Antimicrobial Resistance and Putative Virulence Genes of Pseudomonas aeruginosa Isolates from Patients With Respiratory Tract Infection
Abstract
Introduction
Methods
Patients
Culture, isolation and bacterial identification
Antimicrobial susceptibility
Plasmid DNA extraction and PCR procedures
Identification of P. aeruginosa by PCR
Detection of virulence genes by PCR
Detection of genes of ESBLs, MBLs and blaKPC
Gel electrophoresis
RAPD-PCR to detect genotypic variations among P. aeruginosa isolates
Statistical analysis
Results
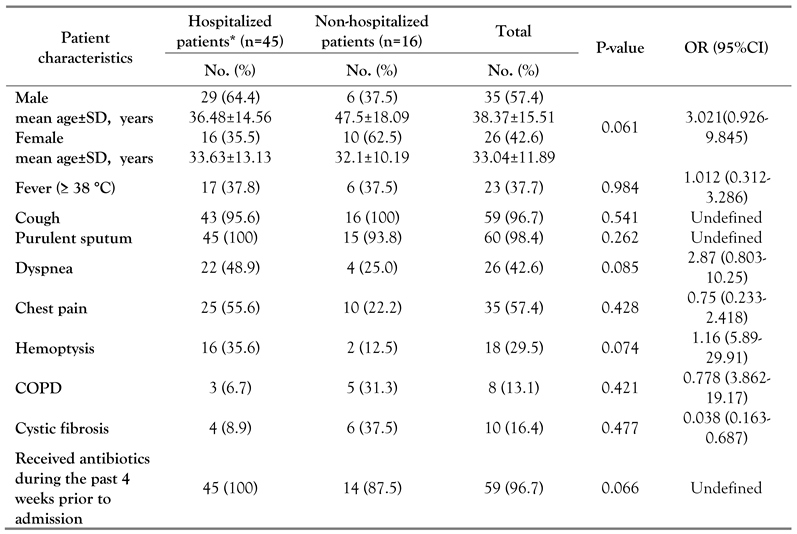
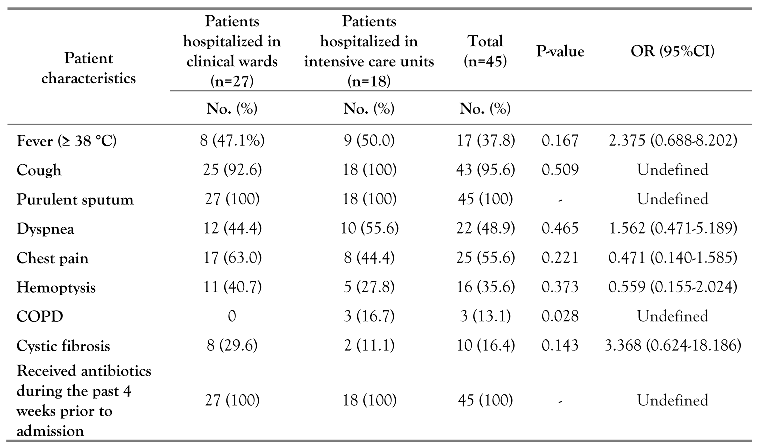
Patients
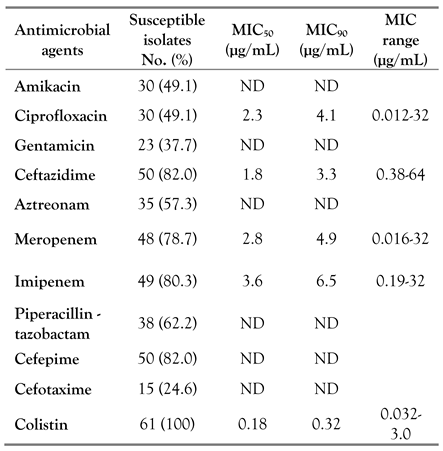
Occurrence of P. aeruginosa and antimicrobial susceptibility profiles
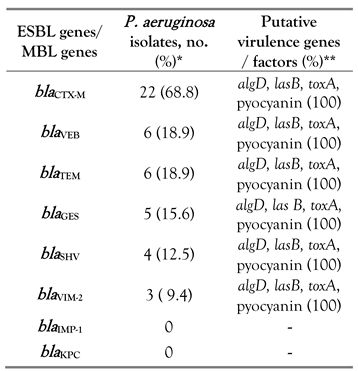
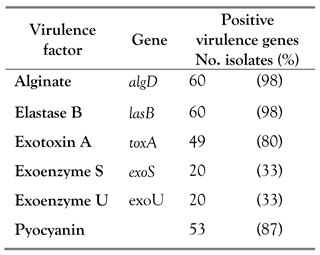
MDR and putative virulence genes
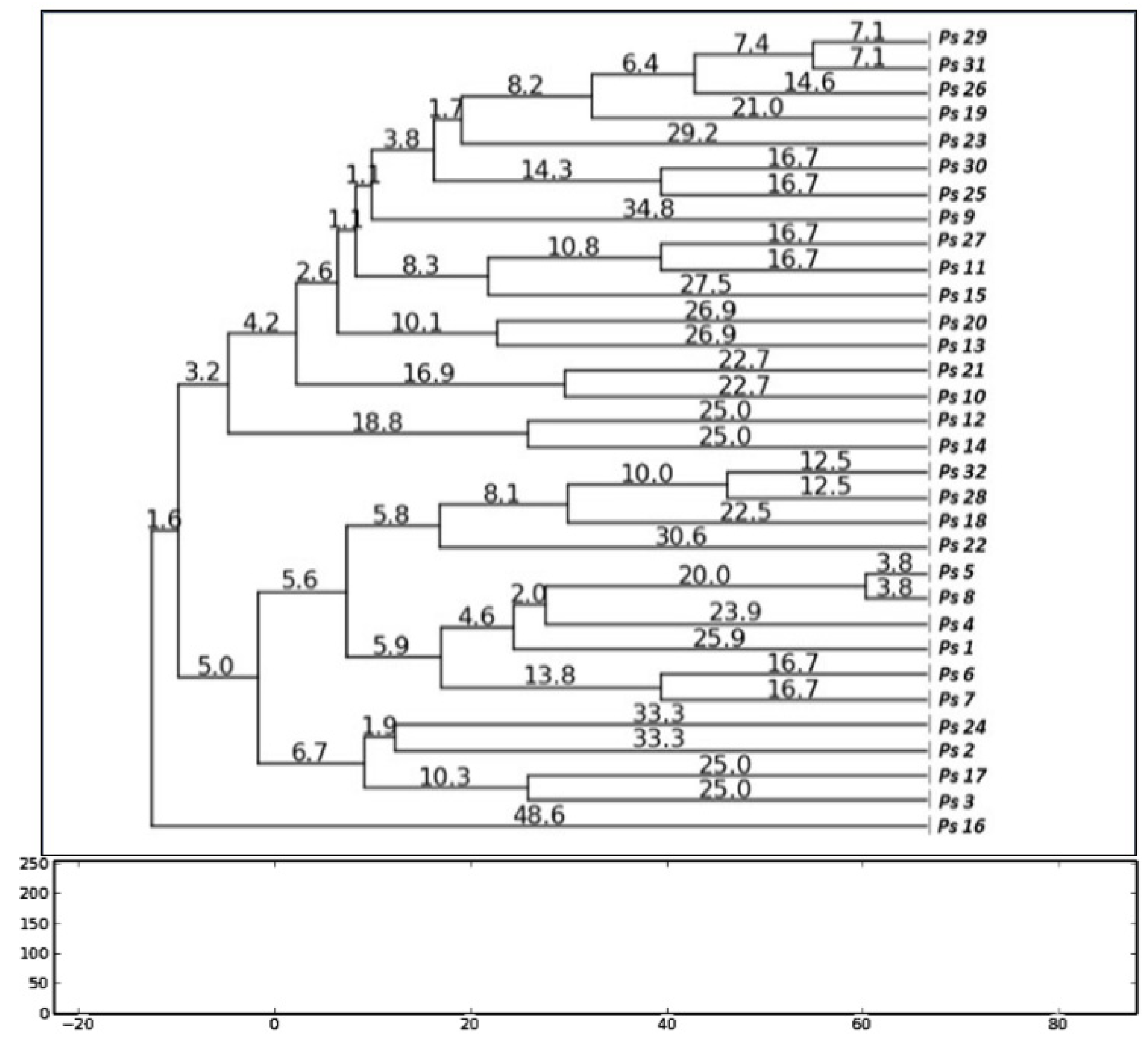
Genotyping
Discussion
Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Nathwani, D.; Raman, G.; Sulham, K.; Gavaghan, M.; Menon, V. Clinical and economic consequences of hospital-acquired resistant and multidrug-resistant Pseudomonas aeruginosa infections: a systematic review and meta-analysis. Antimicrob Resist Infect Control 2014, 3, 32. [Google Scholar] [CrossRef]
- Mitov, I.; Strateva, T.; Markova, B. Prevalence of virulence genes among Bulgarian nosocomial and cystic fibrosis isolates of Pseudomonas aeruginosa. Braz J Microbiol 2010, 41, 588–595. [Google Scholar] [CrossRef]
- Lanotte, P.; Watt, S.; Mereghetti, L.; et al. Genetic features of Pseudomonas aeruginosa isolates from cystic fibrosis patients compared with those of isolates from other origins. J Med Microbiol 2004, 53, 73–81. [Google Scholar] [CrossRef]
- Nikbin, V.S.; Aslani, M.M.; Sharafi, Z.; Hashemipour, M.; Shahcheraghi, F.; Ebrahimipour, G.H. Molecular identification and detection of virulence genes among Pseudomonas aeruginosa isolated from different infectious origins. Iran J Microbiol 2012, 4, 118–123. [Google Scholar]
