Translating Features to Findings: Deep Learning for Melanoma Subtype Prediction
Abstract
1. Introduction
Search Strategy and Scope
2. Background: Melanoma Subtypes and Diagnostic Complexities
2.1. Clinical and Histologic Diversity of Melanoma Subtypes
2.2. Diagnostic Challenges and the Need for Decision Support
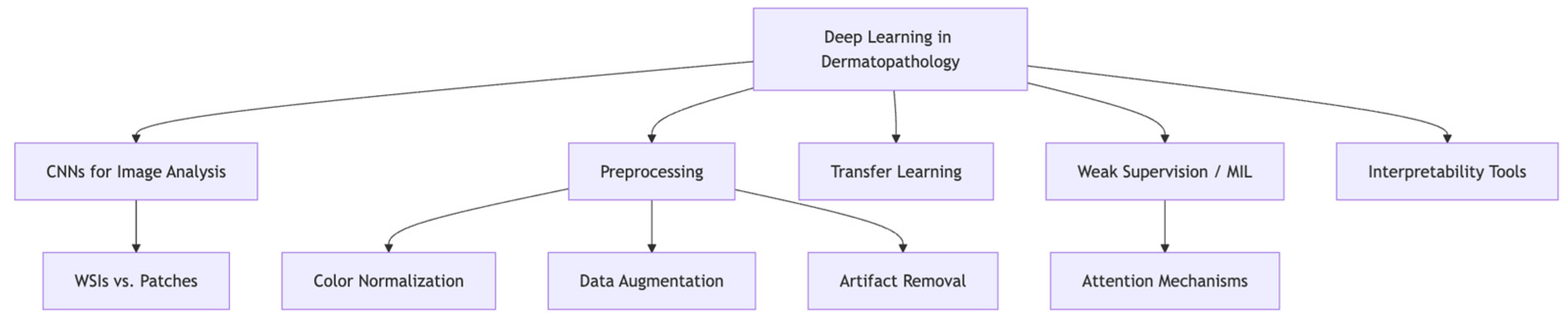
3. Deep Learning Foundations in Dermatopathology
3.1. CNNs, Whole-Slide Images, and the Role of Preprocessing
3.2. Transfer Learning, Weak Supervision, and Model Interpretability
4. Applications of Deep Learning to Melanoma Subtyping
4.1. Overview of Approaches and Input Modalities
4.2. Subtype-Specific Performance and Limitations
4.3. Generalizability and Technical Barriers
5. Limitations and Challenges
5.1. Dataset Imbalance and Lack of Representation
5.2. Model Explainability and Clinical Trust
5.3. Technical and Operational Barriers to Clinical Integration
6. Future Directions and Research Opportunities
6.1. Multimodal Modeling and Personalized Predictions
6.2. Federated Learning and Data Privacy
6.3. Explainable AI and Clinician Confidence
6.4. Clinical Validation and Standardization
7. Discussion
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bertolotto, C. Melanoma: From melanocyte to genetic alterations and clinical options. Scientifica 2013, 2013, 635203. [Google Scholar] [CrossRef]
- Dunn, C.; Brettle, D.; Hodgson, C.; Hughes, R.; Treanor, D. An international study of stain variability in histopathology using qualitative and quantitative analysis. J. Pathol. Inform. 2025, 17, 100423. [Google Scholar] [CrossRef]
- Mienye, I.D.; Swart, T.G.; Obaido, G.; Jordan, M.; Ilono, P. Deep Convolutional Neural Networks in Medical Image Analysis: A Review. Information 2025, 16, 195. [Google Scholar] [CrossRef]
- Hekler, A.; Utikal, J.S.; Enk, A.H.; Solass, W.; Schmitt, M.; Klode, J.; Schadendorf, D.; Sondermann, W.; Franklin, C.; Bestvater, F.; et al. Deep learning outperformed 11 pathologists in the classification of histopathological melanoma images. Eur. J. Cancer 2019, 118, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Naseri, H.; Safaei, A.A. Diagnosis and prognosis of melanoma from dermoscopy images using machine learning and deep learning: A systematic literature review. BMC Cancer 2025, 25, 75. [Google Scholar] [CrossRef] [PubMed]
- Swetter, S.M.; Kashani-Sabet, M.; Johannet, P.; Reddy, S.A.; Phillips, T.L. 67—Melanoma. In Leibel and Phillips Textbook of Radiation Oncology, 3rd ed.; Hoppe, R.T., Phillips, T.L., Roach, M., Eds.; W.B. Saunders: Philadelphia, PA, USA, 2010; pp. 1459–1472. [Google Scholar]
- Liu, V.; Mihm, M.C. Pathology of malignant melanoma. Surg. Clin. N. Am. 2003, 83, 31–60. [Google Scholar] [CrossRef]
- Susok, L.; Gambichler, T. Caucasians with acral lentiginous melanoma have the same outcome as patients with stage- and limb-matched superficial spreading melanoma. J. Cancer Res. Clin. Oncol. 2022, 148, 497–502. [Google Scholar] [CrossRef]
- Cohen, L.M. Lentigo maligna and lentigo maligna melanoma. J. Am. Acad. Dermatol. 1995, 33, 923–936. [Google Scholar] [CrossRef]
- Chopra, A.; Sharma, R.; Rao, U.N.M. Pathology of Melanoma. Surg. Clin. N. Am. 2020, 100, 43–59. [Google Scholar] [CrossRef]
