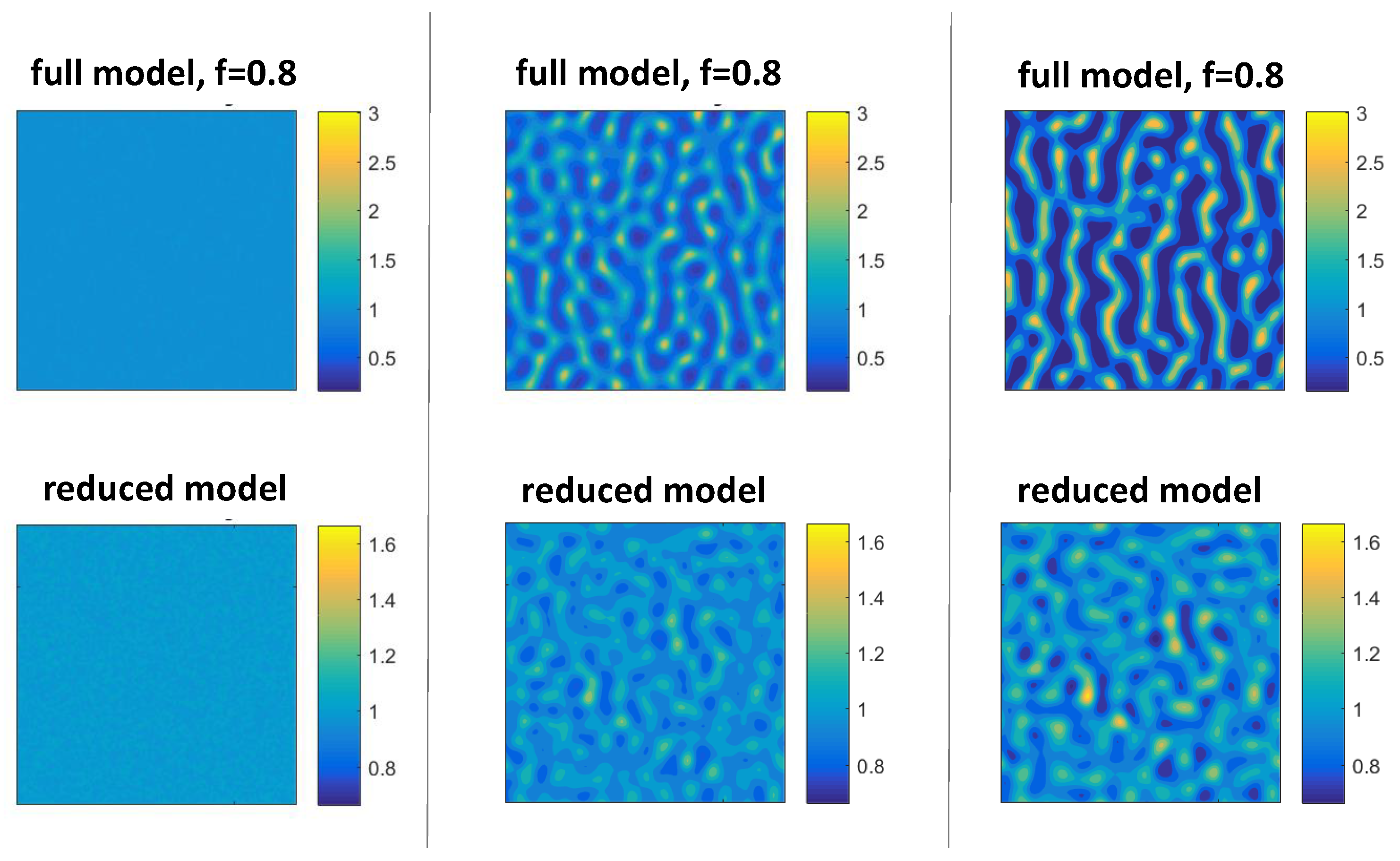
3.4. Comparison of Solutions to the Full and Reduced Models
4. Conclusions and Future Investigations
Author Contributions
Funding
Conflicts of Interest
References
- Forgacs, G.; Newman, S. Biological Physics of the Developing Embryo; Cambridge University Press: Cambridge, UK, 2005. [Google Scholar]
- Wolpert, L.; Tickle, C.; Arias, A.M. Principles of Development; Oxford University Press: Oxford, UK, 2015. [Google Scholar]
- Rejniak, K.A.; McCawley, L.J. Current trends in mathematical modeling of tumor–microenvironment interactions: A survey of tools and applications. Exp. Biol. Med. 2010, 235, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Altrock, P.M.; Liu, L.L.; Michor, F. The mathematics of cancer: Integrating quantitative models. Nat. Rev. Cancer 2015, 15, 730–745. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, N.J.; Painter, K.J.; Sherratt, J.A. A continuum approach to modelling cell-cell adhesion. J. Theor. Biol. 2006, 243, 98–113. [Google Scholar] [CrossRef] [PubMed]
- Anguige, K.; Schmeiser, C. A one-dimensional model of cell diffusion and aggregation, incorporating volume filling and cell-to-cell adhesion. J. Math. Biol. 2008, 58, 395–427. [Google Scholar] [CrossRef]
- Murakawa, H.; Togashi, H. Continuous models for cell-cell adhesion. J. Theor. Biol. 2015, 374, 1–12. [Google Scholar] [CrossRef]
- Buttenschön, A.; Hillen, T.; Gerisch, A.; Painter, K.J. A space-jump derivation for non-local models of cell–cell adhesion and non-local chemotaxis. J. Math. Biol. 2018, 76, 429–456. [Google Scholar] [CrossRef]
- Gerisch, A.; Chaplain, M.A.J. Mathematical modelling of cancer cell invasion of tissue: Local and non-local models and the effect of adhesion. J. Theor. Biol. 2008, 250, 684–704. [Google Scholar] [CrossRef]
- Engwer, C.; Stinner, C.; Surulescu, C. On a structured multiscale model for acid-mediated tumor invasion: The effects of adhesion and proliferation. Math. Models Methods Appl. Sci. 2017, 27, 1355–1390. [Google Scholar] [CrossRef]
- Painter, K.J.; Armstrong, N.J.; Sherratt, J.A. The impact of adhesion on cellular invasion processes in cancer and development. J. Theor. Biol. 2010, 264, 1057–1067. [Google Scholar] [CrossRef]
- Chaplain, M.a.J.; Lachowicz, M.; Szymańska, Z.; Wrzosek, D. Mathematical modelling of cancer invasion: The importance of cell–cell adhesion and cell–matrix adhesion. Math. Models Methods Appl. Sci. 2011, 21, 719–743. [Google Scholar] [CrossRef]