- Michalska, M.; Wolf, P. Pseudomonas exotoxin A: optimized by evolution for effective killing. Front Microbiol 2015, 6, 963. [Google Scholar] [CrossRef]
- Shaver, C.M.; Hauser, A.R. Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infect Immun 2004, 72, 6969–6977. [Google Scholar] [CrossRef] [PubMed]
- Farshadzadeh, Z.; Khosravi, A.D.; Alavi, S.M.; Parhizgari, N.; Hoveizavi, H. Spread of extended-spectrum β-lactamase genes of blaOXA-10, blaPER-1 and blaCTX-M in Pseudomonas aeruginosa strains isolated from burn patients. Burns 2014, 40, 1575–1580. [Google Scholar] [CrossRef]
- Poirel, L.; Nordmann, P.; Lagrutta, E.; Cleary, T.; Munoz-Price, L.S. Emergence of KPC-producing Pseudomonas aeruginosa in the United States. Antimicrob Agents Chemother 2010, 54, 3072. [Google Scholar] [CrossRef] [PubMed]
- Mansour, S.A.; Eldaly, O.; Jiman-Fatani, A.; Mohamed, M.L.; Ibrahim, E.M. Epidemiological characterization of P. aeruginosa isolates of intensive care units in Egypt and Saudi Arabia. East Mediterr Health J 2013, 19, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Hamze, M.; Mallat, H.; Dabboussi, F.; Achkar, M. Antibiotic susceptibility and serotyping of clinical Pseudomonas aeruginosa isolates in northern Lebanon. Int Arabic J Antimicrob Agents 2012, 2, 1–6. [Google Scholar]
- Clinical Laboratory and Standards Institute (CLSI). Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard – tenth edition. CLSI document M07-A10 Villanova, PA, USA: CLSI. 2015. [Google Scholar]
- Spilker, T.; Coenye, T.; Vandamme, P.; LiPuma, J.J. PCR-based assay for differentiation of Pseudomonas aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. J Clin Microbiol 2004, 42, 2074–2079. [Google Scholar] [CrossRef]
