Staphylococcus aureus is commonly isolated in hospital wards and easily transmitted via human-to-human contact. SmaI macro-restriction of S. aureus genomic DNA followed by pulsed-field gel electrophoresis (PFGE) typing has been reported to be the gold standard for S. aureus typing and for investigating the possibility of outbreaks in hospitals [1]. However, PFGE typing requires specific equipment, technical skills, is time consuming and expensive. Sabat et al. established the gel-based multiple locus variable number of tandem repeats analysis (MLVA) for S. aureus typing and Roussel et al., reported the efficiency of this approach, showing that strain clustering is concordant to PFGE typing [2,3]. The MLVA method involves PCR amplification of five variable number of tandem repeats (VNTR) loci specific for S. aureus, namely sdr, clfA, clfB, ssp, and spa in a multiplex-PCR mixture, separation of the amplified bands on an agarose gel, and comparison of the band patterns between strains to identify clusters or clones [2,3]. Indeed, the VNTR loci have been used previously to differentiate S. aureus from other bacteria [2].
In a molecular epidemiology study on methicillin-resistant S. aureus (MRSA) isolated from our medical center carried out in 2009, the strains were typed using PFGE and four major MRSA pulsotypes were identified [4]. We find the PFGE labor-intensive and the technique requires a long turnaround-time (TAT) before final results could be obtained. This will not be ideal in outbreak investigations as techniques with rapid TAT and simpler workflow may help reduce morbidity and mortality in infections [2,5]. As MLVA strain clustering is concordant to PFGE typing, we attempted to type MRSA strains representative of PFGE pulsotypes and also some singletons identified in a separate experiment using MLVA, with the intention of substituting PFGE with MLVA for future MRSA epidemiological studies in the medical center.
MLVA was carried out according to the published protocol [2]. However, no amplification was observed, even after attempts to optimize the protocol. Single-plex amplification of the MLVA primers revealed that, for some reason, primers for the ssp loci were not working. We wondered if the MLVA could still be discriminatory without the ssp loci primers, and proceeded to exclude these primers from the MLVA multiplex reaction in the following experiment, while using the established amplification and gel electrophoresis protocols [2].