- Frieboes, H.B.; Jin, F.; Chuang, Y.L.; Wise, S.M.; Lowengrub, J.S.; Cristini, V. Three-dimensional multispecies nonlinear tumor growth—II: Tumor invasion and angiogenesis. J. Theor. Biol. 2010, 264, 1254–1278. [Google Scholar] [CrossRef]
- Bitsouni, V.; Trucu, D.; Chaplain, M.A.J.; Eftimie, R. Aggregation and travelling wave dynamics in a two-population model of cancer cell growth and invasion. Math. Med. Biol. J. IMA 2018, 35, 541–577. [Google Scholar] [CrossRef] [PubMed]
- Domschke, P.; Trucu, D.; Gerisch, A.; Chaplain, M.A. Mathematical modelling of cancer invasion: Implications of cell adhesion variability for tumour infiltrative growth patterns. J. Theor. Biol. 2014, 361, 41–60. [Google Scholar] [CrossRef] [PubMed]
- Green, J.E.F.; Waters, S.L.; Whiteley, J.P.; Edelstein-Keshet, L.; Shakesheff, K.M.; Byrne, H.M. Non-local models for the formation of hepatocyte–stellate cell aggregates. J. Theor. Biol. 2010, 267, 106–120. [Google Scholar] [CrossRef] [PubMed]
- Glimm, T.; Bhat, R.; Newman, S.A. Modeling the morphodynamic galectin patterning network of the developing avian limb skeleton. J. Theor. Biol. 2014, 346, 86–108. [Google Scholar] [CrossRef]
- Dyson, J.; Webb, G.F. A Cell Population Model Structured by Cell Age Incorporating Cell–Cell Adhesion. In Mathematical Oncology 2013; d’Onofrio, A., Gandolfi, A., Eds.; Springer: New York, NY, USA, 2014; pp. 109–149. [Google Scholar] [CrossRef]
- Gandolfi, A.; Iannelli, M.; Marinoschi, G. An age-structured model of epidermis growth. J. Math. Biol. 2011, 62, 111–141. [Google Scholar] [CrossRef]
- Dyson, J.; Villella-Bressan, R.; Webb, G. A Spatial Model of Tumor Growth with Cell Age, Cell Size, and Mutation of Cell Phenotypes. Math. Model. Nat. Phenom. 2007, 2, 69–100. [Google Scholar] [CrossRef]
- Zwilling, E. Development of fragmented and dissociated limb bud mesoderm. Dev. Biol. 1964, 89, 20–37. [Google Scholar] [CrossRef]
- Ros, M.A.; Lyons, G.E.; Mackem, S.; Fallon, J.F. Recombinant Limbs as a Model to Study Homeobox Gene Regulation during Limb Development. Dev. Biol. 1994, 166, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Downie, S.A.; Newman, S.A. Morphogenetic differences between fore and hind limb precartilage mesenchyme: Relation to mechanisms of skeletal pattern formation. Dev. Biol. 1994, 162, 195–208. [Google Scholar] [CrossRef]