- Hosler, G.A.; Goldberg, M.S.; Estrada, S.I.; O’Neil, B.; Amin, S.M.; Plaza, J.A. Diagnostic discordance among histopathological reviewers of melanocytic lesions. J. Cutan. Pathol. 2024, 51, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Taylor, L.A.; Eguchi, M.M.; Reisch, L.M.; Radick, A.C.; Shucard, H.; Kerr, K.F.; Piepkorn, M.W.; Knezevich, S.R.; Elder, D.E.; Barnhill, R.L.; et al. Histopathologic synoptic reporting of invasive melanoma: How reliable are the data? Cancer 2021, 127, 3125–3136. [Google Scholar] [CrossRef]
- Li, Z.; Cong, Y.; Chen, X.; Qi, J.; Sun, J.; Yan, T.; Yang, H.; Liu, J.; Lu, E.; Wang, L.; et al. Vision transformer-based weakly supervised histopathological image analysis of primary brain tumors. iScience 2023, 26, 105872. [Google Scholar] [CrossRef]
- Cassalia, F.; Danese, A.; Cocchi, E.; Danese, E.; Ambrogio, F.; Cazzato, G.; Mazza, M.; Zambello, A.; Belloni Fortina, A.; Melandri, D. Misdiagnosis and Clinical Insights into Acral Amelanotic Melanoma—A Systematic Review. J. Pers. Med. 2024, 14, 518. [Google Scholar] [CrossRef]
- Lepakshi, V.A. Machine Learning and Deep Learning based AI Tools for Development of Diagnostic Tools. In Computational Approaches for Novel Therapeutic and Diagnostic Designing to Mitigate SARS-CoV2 Infection; Elsevier: Amsterdam, The Netherlands, 2022. [Google Scholar]
- Requa, J.; Godard, T.; Mandal, R.; Balzer, B.; Whittemore, D.; George, E.; Barcelona, F.; Lambert, C.; Lee, J.; Lambert, A.; et al. High-fidelity detection, subtyping, and localization of five skin neoplasms using supervised and semi-supervised learning. J. Pathol. Inf. 2023, 14, 100159. [Google Scholar] [CrossRef] [PubMed]
- Shahamatdar, S.; Saeed-Vafa, D.; Linsley, D.; Khalil, F.; Lovinger, K.; Li, L.; McLeod, H.T.; Ramachandran, S.; Serre, T. Deceptive learning in histopathology. Histopathology 2024, 85, 116–132. [Google Scholar] [CrossRef] [PubMed]
- Sauter, D.; Lodde, G.; Nensa, F.; Schadendorf, D.; Livingstone, E.; Kukuk, M. A Systematic Comparison of Task Adaptation Techniques for Digital Histopathology. Bioengineering 2023, 11, 19. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Rezapour, M.; Sajjad, U.; Gurcan, M.N.; Niazi, M.K.K. Attention2Minority: A salient instance inference-based multiple instance learning for classifying small lesions in whole slide images. Comput. Biol. Med. 2023, 167, 107607. [Google Scholar] [CrossRef]
- Sadeghi, Z.; Alizadehsani, R.; Cifci, M.A.; Kausar, S.; Rehman, R.; Mahanta, P.; Bora, P.K.; Almasri, A.; Alkhawaldeh, R.S.; Hussain, S.; et al. A review of Explainable Artificial Intelligence in healthcare. Comput. Electr. Eng. 2024, 118, 109370. [Google Scholar] [CrossRef]
- Li, S.; Li, T.; Sun, C.; Yan, R.; Chen, X. Multilayer Grad-CAM: An effective tool towards explainable deep neural networks for intelligent fault diagnosis. J. Manuf. Syst. 2023, 69, 20–30. [Google Scholar] [CrossRef]
- Tehrani, K.F.; Park, J.; Chaney, E.J.; Tu, H.; Boppart, S.A. Nonlinear Imaging Histopathology: A Pipeline to Correlate Gold-Standard Hematoxylin and Eosin Staining With Modern Nonlinear Microscopy. IEEE J. Sel. Top. Quantum Electron. 2023, 29, 6800608. [Google Scholar] [CrossRef]
- Raza, M.; Awan, R.; Bashir, R.M.S.; Qaiser, T.; Rajpoot, N.M. Dual attention model with reinforcement learning for classification of histology whole-slide images. Comput. Med. Imaging Graph. 2024, 118, 102466. [Google Scholar] [CrossRef]
- Greenwald, H.S.; Friedman, E.B.; Osman, I. Superficial spreading and nodular melanoma are distinct biological entities: A challenge to the linear progression model. Melanoma Res. 2012, 22, 1–8. [Google Scholar] [CrossRef]