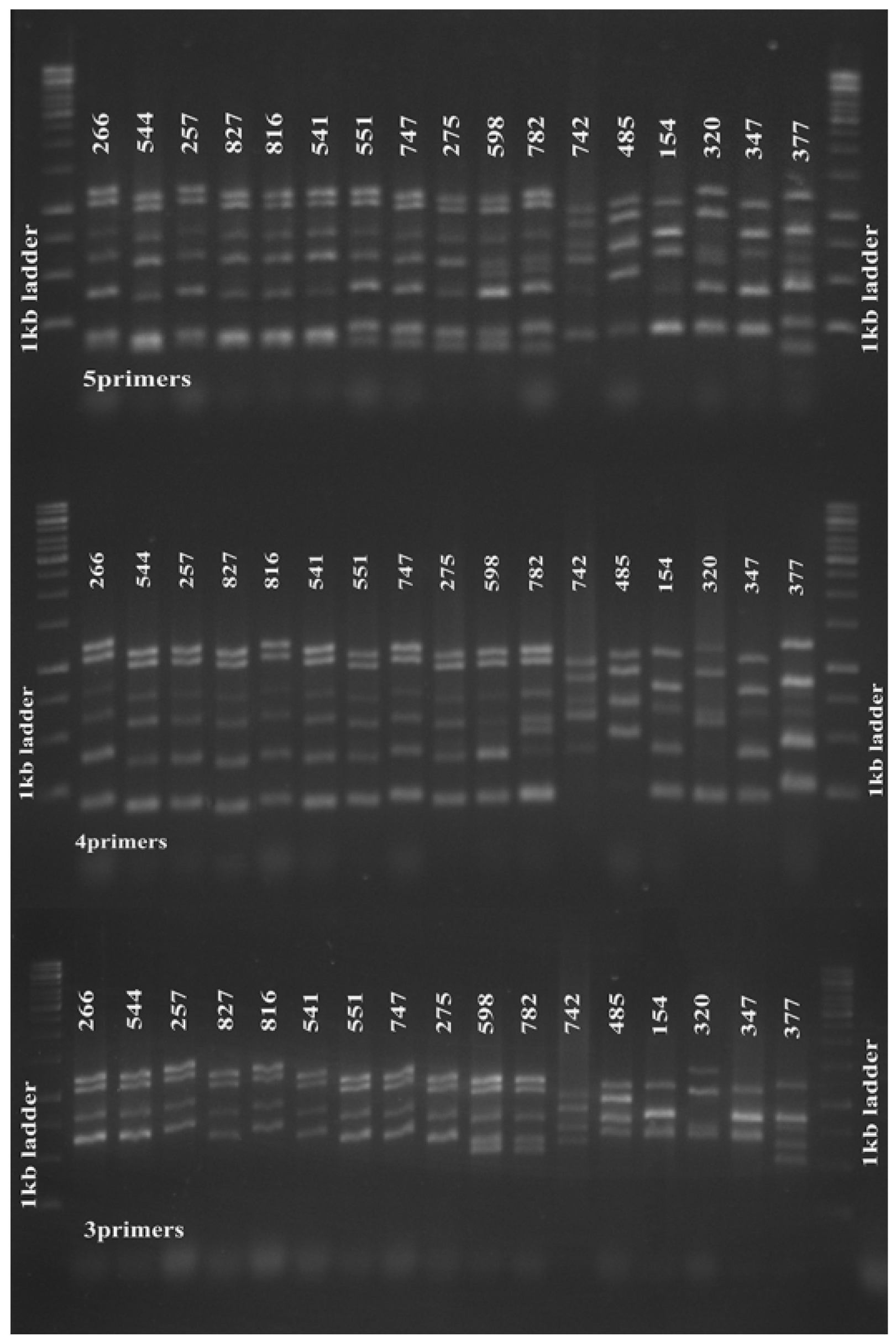
Interestingly, amplification using the four primer pairs of clfA, clfB, sdr and spa were able to distinguish the tested strains according to their corresponding PFGE pulsotypes (sensitivity, specificity, PPV and NPV = 100%) (Figure 1). In addition, when we used newly re-synthesized ssp primers for amplification, we observed that the 4-primer-pair MLVA amplification clustered the strains in a similar manner with the established 5-primer-pair protocol (Figure 1). When we further excluded the spa primers from the MLVA multiplex, it was interesting to note that this 3-primer-pair amplification (clfA, clfB and sdr) also distinguished the strains in concordance to their PFGE pulsotypes and 5-primer-pair MLVA type (sensitivity, specificity, PPV and NPV = 100%) (Figure 1). Cluster differentiation was also easier due to fewer amplification bands.
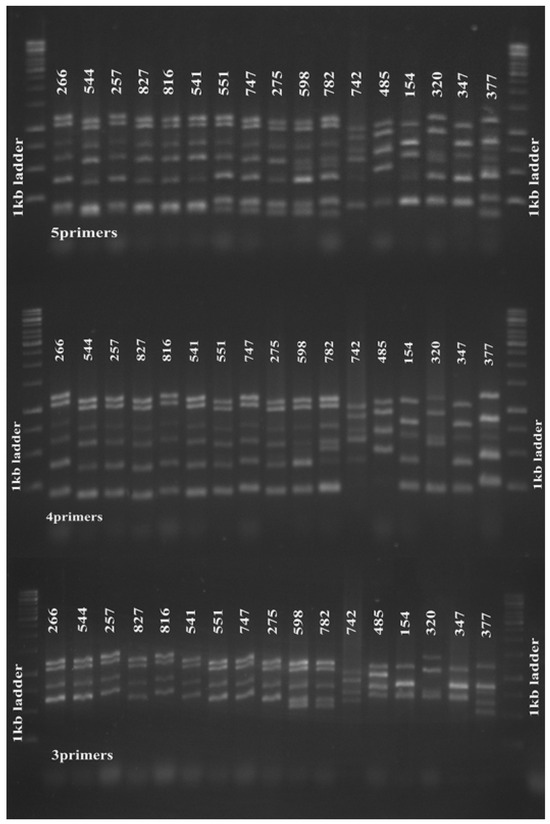
Figure 1.
Five-primer-pair MLVA amplification of S. aureus. Five-primer-pair MLVA amplification clustered 17 tested MRSA strains isolated in 2009 according to their PFGE pulsotypes (upper panel). The 4-(middle panel) and 3-primer pair (lower panel) amplifications also clustered the strains accordingly. From left, strains 266–275 were of PFGE pulsotype P1, strains 598 and 782 were pulsotype P2, strains 742 and 458 were pulsotype P3; strains 154, 320, 347 and 377 were singletons. The 1 kb DNA Ladder (Promega Corporation) was used as a molecular weight marker.clustered 17 tested MRSA strains isolated in 2009 according to their PFGE pulsotypes (upper panel). The 4-(middle panel) and 3-primer pair (lower panel) amplifications also clustered the strains accordingly. From left, strains 266–275 were of PFGE pulsotype P1, strains 598 and 782 were pulsotype P2, strains 742 and 458 were pulsotype P3; strains 154, 320, 347 and 377 were singletons. The 1 kb DNA Ladder (Promega Corporation) was used as a molecular weight marker.