- Bokaeian, M.; Shahraki Zahedani, S.; Soltanian Bajgiran, M.; Ansari Moghaddam, A. Frequency of PER, VEB, SHV, TEM and CTX-M genes in resistant strains of Pseudomonas aeruginosa producing extended spectrum β-lactamases. Jundishapur J Microbiol 2014, 8, e13783. [Google Scholar] [CrossRef]
- Pitout, J.D.D.; Gregson, D.B.; Poirel, L.; McClure, J.A.; Le, P.; Church, D.L. Detection of Pseudomonas aeruginosa producing metallo-β-lactamases in a large centralized laboratory. J Clin Microbiol 2005, 43, 3129–3135. [Google Scholar] [CrossRef]
- Jiang, X.; Zhang, Z.; Li, M.; Zhou, D.; Ruan, F.; Lu, Y. Detection of extended-spectrum β-lactamases in clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother 2006, 50, 2990–2995. [Google Scholar] [CrossRef] [PubMed]
- Fazeli, N.; Momtaz, H. Virulence gene profiles of multidrug resistant Pseudomonas aeruginosa isolated from Iranian hospital infections. Iran Red Crescent Med J 2014, 16, e15722. [Google Scholar] [CrossRef]
- Monteiro, J.; Widen, R.H.; Pignatari, A.C.; Kubasek, C.; Silbert, S. Rapid detection of carbapenemase genes by multiplex real-time PCR. J Antimicrob Chemother 2012, 67, 906–909. [Google Scholar] [CrossRef]
- Akpaka, P.E.; Swanston, W.H.; Ihemere, H.N.; et al. Emergence of KPC-producing Pseudomonas aeruginosa in Trinidad and Tobago. J Clin Microbiol 2009, 47, 2670–2671. [Google Scholar] [CrossRef] [PubMed]
- Wolska, K.; Szweda, P. Genetic features of clinical Pseudomonas aeruginosa strains. Pol J Microbiol 2009, 58, 255–260. [Google Scholar] [PubMed]
- Mitov, I.; Strateva, T.; Markova, B. Prevalence of virulence genes among Bulgarian nosocomial and cystic fibrosis isolates of Pseudomonas aeruginosa. Braz J Microbiol 2010, 41, 588–595. [Google Scholar] [CrossRef]
- Nanvazadeh, F.; Khosravi, A.D.; Zolfaghari, M.R.; Parhizgari, N. Genotyping of Pseudomonas aeruginosa strains isolated from burn patients by RAPD-PCR. Burns 2013, 39, 1409–1413. [Google Scholar] [CrossRef]