- Kulkarni, P.M.; Robinson, E.J.; Sarin Pradhan, J.; Gartrell-Corrado, R.D.; Rohr, B.R.; Trager, M.H.; Geskin, L.J.; Kluger, H.M.; Wong, P.F.; Acs, B.; et al. Deep Learning Based on Standard H&E Images of Primary Melanoma Tumors Identifies Patients at Risk for Visceral Recurrence and Death. Clin. Cancer Res. 2020, 26, 1126–1134. [Google Scholar] [CrossRef]
- Phillips, A.; Teo, I.; Lang, J. Segmentation of Prognostic Tissue Structures in Cutaneous Melanoma Using Whole Slide Images. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW 2019), Long Beach, CA, USA, 16–17 June 2019; Institute of Electrical and Electronics Engineers (IEEE): New York, NY, USA, 2019; pp. 2738–2747. [Google Scholar]
- Stacke, K.; Eilertsen, G.; Unger, J.; Lundstrom, C. Measuring Domain Shift for Deep Learning in Histopathology. IEEE J. Biomed. Health Inform. 2020, 25, 325–336. [Google Scholar] [CrossRef]
- Dolezal, J.; Kochanny, S.; Dyer, E.; Ramesh, S.; Srisuwananukorn, A.; Sacco, M.; Howard, F.; Li, A.; Mohan, P.; Pearson, A. Slideflow: Deep learning for digital histopathology with real-time whole-slide visualization. BMC Bioinform. 2024, 25, 134. [Google Scholar] [CrossRef]
- Li, H.; Rajbahadur, G.K.; Bezemer, C.-P. Studying the Impact of TensorFlow and PyTorch Bindings on Machine Learning Software Quality. ACM Trans. Softw. Eng. Methodol. 2024, 34, 20. [Google Scholar] [CrossRef]
- Comes, M.C.; Fucci, L.; Mele, F.; Bove, S.; Cristofaro, C.; De Risi, I.; Fanizzi, A.; Milella, M.; Strippoli, S.; Zito, A.; et al. A deep learning model based on whole slide images to predict disease-free survival in cutaneous melanoma patients. Sci. Rep. 2022, 12, 20366. [Google Scholar] [CrossRef]
- Druskovich, C.; Kelley, J.; Aubrey, J.; Palladino, L.; Wright, G.P. A Review of Melanoma Subtypes: Genetic and Treatment Considerations. J. Surg. Oncol. 2025, 131, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Cross, J.L.; Choma, M.A.; Onofrey, J.A. Bias in medical AI: Implications for clinical decision-making. PLOS Digit. Health 2024, 3, e0000651. [Google Scholar] [CrossRef] [PubMed]
- Pachetti, E.; Colantonio, S. A systematic review of few-shot learning in medical imaging. Artif. Intell. Med. 2024, 156, 102949. [Google Scholar] [CrossRef] [PubMed]
- Lazarou, E.; Exarchos, T.P. Predicting stress levels using physiological data: Real-time stress prediction models utilizing wearable devices. AIMS Neurosci. 2024, 11, 76–102. [Google Scholar] [CrossRef]
- van der Velden, B.H.M.; Kuijf, H.J.; Gilhuijs, K.G.A.; Viergever, M.A. Explainable artificial intelligence (XAI) in deep learning-based medical image analysis. Med. Image Anal. 2022, 79, 102470. [Google Scholar] [CrossRef]
- Bakoglu, N.; Cesmecioglu, E.; Sakamoto, H.; Yoshida, M.; Ohnishi, T.; Lee, S.-Y.; Smith, L.; Yagi, Y. Artificial intelligence-based automated determination in breast and colon cancer and distinction between atypical and typical mitosis using a cloud-based platform. Pathol. Oncol. Res. POR 2024, 30, 1611815. [Google Scholar] [CrossRef]
- d’Amati, A.; Baldini, G.M.; Difonzo, T.; Santoro, A.; Dellino, M.; Cazzato, G.; Malvasi, A.; Vimercati, A.; Resta, L.; Zannoni, G.F.; et al. Artificial Intelligence in Placental Pathology: New Diagnostic Imaging Tools in Evolution and in Perspective. J. Imaging 2025, 11, 110. [Google Scholar] [CrossRef] [PubMed]