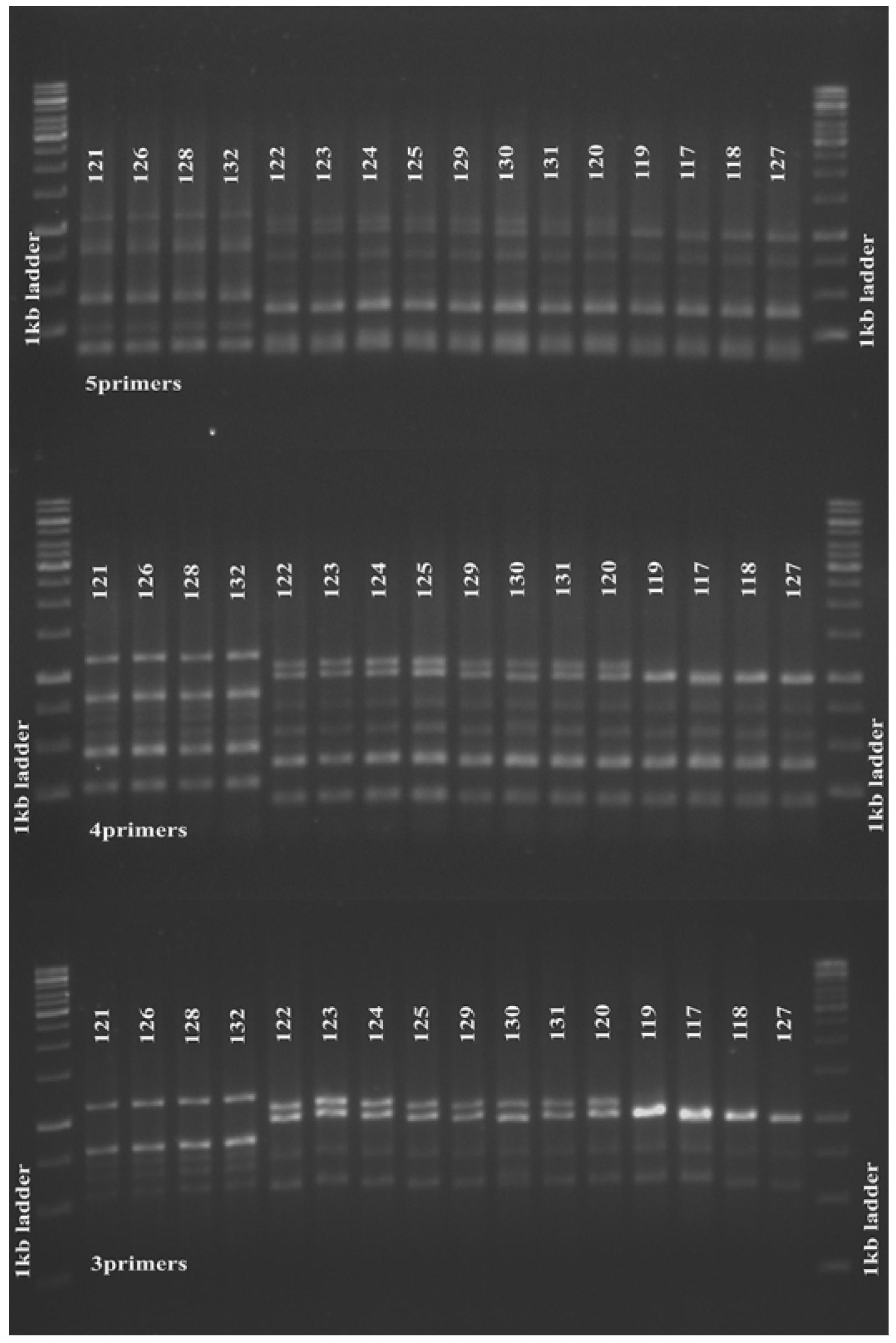
We further investigated the reproducibility of this finding in 16 MRSA strains isolated four years later in 2013, and found that the 3-, 4- and 5-primer MLVA amplification clustered the tested strains in a similar manner (Figure 2). Slower molecular evolution of a certain VNTR locus will reduce its differentiation power in genotyping [2]. In this study, the ssp and spa loci might have slower molecular clocks and thus generate little variation between strains; therefore, exclusion of the primers for these loci in the MLVA experiment did not affect clustering of our study strains.