- Fatima, A.; Naqvi, S.B.; Khaliq, S.A.; Perveen, S.; Jabeen, S. Antimicrobial susceptibility pattern of clinical isolates of Pseudomonas aeruginosa isolated from patients of lower respiratory tract infections. Springerplus 2012, 1, 70. [Google Scholar] [CrossRef]
- Al-Muhairi, S.; Zoubeidi, T.; Ellis, M.; Nicholls, M.G.; Safa, W.; Joseph, J. Demographics and microbiological profile of pneumonia in United Arab Emirates. Monaldi Arch Chest Dis 2006, 65, 13–18. [Google Scholar] [CrossRef]
- Vincent, J.L.; Rello, J.; Marshall, J.; et al. International study of the prevalence and outcomes of infection in intensive care units. JAMA 2009, 302, 2323–2329. [Google Scholar] [CrossRef] [PubMed]
- Al-Hajje, A.; Ezedine, M.; Hammoud, H.; et al. [Current status of nosocomial infections in the Lebanese Hospital Center, Beirut]. East Mediterr Health J 2012, 18, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Wassef, M.; El Mahallawy, H.; Zafer, M.M.; Ghaith, D.; Abdel Hamid, R. Lab based surveillance of multidrug resistant Pseudomonas aeruginosa in Cairo University Hospitals, Egypt. J Microbiol Exp 2015, 2, 00039. [Google Scholar]
- Masaadeh, H.A.; Jaran, A.S. Incident of Pseudomonas aeruginosa in post-operative wound infection. Am J Infect Dis 2009, 5, 1–6. [Google Scholar] [CrossRef]
- Voor In ‘t Holt, A.F.; Severin, J.A.; Lesaffre, E.M.; Vos, M.C. A systematic review and meta-analyses show that carbapenem use and medical devices are the leading risk factors for carbapenem-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother 2014, 58, 2626–2637. [Google Scholar] [CrossRef]
- Zhao, W.H.; Hu, Z.Q. Acquired metallo-β-lactamases and their genetic association with class 1 integrons and ISCR elements in Gram-negative bacteria. Future Microbiol 2015, 10, 873–887. [Google Scholar] [CrossRef] [PubMed]
- Obeidat, N.; Jawdat, F.; Al-Bakri, G.A.; Shehabi, A.A. Major biologic characteristics of Acinetobacter baumannii isolates from hospital environmental and patients’ respiratory tract sources. Am J Infect Control 2014, 42, 401–404. [Google Scholar] [CrossRef]
