Molecular Characterization of Antimicrobial Resistance in Clinical Shigella Isolates During 2014 and 2015: Trends in South India
Abstract
Introduction
Methods
Bacterial strains
Antimicrobial susceptibility testing
Molecular analysis of antibiotic resistance genes
Statistical analysis
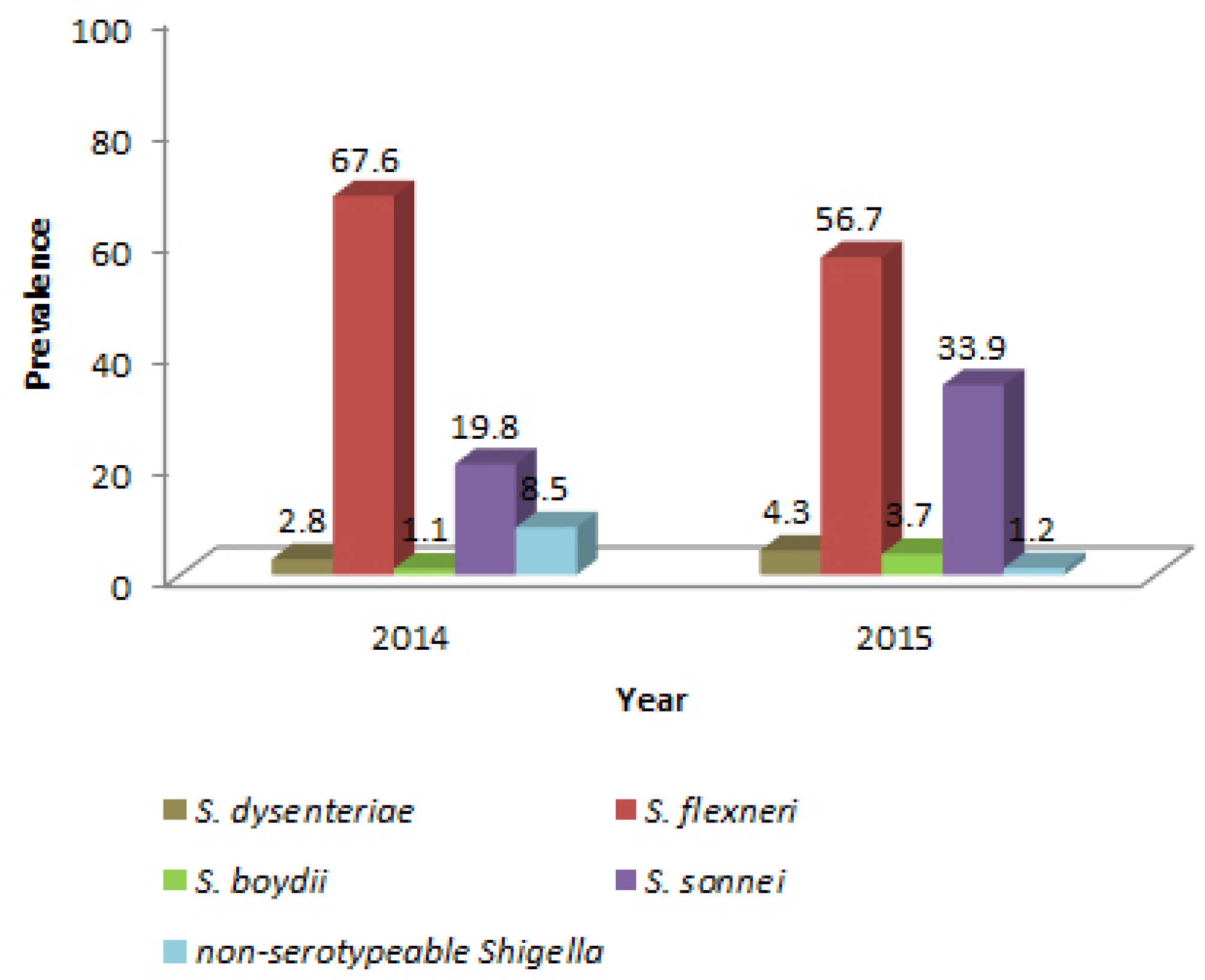
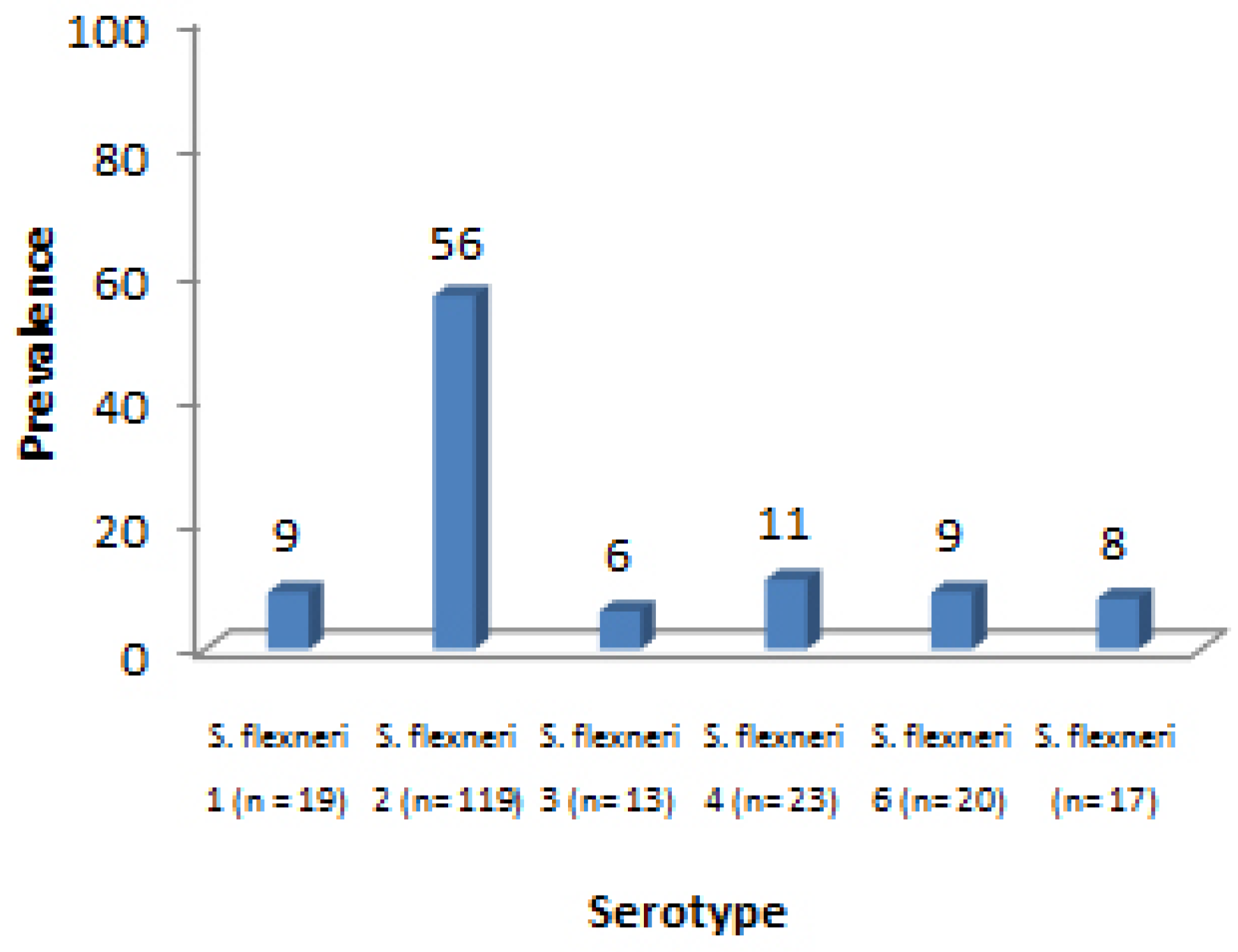
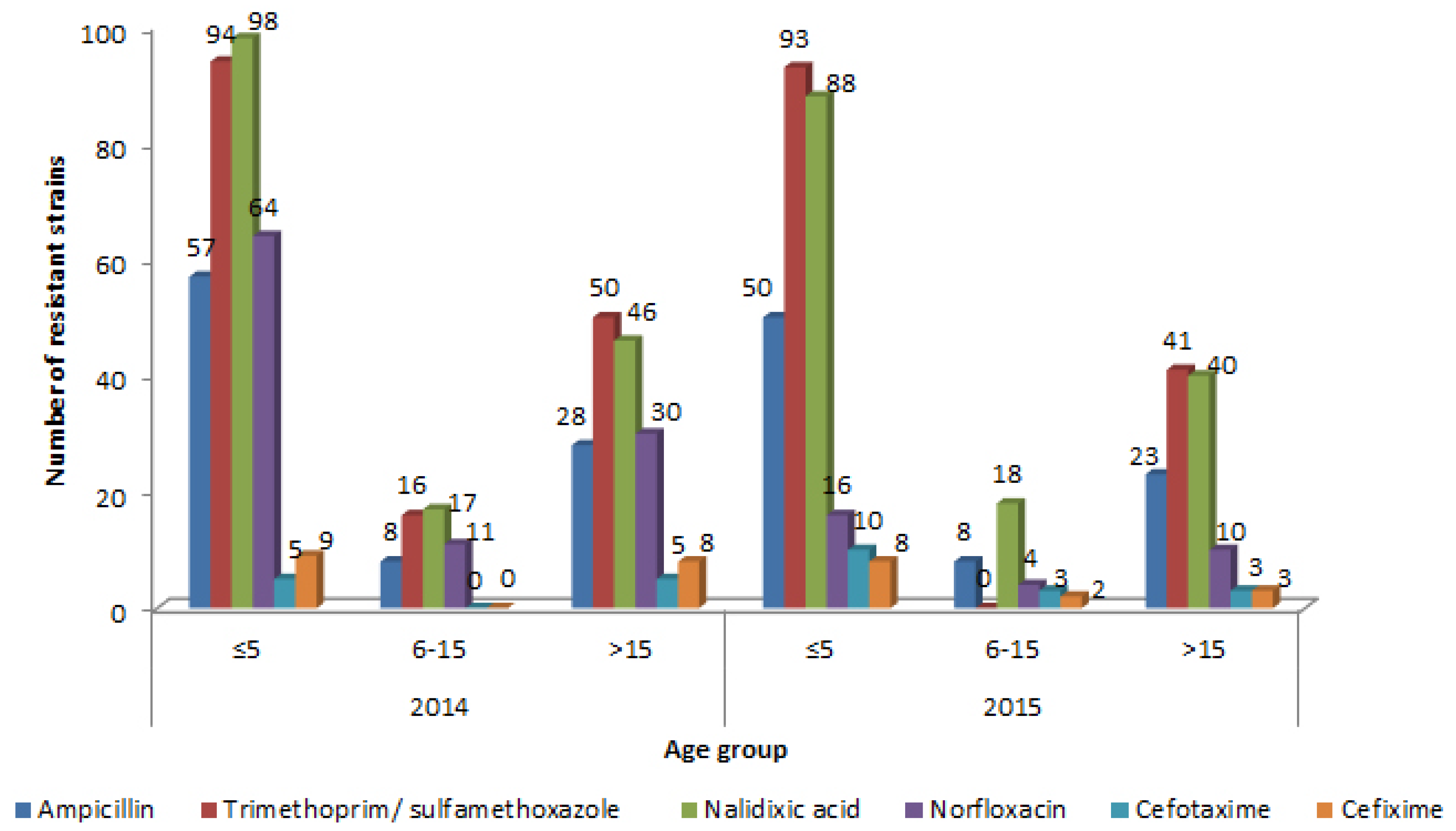
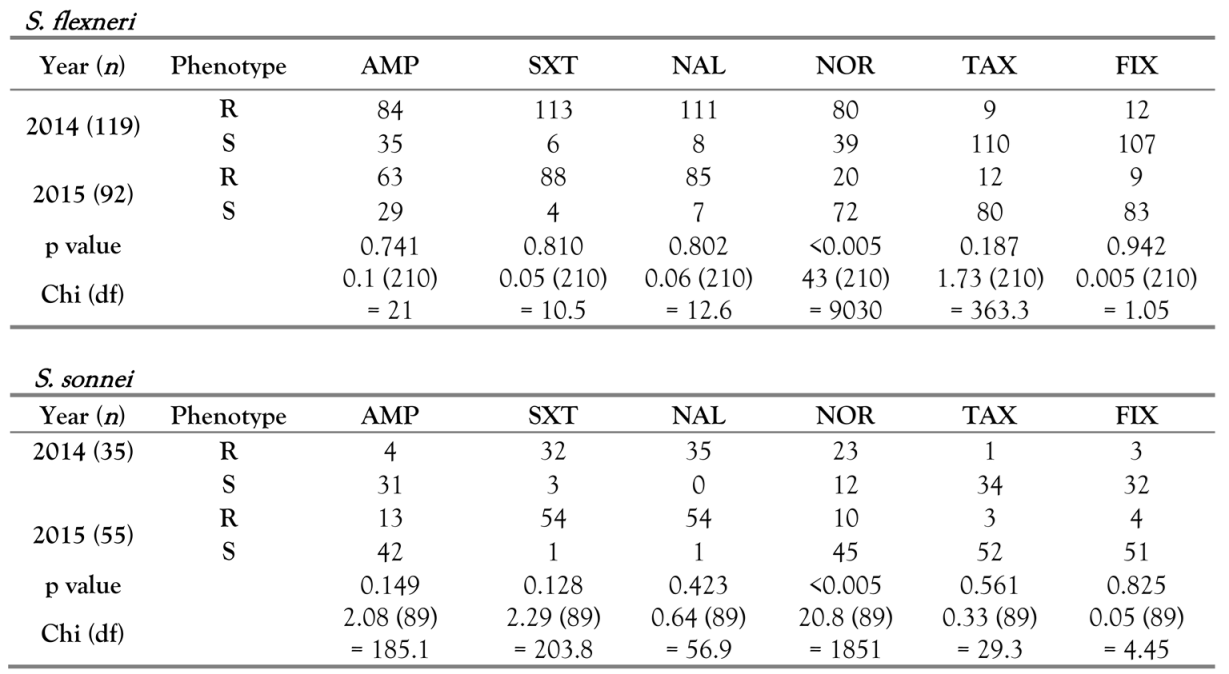
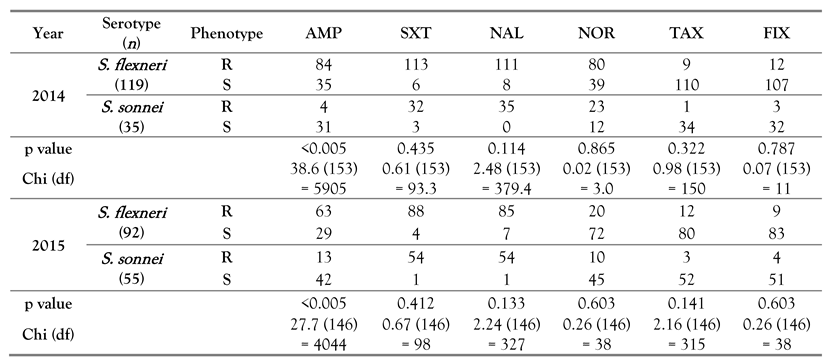
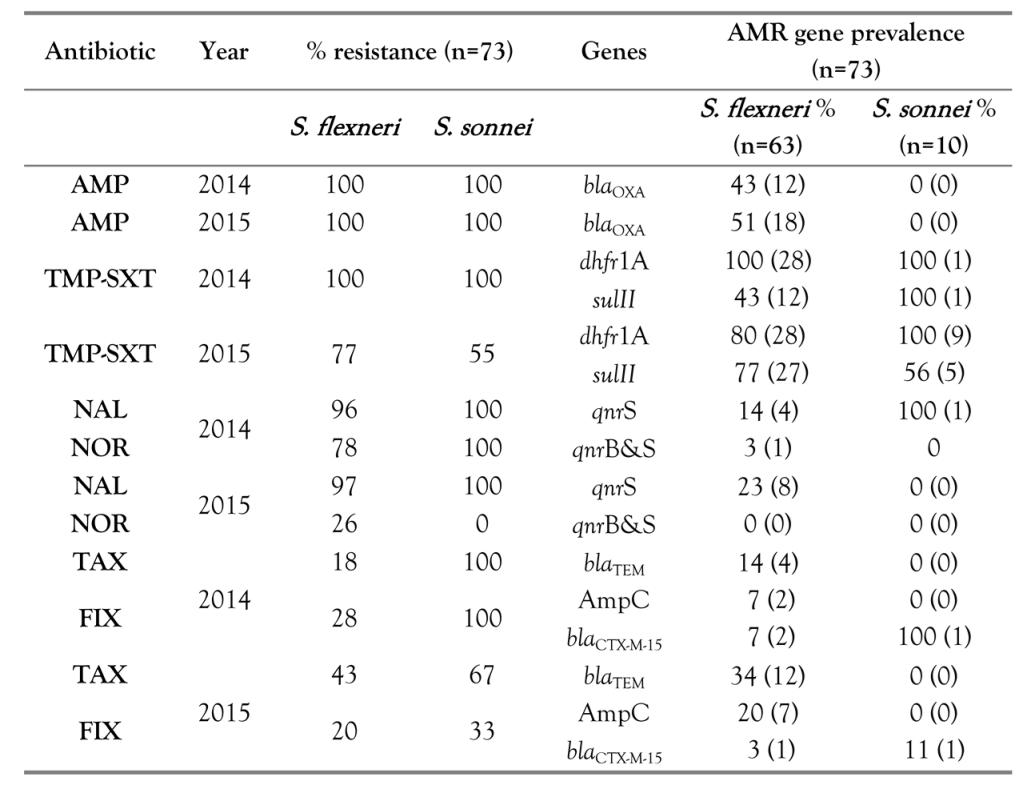
Results
Discussion
Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Khaghani, S.; Shamsizadeh, A.; Nikfar, R.; Hesami, A. Shigella flexneri: A three-year antimicrobial resistance monitoring of isolates in a Children Hospital, Ahvaz, Iran. Iran J Microbiol 2014, 6, 225–229. [Google Scholar]
- Kotloff, K.L.; Nataro, J.P.; Blackwelder, W.C.; et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): A prospective, case-control study. Lancet 2013, 382, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Nüesch-Inderbinen, M.; Heini, N.; Zurfluh, K.; Althaus, D.; Hächler, H.; Stephan, R. Shigella antimicrobial drug resistance mechanisms, 2004-2014. Emerg Infect Dis 2016, 22, 1083–1085. [Google Scholar] [CrossRef]
