Abstract
Background: Enterococcus spp. belongs to a group of pathogens which are responsible for serious infections. This study aims at highlighting the raw milk microbiological contamination and at providing data for prevalence and antimicrobial resistance of Enterococcus spp. isolated from raw cow's milk marketed (without any pasteurization) by street traders. Methods: During the period of May 2015 through April 2016, 150 cow’s raw milk samples were collected from street traders in Meknes city. They were examined for the identification of Enterococcus spp. using biochemical tests and 16S rRNA gene sequencing. The antimicrobial susceptibility of the isolates was determined. Results: The results showed that 11.3% (17/150) of samples were positive for the presence of Enterococcus spp., of which 64.7% were identified as Enterococcus faecalis, 17.6% as Enterococcus faecium, 11.8% as Enterococcus durans and 5.9% as Enterococcus hirae. The antimicrobial susceptibility showed that all Enterococcus spp. were resistant to ampicillin. The species E. faecalis, E. faecium, E. durans and E. hirae were resistant to streptomycin, with percentages of 52.9% (9/17), 11.8% (2/17), 11.8% (2/17), and 5.9% (1/17) respectively. All isolated strains of E. faecalis and E. faecium were resistant to tetracycline. The multiple antibiotic resistance index was elevated in the majority of Enterococcus spp., reaching values higher than 0.5, indicating a risk for public health. Conclusion: This study shows that the raw milk consumed by the population is contaminated with strains of Enterococcus resistant to antibiotics used in breeding for prophylactic purposes. This requires raising the awareness of those involved in the production and marketing of milk, so as to take measures to apply good hygienic practices and rationalize the use of zootechnical antibiotics.
Introduction
Bacteria belonging to Enterococcus spp. are Gram-positive, catalase negative, non-spore forming, and facultatively anaerobic. They optimally grow at 35 °C, but tolerate temperatures between 10-45 °C [1]. They are commensals of the gastrointestinal tract of animals and humans. They are responsible for serious infections, such as surgical infection, aortic valve endocarditis, meningitis and bacteremia [2]. While Enterococcus faecalis and Enterococcus faecium are most commonly recovered from humans, Enterococcus hirae is more frequently identified in animal infections [3]. Enterococcus durans is found in the intestines of animals and is responsible for very rare cases of endocarditis in humans [4].
In Morocco, the consumption level of milk and dairy has considerably increased in recent years [5]. However, milk and milk products delivery from the farm to consumer involves two types of marketing channels. The first one is referred to as “organized” and involves industrial collection centers with production plants. The second is known as “street trading” and is based on the sale of milk by the persons who collect it from the farms and sell it to traditional dairies (Mahlabates), coffee houses and urban households, using motorcycles as transportation means. The informally marketed milk can sometimes reach an amount of 30% of the total milk consumed in towns and in cities [6]. The milk marketed by street traders confronts several constraints such as contamination by utensils or by personnel involved in its supply chain and the absence of a cold storage chain. Also, street traders work in illegal conditions without control and without regulatory legislation and are unaware of good hygienic practices. It should be mentioned that the milk marketed by street traders is generally consumed directly by consumers.
The aim of this study was to evaluate the microbiological quality of raw milk and its contamination with Enterococcus spp. in order to determine the infectious risks associated to its consumption. The originality of this work lies in the particularity that this study was performed in the city of Meknes and the demonstration of the different species of Enterococcus spp. identified as well as their profile of resistance to antibiotics.
Methods
Samples collections
The choice of sampling sites is based on the geographical location, with significant population density, and poor hygienic conditions. These sites were in a traditional market (souk) and overcrowded popular neighborhoods (site 1 and site 2 respectively) located in the city of Meknes, Morocco.
During the period of May 2015 through April 2016, a total of 150 raw cow’s milk samples were randomly collected from different street traders with twelve samples per month. Each milk sample consisted 1 L of raw milk collected aseptically into a sterile container. Samples were brought to the Microbiology Laboratory at the Faculty of Science in Meknes, in cool boxes and kept at 4 °C. The analyses were performed the same day.
Isolation and identification of Enterococcus spp.
A quantity of 25 g of each milk sample was aseptically taken, placed in 225 mL of buffered peptone water (Oxoid, Cheshire, UK), and then homogenized through a Stomacher 400 Circulator (Seward, West Sussex, UK) for 3 min at 260 rpm. From this primary enrichment, 1 mL was streaked to Slanetz and Bartely Agar (Oxoid) and incubated for 24±2 h at 37±1 °C. Enterococcus spp. suspected colonies that had a maximum diameter of 1 mm, a dark red and a narrow white border were transferred to Tryptone Soya Agar (Biokar, Beauvais, France) and incubated for 24±2 h at 37±1 °C. Isolates were identified to be Enterococcus spp. using the following biochemical analyses: Gram staining, mobility, pigmentation, hydrolysis of esculin in the presence of bile, absence of catalase and growth in 6.5% NaCl [6].