- Moreau-Marquis, S.; Stanton, B.A.; O’Toole, G.A. Pseudomonas aeruginosa biofilm formation in the cystic fibrosis airway. Pulm Pharmacol Ther 2008, 21, 595–599. [Google Scholar] [CrossRef]
- Schulert, G.S.; Feltman, H.; Rabin, S.D.; Martin, C.G.; Battle, S.E.; Rello, J.; Hauser, A.R. Secretion of the toxin ExoU is a marker for highly virulent Pseudomonas aeruginosa isolates obtained from patients with hospital-acquired pneumonia. J Infect Dis 2003, 188, 1695–1706. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, C.C.; Chen, Y.; Goetzmann, H.S.; et al. Pseudomonas aeruginosa exotoxin pyocyanin causes cystic fibrosis airway pathogenesis. Am J Pathol 2009, 175, 2473–2488. [Google Scholar] [CrossRef] [PubMed]
 |
 |
 |
 |
 |
 |
© GERMS 2017.
Share and Cite
Al Dawodeyah, H.Y.; Obeidat, N.; Abu-Qatouseh, L.F.; Shehabi, A.A. Antimicrobial Resistance and Putative Virulence Genes of Pseudomonas aeruginosa Isolates from Patients With Respiratory Tract Infection. Germs 2018, 8, 31-40. https://doi.org/10.18683/germs.2018.1130
Al Dawodeyah HY, Obeidat N, Abu-Qatouseh LF, Shehabi AA. Antimicrobial Resistance and Putative Virulence Genes of Pseudomonas aeruginosa Isolates from Patients With Respiratory Tract Infection. Germs. 2018; 8(1):31-40. https://doi.org/10.18683/germs.2018.1130
Chicago/Turabian StyleAl Dawodeyah, Heba Y., Nathir Obeidat, Luay F. Abu-Qatouseh, and Asem A. Shehabi. 2018. "Antimicrobial Resistance and Putative Virulence Genes of Pseudomonas aeruginosa Isolates from Patients With Respiratory Tract Infection" Germs 8, no. 1: 31-40. https://doi.org/10.18683/germs.2018.1130
APA StyleAl Dawodeyah, H. Y., Obeidat, N., Abu-Qatouseh, L. F., & Shehabi, A. A. (2018). Antimicrobial Resistance and Putative Virulence Genes of Pseudomonas aeruginosa Isolates from Patients With Respiratory Tract Infection. Germs, 8(1), 31-40. https://doi.org/10.18683/germs.2018.1130