- Ghosh, S.; Pazhani, G.P.; Chowdhury, G.; et al. Genetic characteristics and changing antimicrobial resistance among Shigella spp. isolated from hospitalized diarrhoeal patients in Kolkata, India. J Med Microbiol 2011, 60, 1460–1466. [Google Scholar] [CrossRef] [PubMed]
- Pazhani, G.P.; Niyogi, S.K.; Singh, A.K.; et al. Molecular characterization of multidrug-resistant Shigella species isolated from epidemic and endemic cases of shigellosis in India. J Med Microbiol 2008, 57, 856–863. [Google Scholar] [CrossRef] [PubMed]
- Taneja, N.; Mewara, A.; Kumar, A.; Verma, G.; Sharma, M. Cephalosporin-resistant Shigella flexneri over 9 years (2001-09) in India. J Antimicrob Chemother 2012, 67, 1347–1353. [Google Scholar] [CrossRef]
- Mauldin, P.D.; Salgado, C.D.; Hansen, I.S.; Durup, D.T.; Bosso, J.A. Attributable hospital cost and length of stay associated with health care-associated infections caused by antibiotic-resistant gram-negative bacteria. Antimicrob Agents Chemother 2010, 54, 109–115. [Google Scholar] [CrossRef]
- von Seidlein, L.; Kim, D.R.; Ali, M.; et al. A multicentre study of Shigella diarrhoea in six Asian countries: Disease burden, clinical manifestations, and microbiology. PLoS Med 2006, 3, e353. [Google Scholar] [CrossRef]
- Kotloff, K.L.; Winickoff, J.P.; Ivanoff, B.; et al. Global burden of Shigella infections: Implications for vaccine development and implementation of control strategies. Bull World Health Organ 1999, 77, 651–666. [Google Scholar]
- Bopp, C.A.; Brenner, F.W.; Fields, P.L.; et al. Escherichia, Shigella, and Salmonella. In Manual of Clinical Microbiology, 8th ed.; Murray, P.R., Baron, E.J., Jorgensen, J., Pfaller, M.A., Yolken, R.H., Eds.; American Society for Microbiology: Washington, DC, USA, 2003; pp. 654–671. [Google Scholar]
- Muthuirulandi Sethuvel, D.P.; Anandan, S.; Devanga Ragupathi, N.K.; Veeraraghavan, B. Identification of typical class 1 and class 2 integron gene cassettes in clinical isolates of MDR Shigella flexneri in South Indian population. Br Microbiol Res J 2015, 10, 1–6. [Google Scholar]
- Sambe-Ba, B.; Espié, E.; Faye, M.E.; Timbiné, L.G.; Sembene, M.; Gassama-Sow, A. Community-acquired diarrhea among children and adults in urban settings in Senegal: Clinical, epidemiological and microbiological aspects. BMC Infect Dis 2013, 13, 580. [Google Scholar] [CrossRef]