PCR amplification and sequencing of bacterial 16S rRNA
Seventeen isolates showed the same phenotypical characteristics for Enterococcus spp. as described by Castillo et al. in 2013 [6]. To confirm this result, the DNA was extracted from each strain using the Isolate II Genomic DNA kit (Bioline, Taunton, MA, USA), following the manufacturer instructions. The quality of the DNA was controlled by a 1% agarose gel; and its quantification was determined by the Nanodrop 8000 (ThermoFisher Scientific, Waltham, MA, USA). The 16S rRNA for each strain was amplified using FD1 (5′- AGAGTTTGATCCTGGCTCAG-3′) and RP2 (5′-ACGGCTACCTTGTTACGACTT-3′) [7]. PCR was performed in 25 µL reaction mixture containing 15.3 µL of ddH2O, 2.5 µL of buffer (10×) (KAPA Biosystems, Wilmington, MA, USA), 0.75 µL of 50 mM MgCl2, 2 µL of template DNA (50 ng/µL-1), 0.45 µL of Taq DNA polymerase (5 U/µL KAPA Biosystems), 0.5 µL of 10 Mm dNTPs mix (KAPA Biosystems) and 1.75 µL of 50 pmol from each primer.
The PCR temperature profile (Verity Machin) comprises an initial denaturation at 95 °C for 4 min, 35 cycles of denaturation at 95 °C for 30 s, annealing at 50 °C for 1 min and extension at 72 °C for 1 min, with a final extension at 72 °C for 7 min. The PCR products were separated by gel electrophoresis using 1.5 % (w/v) agarose (FMC, Products, Mississauga, Canada) in 0.5 TBE buffer at 100V; the gel was stained with ethidium bromide and visualized under UV light. All PCR products were sequenced by the platform of molecular biology and functional genomic in the National Center for Scientific and Technical Research (CNRST) of Rabat city (Morocco), using Analyzer ABI-3130 XL (ThermoFisher Scientific). The sequences were trimmed, analyzed through the National Center of Biotechnology Information (NCBI) server using Blast N. The sequences were deposited in GenBank under accession numbers: MF185235-MF185251.
Antimicrobial susceptibility testing
Enterococcus spp. isolates were tested for susceptibility to 14 antimicrobial compounds, which were selected on the basis of their use against infections and for prophylactic purposes. The disk diffusion method described by the Clinical and Laboratory Standards Institute was used [8]. The antimicrobial disks examined in this study are presented in Table 1. The measured inhibition zones were interpreted as sensitive, intermediate and resistant according to the CLSI guidelines. Enterococcus faecalis 2921 was used as a reference strain.

Table 1.
Antimicrobial agents and the range of concentrations tested.
Phylogenetic analysis
The phylogenetic and molecular evolutionary analyses were conducted using MEGA 7 of our sequences and the references Enterococcus spp. obtained from the NCBI databases. The 16S rRNA sequences were first aligned using the Clustal W [9]. The evolutionary trees for the dataset were inferred using the neighbor joining method [9,10]. The stability of the relationships was assessed by 1000 bootstrap replications. Escherichia coli ATCC8739 (NC_010468.1) was used as outgroup.
Statistical analysis
Data were analyzed to assess the existence of statistical differences between the parameters evaluated by season and site regarding the prevalence and antimicrobial resistances of Enterococcus spp. The study of the effect of season and site of sampling was made by ANOVA with repeated measures using software (SPSS version 20, IBM Corp, Armonk, NY, USA). Differences were considered statistically significant at p <0.05.
Results
Isolation and identification of Enterococcus spp.
The prevalence of Enterococcus spp. isolated from 150 raw cow’s milk samples traded in Meknes city was calculated from May 2015 until April 2016. Enterococcus spp. were found in 17 (11.3%) of the samples. This prevalence was significantly different by season (p=0.001) and it was not significant among the sites sampled (p=0.281). The highest prevalence of Enterococcus spp. was recorded during the summer (p=0.013) and the lowest in winter (p=0.273). Indeed, 64% of contamination was observed in summer and 5.8% in winter. The most frequently identified species of Enterococcus were E. faecalis 11 (64.7%), E. faecium 3 (17.6%), E. durans 2 (11.8%) and E. hirae 1 (5.9%).