- De Lise, A.M.; Stringa, E.; Woodward, W.A.; Mello, M.A.; Tuan, R.S. Embryonic Limb Mesenchyme Micromass Culture as an In Vitro Model for Chondrogenesis and Cartilage Maturation. In Developmental Biology Protocols; Tuan, R.S., Lo, C.W., Eds.; Humana Press: Totowa, NJ, USA, 2000; pp. 359–375. [Google Scholar] [CrossRef]
- Zeng, W.; Thomas, G.L.; Glazier, J.A. Non-Turing stripes and spots: A novel mechanism for biological cell clustering. Phys. A Stat. Mech. Appl. 2004, 341, 482–494. [Google Scholar] [CrossRef]
- Saha, A.; Rolfe, R.; Carroll, S.; Kelly, D.J.; Murphy, P. Chondrogenesis of embryonic limb bud cells in micromass culture progresses rapidly to hypertrophy and is modulated by hydrostatic pressure. Cell Tissue Res. 2017, 368, 47–59. [Google Scholar] [CrossRef]
- Miura, T.; Komori, M.; Shiota, K. A novel method for analysis of the periodicity of chondrogenic patterns in limb bud cell culture: Correlation of in vitro pattern formation with theoretical models. Anat. Embryol. 2000, 201, 419–428. [Google Scholar] [CrossRef]
- Murray, J.D. Mathematical Biology. II, 3rd ed.; Springer: New York, NY, USA, 2002; Volume 18. [Google Scholar]
- Glimm, T.; Headon, D.; Kiskowski, M.A. Computational and mathematical models of chondrogenesis in vertebrate limbs. Birth Defects Res. C Embryo Today 2012, 96, 176–192. [Google Scholar] [CrossRef]
- Newman, S.A.; Glimm, T.; Bhat, R. The vertebrate limb: An evolving complex of self-organizing systems. Prog. Biophys. Mol. Biol. 2018, 137, 12–24. [Google Scholar] [CrossRef]
- Glimm, T.; Bhat, R.; Newman, S.A. Multiscale modeling of vertebrate limb development. WIREs Syst. Biol. Med. 2020, 12, e1485. [Google Scholar] [CrossRef]
- Chatterjee, P.; Glimm, T.; Kaźmierczak, B. Mathematical modeling of chondrogenic pattern formation during limb development: Recent advances in continuous models. Math. Biosci. 2020, 322, 108319. [Google Scholar] [CrossRef]
- Raspopovic, J.; Marcon, L.; Russo, L.; Sharpe, J. Modeling digits. Digit patterning is controlled by a Bmp-Sox9-Wnt Turing network modulated by morphogen gradients. Science 2014, 345, 566–570. [Google Scholar] [CrossRef]
- Bhat, R.; Lerea, K.M.; Peng, H.; Kaltner, H.; Gabius, H.J.; Newman, S.A. A regulatory network of two galectins mediates the earliest steps of avian limb skeletal morphogenesis. BMC Dev. Biol. 2011, 11. [Google Scholar] [CrossRef]
- Aranson, I.S. Physical Models of Cell Motility; Springer: New York, NY, USA, 2015. [Google Scholar]