- Taneja, N.; Mewara, A. Shigellosis: Epidemiology in India. Indian J Med Res 2016, 143, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Jesudason, M.V. Shigella isolation in Vellore, south India (1997-2001). Indian J Med Res 2002, 115, 11–13. [Google Scholar] [PubMed]
- De Lappe, N.; O’Connor, J.; Garvey, P.; McKeown, P.; Cormican, M. Ciprofloxacin-resistant Shigella sonnei associated with travel to India. Emerg Infect Dis 2015, 21, 894–896. [Google Scholar] [CrossRef]
- Das, A.; Natarajan, M.; Mandal, J. The emergence of quinolone resistant Shigella sonnei, Pondicherry, India. PLoS ONE 2016, 11, e0160290. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.N.; Duy, P.T.; Baker, S. The rising dominance of Shigella sonnei: An intercontinental shift in the etiology of bacillary dysentery. PLoS Negl Trop Dis 2015, 9, e0003708. [Google Scholar] [CrossRef]
- Qu, M.; Zhang, X.; Liu, G.; et al. An eight-year study of Shigella species in Beijing, China: Serodiversity, virulence genes, and antimicrobial resistance. J Infect Dev Ctries 2014, 8, 904–908. [Google Scholar] [CrossRef][Green Version]
- Khan, E.; Jabeen, K.; Ejaz, M.; Siddiqui, J.; Shezad, M.F.; Zafar, A. Trends in antimicrobial resistance in Shigella species in Karachi, Pakistan. J Infect Dev Ctries 2009, 3, 798–802. [Google Scholar] [CrossRef]
- Dutta, S.; Jain, P.; Nandy, S.; Matsushita, S.; Yoshida, S. Molecular characterization of serologically atypical provisional serovars of Shigellaisolates from Kolkata, India. J Med Microbiol 2014, 63, 1696–1703. [Google Scholar] [CrossRef]
- Nair, G.B.; Ramamurthy, T.; Bhattacharya, M.K.; et al. Emerging trends in the etiology of enteric pathogens as evidenced from an active surveillance of hospitalized diarrhoeal patients in Kolkata, India. Gut Pathog 2010, 2, 4. [Google Scholar] [CrossRef]
- Mamatha, B.; Rituparna, C. Decreased susceptibility to antimicrobials among Shigella flexneri isolates in Manipal, South India--a 5 year hospital based study. Southeast Asian J Trop Med Public Health 2012, 43, 1447–1451. [Google Scholar]
- Navia, M.M.; Capitano, L.; Ruiz, J.; et al. Typing and characterization of mechanisms of resistance of Shigella spp. isolated from feces of children under 5 years of age from Ifakara, Tanzania. J Clin Microbiol 1999, 37, 3113–3117. [Google Scholar] [CrossRef]