Antimicrobial susceptibility testing
The susceptibility of the isolates to different antibiotics showed that 100% of Enterococcus spp. isolates (17/17) were resistant to at least one antimicrobial, 82.3%(14/17) were resistant to two or more antimicrobials and 17.6% (3/17) were resistant to three or more antimicrobials (Table 2).

Table 2.
Antimicrobial resistance profiles of Enterococcus spp. isolated from raw cow’s milk.
Furthermore, all Enterococcus spp. were resistant to ampicillin. E. faecalis, E. faecium, E. durans and E. hirae were resistant to streptomycin, with percentages of 52.9 % (9/17), 11.8% (2/17), 11.8% (2/17), and 5.9% (1/17) respectively. All E. faecalis and E. faecium isolates were resistant to tetracycline. A high percentage of resistance to antibiotics was recorded during summer, as follows: ampicillin (75.0%), streptomycin (50.0%) and tetracycline (50.0%), while during winter this value was lower for ampicillin (8.3%), streptomycin (16.6%) and tetracycline (12.9%). Moreover, this resistance was more pronounced in site 2 for streptomycin (77.8%), tetracycline (50.0%) and ampicillin (46.6%). All isolates were sensitive to vancomycin, nitrofurantoin and gentamicin. The resistance profile established for the different Enterococcus isolates showed 11 different strains of Enterococcus faecalis, 3 strains of Enterococcus faecium, 2 strains of Enterococcus durans and 1 strain of Enterococcus hirae (Table 2 and Table 3).

Table 3.
Antimicrobial resistance percentages of Enterococcus spp. isolated from raw cow’s milk.
Sequencing of the 16S rRNA genes
Recently, new species of Enterococcus have been discovered. In order to understand the phylogenetic relationships within the genus Enterococcus, we constructed a phylogenetic tree based on the 16S rRNA sequences. Results showed two branches, each with a high diversity of Enterococcus spp. isolated from raw cow’s milk samples. The results of the determination of the disparity degree between the studied isolates and their classification according to the phylogenetic tree are illustrated in Figure 1. The first group contains 11 strains of Enterococcus faecalis and 1 Enterococcus faecium. The second group includes 2 Enterococcus faecium, 1 Enterococcus hirae and 2 Enterococcus durans (Figure 1).
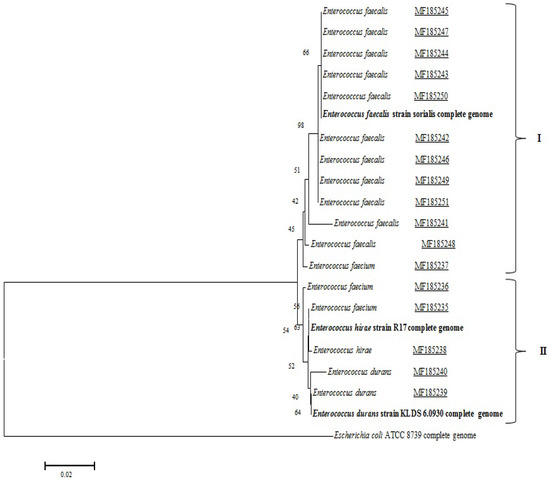
Figure 1.
Phylogenetic tree of 17 Enterococcus isolates based on 16S rRNA sequences.
Discussion
This study indicates that Enterococcus spp. were found in 11.3% of samples of raw milk. These rates are lower than those found in Austria (96%) [11], and than those found in France (8%) in fresh milk cheese [12]. A study carried out in Australia reported a high percentage of E. faecalis (74.3%) and a low percentage of E. hirae (8.5%), E. faecium (8.6%) and E. durans (2.9%) [12]. In Egypt, the overall prevalence of Enterococcus spp. identified in raw milk cheese is reported at 90% [13].
The presence of Enterococcus spp. in animal-derived food may be an indicator of contamination or poor hygienic conditions. In Morocco, the raw milk is mainly produced under conditions of food safety. However, the presence of Enterococcus spp. in food in general and particularly in raw milk consumed increases the risks to public health. Infants, the elderly and patients with immunodeficiency are the populations most vulnerable to poisoning [14]. In addition, antimicrobial resistance poses a major public health problem and can result in an increase of morbidity, disease burden and mortality of the population, especially when these isolated strains are multidrug-resistant. Various studies have shown that the Enterococcus spp. have become resistant to a wide range of antibiotics [15], and are widespread in dairy products, meat product and ready-to-eat foods [16].