- Yanzhen, M.; Song, C.; Wanping, L.; Zufang, Y.; Wang, A. Exploring approaches to tackle cross-domain challenges in brain medical image segmentation: A systematic review. Front. Neurosci. 2024, 18, 1401329. [Google Scholar] [CrossRef]
- Mennella, C.; Maniscalco, U.; De Pietro, G.; Esposito, M. Ethical and regulatory challenges of AI technologies in healthcare: A narrative review. Heliyon 2024, 10, e26297. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.; Collie, C.J. What’s new in dermatopathology 2023: WHO 5th edition updates. J. Pathol. Transl. Med. 2023, 57, 337–340. [Google Scholar] [CrossRef]
- Höhn, J.; Krieghoff-Henning, E.; Jutzi, T.B.; von Kalle, C.; Utikal, J.S.; Meier, F.; Gellrich, F.F.; Hobelsberger, S.; Hauschild, A.; Schlager, J.G.; et al. Combining CNN-based histologic whole slide image analysis and patient data to improve skin cancer classification. Eur. J. Cancer 2021, 149, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Wu, J. Molecular and immune landscape of melanoma: A risk stratification model for precision oncology. Discov. Oncol. 2025, 16, 667. [Google Scholar] [CrossRef]
- Zhang, B.; Wan, Z.; Luo, Y.; Zhao, X.; Samayoa, J.; Zhao, W.; Wu, S. Multimodal integration strategies for clinical application in oncology. Front. Pharmacol. 2025, 16, 1609079. [Google Scholar] [CrossRef]
- Xie, J.; Luo, X.; Deng, X.; Tang, Y.; Tian, W.; Cheng, H.; Zhang, J.; Zou, Y.; Guo, Z.; Xie, X. Advances in artificial intelligence to predict cancer immunotherapy efficacy. Front. Immunol. 2022, 13, 1076883. [Google Scholar] [CrossRef] [PubMed]
- Pawłowski, M.; Wróblewska, A.; Sysko-Romańczuk, S. Effective Techniques for Multimodal Data Fusion: A Comparative Analysis. Sensors 2023, 23, 2381. [Google Scholar] [CrossRef] [PubMed]
- Brussee, S.; Buzzanca, G.; Schrader, A.M.R.; Kers, J. Graph neural networks in histopathology: Emerging trends and future directions. Med. Image Anal. 2025, 101, 103444. [Google Scholar] [CrossRef]
- Cassidy, B.; Kendrick, C.; Brodzicki, A.; Jaworek-Korjakowska, J.; Yap, M.H. Analysis of the ISIC image datasets: Usage, benchmarks and recommendations. Med. Image Anal. 2022, 75, 102305. [Google Scholar] [CrossRef]
- Dzieniszewska, A.; Garbat, P.; Pietkiewicz, P.; Piramidowicz, R. Early-Stage Melanoma Benchmark Dataset. Cancers 2025, 17, 2476. [Google Scholar] [CrossRef]
- Phaphuangwittayakul, A.; Guo, Y.; Ying, F. Fast Adaptive Meta-Learning for Few-Shot Image Generation. IEEE Trans. Multimed. 2022, 24, 2205–2217. [Google Scholar] [CrossRef]
- Madathil, N.T.; Dankar, F.K.; Gergely, M.; Belkacem, A.N.; Alrabaee, S. Revolutionizing healthcare data analytics with federated learning: A comprehensive survey of applications, systems, and future directions. Comput. Struct. Biotechnol. J. 2025, 28, 217–238. [Google Scholar] [CrossRef]
- Riaz, S.; Naeem, A.; Malik, H.; Naqvi, R.A.; Loh, W.K. Federated and Transfer Learning Methods for the Classification of Melanoma and Nonmelanoma Skin Cancers: A Prospective Study. Sensors 2023, 23, 8457. [Google Scholar] [CrossRef]
- Dembani, R.; Karvelas, I.; Akbar, N.A.; Rizou, S.; Tegolo, D.; Fountas, S. Agricultural data privacy and federated learning: A review of challenges and opportunities. Comput. Electron. Agric. 2025, 232, 110048. [Google Scholar] [CrossRef]
- Mukhtiar, N.; Mahmood, A.; Sheng, Q.Z. Fairness in Federated Learning: Trends, Challenges, and Opportunities. Adv. Intell. Syst. 2025, 7, 2400836. [Google Scholar] [CrossRef]