- Pu, X.Y.; Pan, J.C.; Wang, H.Q.; Zhang, W.; Huang, Z.C.; Gu, Y.M. Characterization of fluoroquinolone-resistant Shigella flexneri in Hangzhou area of China. J Antimicrob Chemother 2009, 63, 917–920. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hu, L.; Pan, Y.; et al. Prevalence of plasmid-mediated quinolone resistance determinants in association with β-lactamases, 16S rRNA methylase genes and integrons amongst clinical isolates of Shigella flexneri. J Med Microbiol 2012, 61, 1174–1176. [Google Scholar] [CrossRef]
- Cui, X.; Yang, C.; Wang, J.; et al. Antimicrobial resistance of Shigella flexneri serotype 1b isolates in China. PLoS ONE 2015, 10, e0129009. [Google Scholar] [CrossRef] [PubMed]
- Auda, I.G. Occurrence of CTX-M-I and CTX-M-III genes on plasmids of Shigella species isolated from cases of diarrhea in Baghdad. World J Pharm Res 2014, 3, 1273–1280. [Google Scholar]
- Muthuirulandi Sethuvel, D.P.; Devanga Ragupathi, N.K.; Anandan, S.; Veeraraghavan, B. Update on: Shigella new serogroups/serotypes and their antimicrobial resistance. Lett Appl Microbiol 2017, 64, 8–18. [Google Scholar] [CrossRef]
 |
 |
 |
© GERMS 2017.
Share and Cite