In our study, the resistance rate was very high for ampicillin, streptomycin, tetracycline, erythromycin and ciprofloxacin and moderate for penicillin (Table 2 and Table 3). It is therefore important to report that these acquired resistances reflect the frequency and importance of antibiotics used in veterinary medicine and animal breeding [13]. Several antibiotics are banned for use in zootechny (for example vancomycin) [13], but are still used clandestinely and haphazardly.
Comparing these results with the literature, we find that they are higher than those found in Egypt for tetracycline (65%), chloramphenicol (1.6%) [13], penicillin (0%) [17] and lower than those found for streptomycin (78%), erythromycin (40%), ciprofloxacin (15%), gentamicin (55%), and vancomycin (5%) in raw milk cheese [13]. Other studies in Morocco have shown that the following Enterococcus spp. were also isolated from milk and dairy products: E. faecalis, E. faecium [18], E. durans [19], E. hirae [20], E. sulfureus, E. italicus and E. bulliens, which were identified in fresh camel milk [21]. Enterococcus spp. have been isolated all over the world, for example in China E. faecalis, E. faecium and E. hirae were isolated in fresh goat milk [22]. In Tunisia, Enterococcus faecium, Enterococcus faecalis, Enterococcus casseliflavus, E. durans and E. mundtii were isolated in fermented dairy products (Lben, Jben and Rayeb) [23], and in Italy E. italicus was found in raw milk [24].
The antibiotic resistance pattern of Enterococcus isolates in this study showed 11 different phenotypes for E. faecalis, 3 for E. faecium, 2 E. durans and 1 E. hirae (Figure 1). The ratio of antimicrobial resistance profile to total number of antibiotics (multiple antibiotic resistance – MAR – index) is high for both of the first species cited. The ratio varied between values of 0.2 and 0.6; more than 65% of the strains had a MAR index greater than or equal to 0.4 (Table 3), indicating a high risk of contamination for the consumer. This MAR index level is lower than that found in Brazil (from 0.25 to 0.87) [25]. For this reason, a set of measures and recommendations are needed to reinforce the application of good practices for the production of raw milk, at the primary collection stage. Awareness raising campaigns for hygiene measures are needed to improve the quality of raw milk and organize the activity of street vendors. A framework for tightening up the regulations governing the activity of traditional dairies as well as imposing traceability rules are essential to fight against this informal problem.
Conclusion
The results of this study can be considered as an alert to the dangers of contamination by multidrug-resistant enterococci isolated from consumed raw milk. Awareness of hygiene and the application of good production practices are necessary to control and prevent the resulting health and socio-economic damage.
Author Contributions
AB prepared the manuscript, conducted the PCR amplification and analyzed the 16S rRNA sequences. FRF edited the manuscript. SO analyzed the 16S rRNA sequences. AE conducted experiment. FB and MM approved the manuscript. KS and JA conducted the PCR amplification and read the manuscript. AEA conducted statistical analysis. BO read the manuscript. All authors read and approved the final version of the manuscript.
Funding
None to declare.
Acknowledgments
The authors would like to acknowledge the responsible of molecular biology and functional genomics platform (National Center for Scientific and Technical Research, Morocco), Dr. El Fahim M., for his great contribution, also the professor Ayoujil Said for reading this manuscript.
Conflicts of interest
All authors – none to disclose.
References
- Guzman Prieto, A.M.; van Schaik, W.; Rogers, M.R.; et al. Global emergence and dissemination of enterococci as nosocomial pathogens: attack of the clones. Front Microbiol 2016, 7, 788. [Google Scholar] [CrossRef]
- Hannaoui, I.; Barguigua, A.; Serray, B.; et al. Intestinal carriage of vancomycin-resistant enterococci in a community setting in Casablanca, Morocco. J Global Antimicrob Resist 2016, 6, 84–87. [Google Scholar] [CrossRef]
- Pasquaroli, S.; Di Cesare, A.; Vignaroli, C.; Conti, G.; Citterio, B.; Biavasco, F. Erythromycin- and copper-resistant Enterococcus hirae from marine sediment and co-transfer of erm(B) and tcrB to human Enterococcus faecalis. Diagn Microbiol Infect Dis 2014, 80, 26–28. [Google Scholar] [CrossRef]