- Leventer, I.; Card, K.R.; Shields, C.L. Fitzpatrick skin type and relationship to ocular melanoma. Clin. Dermatol. 2025, 43, 56–63. [Google Scholar] [CrossRef]
- Asadi-Aghbolaghi, M.; Darbandsari, A.; Zhang, A.; Contreras-Sanz, A.; Boschman, J.; Ahmadvand, P.; Köbel, M.; Farnell, D.; Huntsman, D.G.; Churg, A.; et al. Learning generalizable AI models for multi-center histopathology image classification. npj Precis. Oncol. 2024, 8, 151. [Google Scholar] [CrossRef]
- Chen, F.; Wang, L.; Hong, J.; Jiang, J.; Zhou, L. Unmasking bias in artificial intelligence: A systematic review of bias detection and mitigation strategies in electronic health record-based models. arXiv 2024, arXiv:2310.19917. [Google Scholar] [CrossRef] [PubMed]
- Wil-Trenchard, K. Melanoma 2010 Congress. Pigment Cell Melanoma Res. 2010, 23, 874–1004. [Google Scholar] [CrossRef]
- Calvert, M.; King, M.; Mercieca-Bebber, R.; Aiyegbusi, O.; Kyte, D.; Slade, A.; Chan, A.W.; Basch, E.; Bell, J.; Bennett, A.; et al. SPIRIT-PRO Extension explanation and elaboration: Guidelines for inclusion of patient-reported outcomes in protocols of clinical trials. BMJ Open 2021, 11, e045105. [Google Scholar] [CrossRef]
- Carvalho, M.; Pinho, A.J.; Brás, S. Resampling approaches to handle class imbalance: A review from a data perspective. J. Big Data 2025, 12, 71. [Google Scholar] [CrossRef]
- Alipour, N.; Burke, T.; Courtney, J. Skin Type Diversity in Skin Lesion Datasets: A Review. Curr. Dermatol. Rep. 2024, 13, 198–210. [Google Scholar] [CrossRef]
- Naved, B.A.; Luo, Y. Contrasting rule and machine learning based digital self triage systems in the USA. NPJ Digit. Med. 2024, 7, 381. [Google Scholar] [CrossRef]
- Griffiths, P.; Saville, C.; Ball, J.; Jones, J.; Pattison, N.; Monks, T. Nursing workload, nurse staffing methodologies and tools: A systematic scoping review and discussion. Int. J. Nurs. Stud. 2020, 103, 103487. [Google Scholar] [CrossRef]
- Salehi, F.; Salin, E.; Smarr, B.; Bayat, S.; Kleyer, A.; Schett, G.; Fritsch-Stork, R.; Eskofier, B.M. A robust machine learning approach to predicting remission and stratifying risk in rheumatoid arthritis patients treated with bDMARDs. Sci. Rep. 2025, 15, 23960. [Google Scholar] [CrossRef]
- Vickers, A.J.; Holland, F. Decision curve analysis to evaluate the clinical benefit of prediction models. Spine J. 2021, 21, 1643–1648. [Google Scholar] [CrossRef]
- Kiran, A.; Narayanasamy, N.; Ramesh, J.V.N.; Ahmad, M.W. A novel deep learning framework for accurate melanoma diagnosis integrating imaging and genomic data for improved patient outcomes. Ski. Res. Technol. 2024, 30, e13770. [Google Scholar] [CrossRef]
- Lalmalani, R.M.; Lim, C.X.Y.; Oh, C.C. Artificial intelligence in dermatopathology: A systematic review. Clin. Exp. Dermatol. 2025, 50, 251–259. [Google Scholar] [CrossRef]
- Chen, L.L.; Jaimes, N.; Barker, C.A.; Busam, K.J.; Marghoob, A.A. Desmoplastic melanoma: A review. J. Am. Acad. Dermatol. 2013, 68, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Davri, A.; Birbas, E.; Kanavos, T.; Ntritsos, G.; Giannakeas, N.; Tzallas, A.T.; Batistatou, A. Deep Learning for Lung Cancer Diagnosis, Prognosis and Prediction Using Histological and Cytological Images: A Systematic Review. Cancers 2023, 15, 3981. [Google Scholar] [CrossRef]