- Wang, S.E.; Hinow, P.; Bryce, N.; Weaver, A.M.; Estrada, L.; Arteaga, C.L.; Webb, G.F. A mathematical model quantifies proliferation and motility effects of TGF-β on cancer cells. Comput. Math. Methods Med. 2009, 10, 71–83. [Google Scholar] [CrossRef]
- Graner, F.; Glazier, J.A. Simulation of biological cell sorting using a two-dimensional extended Potts model. Phys. Rev. Lett. 1992, 69, 2013–2016. [Google Scholar] [CrossRef]
- Guisoni, N.; Mazzitello, K.I.; Diambra, L. Modeling Active Cell Movement with the Potts Model. Front. Phys. 2018, 6. [Google Scholar] [CrossRef]
- Jiang, G.S.; Shu, C.W. Efficient Implementation of Weighted ENO Schemes. J. Comput. Phys. 1996, 126, 202–228. [Google Scholar] [CrossRef]
- Jiang, T.; Zhang, Y.T. Krylov single-step implicit integration factor WENO methods for advection-diffusion-reaction equations. J. Comput. Phys. 2016, 311, 22–44. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, Y.T.; Newman, S.A.; Alber, M.S. A Finite Element Model Based on Discontinuous Galerkin Methods on Moving Grids for Vertebrate Limb Pattern Formation. Math. Model. Nat. Phenom. 2009, 4, 131–148. [Google Scholar] [CrossRef][Green Version]
- Fornberg, B.; Flyer, N. Solving PDEs with radial basis functions. Acta Numer. 2015, 24, 215–258. [Google Scholar] [CrossRef]
- Nie, Q.; Wan, F.Y.M.; Zhang, Y.T.; Liu, X.F. Compact integration factor methods in high spatial dimensions. J. Comput. Phys. 2008, 227, 5238–5255. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, L.; Nie, Q. Array-representation integration factor method for high-dimensional systems. J. Comput. Phys. 2014, 258, 585–600. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, Y.T. Krylov implicit integration factor methods for spatial discretization on high dimensional unstructured meshes: Application to discontinuous Galerkin methods. J. Comput. Phys. 2011, 230, 4336–4352. [Google Scholar] [CrossRef]
- Jiang, T.; Zhang, Y.T. Krylov implicit integration factor WENO methods for semilinear and fully nonlinear advection-diffusion-reaction equations. J. Comput. Phys. 2013, 253, 368–388. [Google Scholar] [CrossRef]
- Cox, S.M.; Matthews, P.C. Exponential time differencing for stiff systems. J. Comput. Phys. 2002, 176, 430–455. [Google Scholar] [CrossRef]
- Bhat, R.; Glimm, T.; Linde-Medina, M.; Cui, C.; Newman, S.A. Synchronization of Hes1 oscillations coordinate and refine condensation formation and patterning of the avian limb skeleton. Mech. Dev. 2019, 156, 41–54. [Google Scholar] [CrossRef] [PubMed]