- Rajakrishnan, V.; Alwyn, R. Fatal Enterococcus durans aortic valve endocarditis: a case report and review of the literature. BMJ Case Rep 2012, 2012, bcr0220125855. [Google Scholar] [CrossRef]
- Bertrand, H. International Center for Advanced Mediterranean Agronomic Studies (CIHEAM), 1st ed.; Mediterra: Paris, 2007. [Google Scholar]
- Castillo-Rojas, G.; Mazari-Hiríart, M.; Ponce de León, S.; et al. Comparison of Enterococcus faecium and Enterococcus faecalis strains isolated from water and clinical samples: antimicrobial susceptibility and genetic relationships. PLoS One 2013, 8, e59491. [Google Scholar] [CrossRef] [PubMed]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing; twenty-fifth informational supplement. CLSI document M100-S25; Clinical and Laboratory Standards Institute: Wayne, PA, 2015. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011, 28, 2731–2739. [Google Scholar] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987, 4, 406–425. [Google Scholar]
- McAuley, C.M.; Britz, M.L.; Gobius, K.S.; Craven, H.M. Prevalence, seasonality, and growth of enterococci in raw and pasteurized milk in Victoria, Australia. J Dairy Sci 2015, 98, 8348–8358. [Google Scholar] [CrossRef]
- Jamet, E.; Akary, E.; Poisson, M.A.; Chamba, J.F.; Bertrand, X.; Serror, P. Prevalence and characterization of antibiotic resistant Enterococcus faecalis in French cheeses. Food Microbiol 2012, 31, 191–198. [Google Scholar] [CrossRef]
- Hammad, A.M.; Hamdy, A.H.; Tadashi, S. Prevalence, antibiotic resistance and virulence of Enterococcus spp. in Egyptian fresh raw milk cheese. Food Control 2015, 50, 815–820. [Google Scholar] [CrossRef]
- World Health Organization. WHO Estimates of the Global Burden of Foodborne Diseases; WHO: Geneva, 2015. [Google Scholar]
- Iweriebor, B.C.; Gaqavu, S.; Obi, L.C.; Nwodo, U.U.; Okoh, A.I. Antibiotic susceptibilities of Enterococcus species isolated from hospital and domestic wastewater effluents in Alice, Eastern Cape Province of South Africa. Int J Environ Res Public Health 2015, 12, 4231–4246. [Google Scholar] [CrossRef] [PubMed]
- Sanders, P.; Perrin-Guyomard, A.; Moulin, G. Évolution de l’utilisation des antibiotiques en production animale. Cah Nutr Diet 2017, 52, 301–11. [Google Scholar] [CrossRef]
- Boussouar, N.; Boumediene, M.B. Antibiotic resistance of enterococci isolated from raw camel milk in the South West of Algeria. Afr J Microbiol Res 2016, 10, 420–427. [Google Scholar] [CrossRef]
- Valenzuela, A.S.; Omar, N.B.; Abriouel, H.; et al. Risk factors in enterococci isolated from foods in Morocco: determination of antimicrobial resistance and incidence of virulence traits. Food Chem Toxicol 2008, 46, 2648–2652. [Google Scholar] [CrossRef]
- Jamaly, N.; Benjouad, A.; Comunian, R.; Daga, E.; Bouksaim, M. Characterization of enterococci isolated from Moroccan dairy products. Afr J Microbiol Res 2010, 4, 1768–1774. [Google Scholar]
- Achemchem, F.; Cebrián, R.; Abrini, J.; Martínez-Bueno, M.; Valdivia, E.; Maqueda, M. Antimicrobial characterization and safety aspects of the bacteriocinogenic Enterococcus hirae F420 isolated from Moroccan raw goat milk. Can J Microbiol 2012, 58, 596–604. [Google Scholar] [CrossRef]
- Kadri, Z.; Spitaels, F.; Cnockaert, M.; et al. Enterococcus bulliens sp. nov., a novel lactic acid bacterium isolated from camel milk. Antonie Van Leeuwenhoek 2015, 108, 1257–1265. [Google Scholar] [CrossRef]
- Zhang, F.; Zhang, W.; Lei, F.; et al. Predominant species of lactic acid bacteria in fresh goat milk from the Guanzhong area of China. SF Food Dairy Tech J 2017, 1, 1–8. [Google Scholar]
- Amel, R.; Imène, F.; Amine, F.S.; Ilhem, B.B.; Abdellatif, B.C.; Hadda-Imène, O. Prevalence, acquired antibiotic resistance and bacteriocin production of Enterococcus spp. isolated from tunisian fermented food products. Food Control 2016, 63, 259–266. [Google Scholar] [CrossRef]
- Fornasari, M.F.; Rossetti, L.; Remagni, C.; Giraffa, G. Quantification of Enterococcus italicus in traditional Italian cheeses by fluorescence whole-cell hybridization. Syst App Microbiol 2008, 31, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, E.M.R.; Costa, R.A.; Araújo, A.J.G.; et al. Multiple antibiotic-resistance of Enterococcus isolated from coastal water near an outfall in Brazil. Afr J Microbiol Res 2014, 8, 1825–1831. [Google Scholar]
© GERMS 2018.