- Witkowski, A.M.; Burshtein, J.; Christopher, M.; Cockerell, C.; Correa, L.; Cotter, D.; Ellis, D.L.; Farberg, A.S.; Grant-Kels, J.M.; Greiling, T.M.; et al. Clinical Utility of a Digital Dermoscopy Image-Based Artificial Intelligence Device in the Diagnosis and Management of Skin Cancer by Dermatologists. Cancers 2024, 16, 3529. [Google Scholar] [CrossRef]
- Mevorach, L.; Farcomeni, A.; Pellacani, G.; Cantisani, C. A Comparison of Skin Lesions’ Diagnoses Between AI-Based Image Classification, an Expert Dermatologist, and a Non-Expert. Diagnostics 2025, 15, 1115. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, X.; Tang, Q.; Bi, Y.; Yuan, L.; Yang, B.; Cai, M.; Zhang, J.; Deng, D.; Cao, W. The correlation between dermoscopy and clinical and pathological tests in the evaluation of skin photoaging. Ski. Res. Technol. 2024, 30, e13578. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, C.; Peng, L.; Lin, K.; Xue, Y.; Chen, X.; Bao, L.; Liu, C.; Zhang, J.; Xie, Y. Radiomic and deep learning analysis of dermoscopic images for skin lesion pattern decoding. Sci. Rep. 2024, 14, 19781. [Google Scholar] [CrossRef] [PubMed]
- Kuziemsky, C.E.; Chrimes, D.; Minshall, S.; Mannerow, M.; Lau, F. AI Quality Standards in Health Care: Rapid Umbrella Review. J. Med. Internet Res. 2024, 26, e54705. [Google Scholar] [CrossRef] [PubMed]
| Subtype | Key Features | Common Sites | Challenges |
|---|---|---|---|
| SSM | Radial growth, pagetoid spread | Trunk, limbs | Common, but overlaps possible |
| NM | Vertical growth, rapid progression | Anywhere | Aggressive, sometimes missed early |
| ALM | Acral sites, lentiginous growth | Palms, soles, nail beds | Often diagnosed late; rare |
| LMM | Chronic sun exposure, slow-growing | Face, scalp | Long in situ phase; can become invasive |
| DM | Fibrous stroma, bland histology | Sun-exposed or scar-like areas | Mimics scarring; difficult to identify |
| Study (Year) | Subtypes Covered/Task | Dataset | N (Patients/WSIs) | External Validation | Key Limitations |
|---|---|---|---|---|---|
| Requa et al., 2023, J Pathol Inform [16] | Detection of 5 lesion types with subtyping and localization (melanocytic lesions are subtyped as benign/mildly/severely atypical; BCC and ASL also subtyped). | Single organization (PathologyWatch); multiple PW labs; private | Supervised subset: 2188 WSIs; Weakly supervised subset: 5161 WSIs; Validation: 250 WSIs (+50 mimickers); Testing: 3821 WSIs (daily cases). | No true external institutional cohort; testing was non-curated PW daily cases. | Industry dataset; internal test only; subtype targets differ from WHO melanoma subtypes (focus on severity/phenotype classes); label noise risk in SSL. |
| Su et al., 2023, Comp Biol Med (Attention2Minority/SiiMIL) [19] | MIL method targeting small/rare lesion instances—relevant to under-represented melanoma subtypes (e.g., ALM/DM) in principle. | Method paper; datasets vary; largely private. | NR | Usually no melanoma-specific subtype benchmark; focuses on algorithmic advance; needs melanoma-task validation. | |
| Raza et al., 2024, CMIG (Dual-attention + RL for WSI) [23] | Method for slide- and tile-level attention; applicable to histology classification (not melanoma-specific). | Varies by experiment; typically private; indication toward IHC WSIs in examples. | NR | Often no external medical-domain validation reported; not melanoma-specific; demonstrates technique rather than a melanoma study. | |