- Frank, S.A. The Common Patterns of Nature. J. Evol. Biol. 2009, 22, 1563–1585. [Google Scholar] [CrossRef]
- Chen, Y.; Munteanu, A.C.; Huang, Y.F.; Phillips, J.; Zhu, Z.; Mavros, M.; Tan, W. Mapping Receptor Density on Live Cells by Using Fluorescence Correlation Spectroscopy. Chemistry 2009, 15, 5327–5336. [Google Scholar] [CrossRef]
- Dillon, R.; Othmer, H.G. A mathematical model for outgrowth and spatial patterning of the vertebrate limb bud. J. Theor. Biol. 1999, 197, 295–330. [Google Scholar] [CrossRef]
- Boehm, B.; Westerberg, H.; Lesnicar-Pucko, G.; Raja, S.; Rautschka, M.; Cotterell, J.; Swoger, J.; Sharpe, J. The Role of Spatially Controlled Cell Proliferation in Limb Bud Morphogenesis. PLoS Biol. 2010, 8, e1000420. [Google Scholar] [CrossRef]
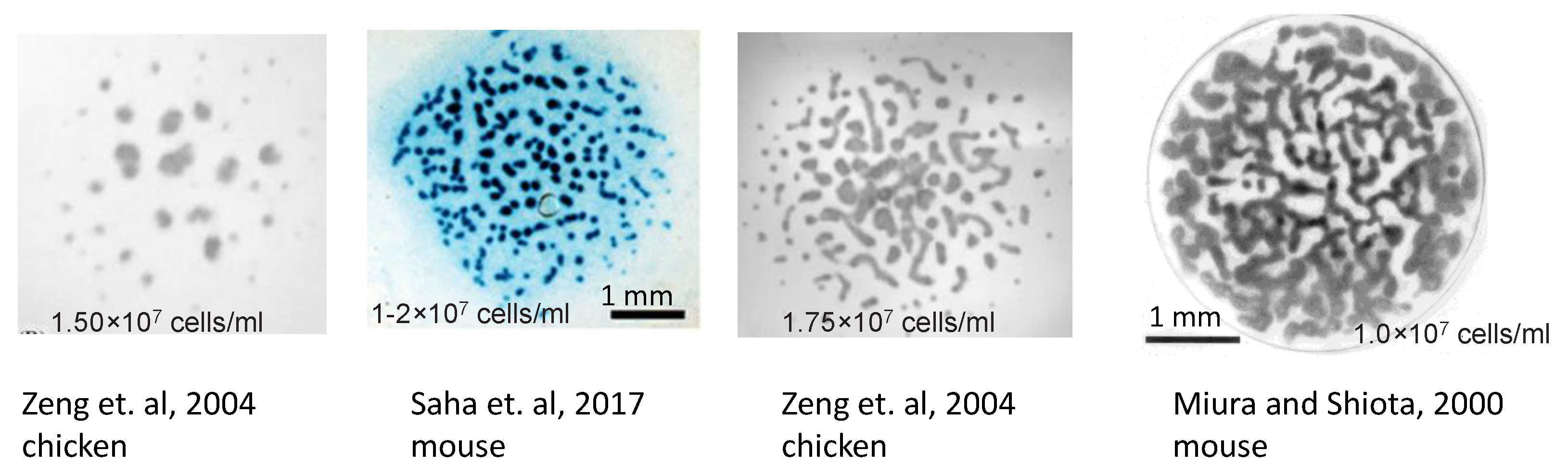
| Variable | Meaning |
|---|---|
| space | |
| t | time |
| density of counterreceptor CR-1 (membrane-bound) | |
| density of counterreceptor CR-8 (membrane-bound) | |
| cell density as a function of time, space, counterreceptor densities | |
| CG-1A concentration | |
| CG-8 concentration |
| 1 | 2 | ||||
| cond(A) | |||||
| 3 | |||||
| cond(A) |
| 1 | |||
| 2 | |||
| 3 | |||
| 4 | |||
| 5 | |||
| 6 | |||
| 7 | |||
| 8 | |||
| 9 | |||
| 10 |
| Center of Gaussian | ||||
|---|---|---|---|---|
| f | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0.04 | 1.5 | 6 | 4 | 15 | 0.001 | 0.1 | 0.8 | 1 | 1 | 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Glimm, T.; Zhang, J. Numerical Approach to a Nonlocal Advection-Reaction-Diffusion Model of Cartilage Pattern Formation. Math. Comput. Appl. 2020, 25, 36. https://doi.org/10.3390/mca25020036
Glimm T, Zhang J. Numerical Approach to a Nonlocal Advection-Reaction-Diffusion Model of Cartilage Pattern Formation. Mathematical and Computational Applications. 2020; 25(2):36. https://doi.org/10.3390/mca25020036
Chicago/Turabian StyleGlimm, Tilmann, and Jianying Zhang. 2020. "Numerical Approach to a Nonlocal Advection-Reaction-Diffusion Model of Cartilage Pattern Formation" Mathematical and Computational Applications 25, no. 2: 36. https://doi.org/10.3390/mca25020036
APA StyleGlimm, T., & Zhang, J. (2020). Numerical Approach to a Nonlocal Advection-Reaction-Diffusion Model of Cartilage Pattern Formation. Mathematical and Computational Applications, 25(2), 36. https://doi.org/10.3390/mca25020036