Anandan, S.; Sethuvel, D.P.M.; Gajendiren, R.; Verghese, V.P.; Walia, K.; Veeraraghavan, B. Molecular Characterization of Antimicrobial Resistance in Clinical Shigella Isolates During 2014 and 2015: Trends in South India. Germs 2017, 7, 115-122. https://doi.org/10.18683/germs.2017.1116
Anandan S, Sethuvel DPM, Gajendiren R, Verghese VP, Walia K, Veeraraghavan B. Molecular Characterization of Antimicrobial Resistance in Clinical Shigella Isolates During 2014 and 2015: Trends in South India. Germs. 2017; 7(3):115-122. https://doi.org/10.18683/germs.2017.1116
Chicago/Turabian StyleAnandan, Shalini, Dhiviya Prabaa Muthuirulandi Sethuvel, Revathi Gajendiren, Valsan Philip Verghese, Kamini Walia, and Balaji Veeraraghavan. 2017. "Molecular Characterization of Antimicrobial Resistance in Clinical Shigella Isolates During 2014 and 2015: Trends in South India" Germs 7, no. 3: 115-122. https://doi.org/10.18683/germs.2017.1116
APA StyleAnandan, S., Sethuvel, D. P. M., Gajendiren, R., Verghese, V. P., Walia, K., & Veeraraghavan, B. (2017). Molecular Characterization of Antimicrobial Resistance in Clinical Shigella Isolates During 2014 and 2015: Trends in South India. Germs, 7(3), 115-122. https://doi.org/10.18683/germs.2017.1116