| Kulkarni et al., 2020, Clin Cancer Res [25] | Prognostic modeling from H&E (risk of visceral recurrence/death). Not a subtype classifier per se but informs subtype-aware pipelines. | Multi-institution (4 sites); private | Train: 108 pts (4 institutions). External val: Yale 104 pts. Independent test: Geisinger 51 pts. | Yes—Yale (AUC 0.905) and Geisinger (AUC 0.880); KM p < 0.0001. | Prognosis (not subtype labels); slide-level labels; no paired genomics; generalizability beyond studied centers not shown. |
| Phillips et al., 2019, CVPR Workshops (ISIC) [26] | Segmentation of prognostic tissue structures (epidermis/dermis/tumor) in cutaneous melanoma to enable Breslow measurement (enabling feature for subtype-aware/clinical models). | Multi-institution source slides (from TCGA; 9 institutions noted), public images (TCGA) with curated annotations. | 49 pts/50 WSIs, 40× scans; split 36/7/7 (train/val/test). | No (single curated dataset split). | Segmentation (not classification); small test set; TCGA quality variability; no direct subtype labels. |
| Focus Area | Strategies and Technologies | Goals/Expected Outcomes |
|---|---|---|
| Multimodal Modeling | Combine histology with genomics (e.g., BRAF, NRAS) Integrate clinical metadata (e.g., age, location, treatment) Use non-invasive imaging (dermoscopy, confocal) Apply GANs and diffusion models for synthetic data Leverage few-shot/zero-shot learning for rare subtypes | Enhance diagnostic accuracy Enable precision dermatopathology Address data imbalance and rare subtype scarcity |
| Federated Learning and Privacy | Train models across institutions without sharing raw data Use differential privacy techniques | Preserve patient confidentiality Improve generalizability Promote collaboration across centers |
| Explainable AI (XAI) | Concept bottleneck models Counterfactual explanations Attention-based networks | Increase transparency and interpretability Build pathologist trust Support diagnostic quality control |
| Clinical Validation and Standards | Conduct prospective clinical trials Evaluate integration into diagnostic workflows Standardize datasets and benchmarks Follow models such as TCGA and ISIC | Demonstrate real-world utility Secure regulatory approval Promote reproducibility and equity |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Published by MDPI on behalf of the European Society of Dermatopathology. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guermazi, D.; Khemchandani, S.; Wahood, S.; Nguyen, C.; Saliba, E. Translating Features to Findings: Deep Learning for Melanoma Subtype Prediction. Dermatopathology 2025, 12, 42. https://doi.org/10.3390/dermatopathology12040042
Guermazi D, Khemchandani S, Wahood S, Nguyen C, Saliba E. Translating Features to Findings: Deep Learning for Melanoma Subtype Prediction. Dermatopathology. 2025; 12(4):42. https://doi.org/10.3390/dermatopathology12040042
Chicago/Turabian StyleGuermazi, Dorra, Sarina Khemchandani, Samer Wahood, Cuong Nguyen, and Elie Saliba. 2025. "Translating Features to Findings: Deep Learning for Melanoma Subtype Prediction" Dermatopathology 12, no. 4: 42. https://doi.org/10.3390/dermatopathology12040042
APA StyleGuermazi, D., Khemchandani, S., Wahood, S., Nguyen, C., & Saliba, E. (2025). Translating Features to Findings: Deep Learning for Melanoma Subtype Prediction. Dermatopathology, 12(4), 42. https://doi.org/10.3390/dermatopathology12040042