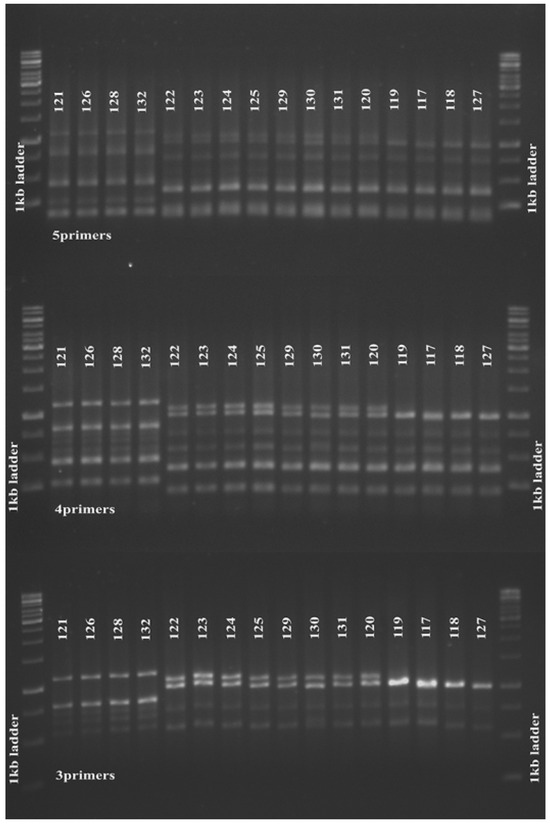
Figure 2.
Five, four and 3 primer pair MLVA amplification of S. aureus. Five (upper panel), 4-(middle panel) and 3-(lower panel) primer pair MLVA amplifications produced similar clustering for 16 MRSA strains isolated from the same medical centre in 2013. The 1 kb DNA Ladder (Promega Corporation) was used as a molecular weight marker.
In addition to being a laborious and time consuming method, many small hospital laboratories might not be able to afford the PFGE equipment, which is space consuming and requires maintenance. The gel-based MLVA provides a good alternative for S. aureus typing and for outbreak investigations in hospitals. Taken together with the results from this small observation, the gel-based MLVA could be further simplified into a 3-primer-pair multiplex PCR reaction, whereby only a thermal cycler, gel electrophoresis and a viewing apparatus will be needed for this procedure. Nevertheless, this observation needs to be validated in a separate study using a larger number of samples. This simplified gel-based MLVA may provide an easy, inexpensive yet accurate approach for S. aureus typing.
Author Contributions
HN conceived and designed the study, analyzed the data and wrote the manuscript. CY and FR performed the experiments and prepared the manuscript figures. SH and RJ participated in the design of the study and provided reagents and laboratory space. All authors read and approved the final manuscript.
Funding
This study was funded by the UKM Medical Centre Research Grant FF-2014-218 and Fundamental Research Grant Scheme FRGS/1/2014/SKK04/UKM/03/1 from the Ministry of Higher Education, Malaysia. The funders had no role in the design of the study, data collection, analysis and interpretation and in writing the manuscript.
Acknowledgments
The authors would like to thank Hassriana Fazilla Sapri, Nurul Azirah Mohamad Sani, Ainihayati Noordin, and Masitah Ismail for their help in MRSA sample collection.
Conflicts of Interest
All authors—none to declare.
References
- Mehndiratta, P.L.; Bhalla, P. Typing of methicillin resistant Staphylococcus aureus: A technical review. Indian J. Med. Microbiol. 2012, 30, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Sabat, A.; Krzyszton-Russjan, J.; Strzalka, W.; et al. New method for typing Staphylococcus aureus strains: Multiple-locus variable-number tandem repeat analysis of polymorphism and genetic relationships of clinical isolates. J. Clin. Microbiol. 2003, 41, 1801–1804. [Google Scholar] [CrossRef] [PubMed]
- Roussel, S.; Felix, B.; Vingadassalon, N.; et al. Staphylococcus aureus strains associated with food poisoning outbreaks in France: Comparison of different molecular typing methods, including MLVA. Front. Microbiol. 2015, 6, 882. [Google Scholar] [CrossRef] [PubMed]
- Noordin, A.; Sapri, H.F.; Mohamad Sani, N.A.; et al. Antimicrobial resistance profiling and molecular typing of methicillin-resistant Staphylococcus aureus isolated from a Malaysian teaching hospital. J. Med. Microbiol. 2016, 65, 1476–1481. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, C.; Kiernan, M.G.; Finnegan, C.; et al. An optimized work-flow to reduce time-to-detection of carbapenemase-producing Enterobacteriaceae (CPE) using direct testing from rectal swabs. Bioengineered 2016, 1–8. [Google Scholar] [CrossRef] [PubMed][Green Version]
© GERMS 2017.