Comparative Phenotypic and Proteomic Analysis of Colistin-Exposed Pseudomonas aeruginosa
Abstract
Introduction
Methods
Bacterial strains
Antibiotic susceptibility testing
Morphology and virulence factor assessment
Colony morphology and cell length observation
Virulence factor assessment
iTRAQ LC-MS/MS proteomic analysis
Quantitative iTRAQ- LC-MS/MS analysis
Quantitative reverse transcription PCR (RT- qPCR) analysis
Data analysis
Results
Antibiotic susceptibility profile of P. aeruginosa under colistin exposure
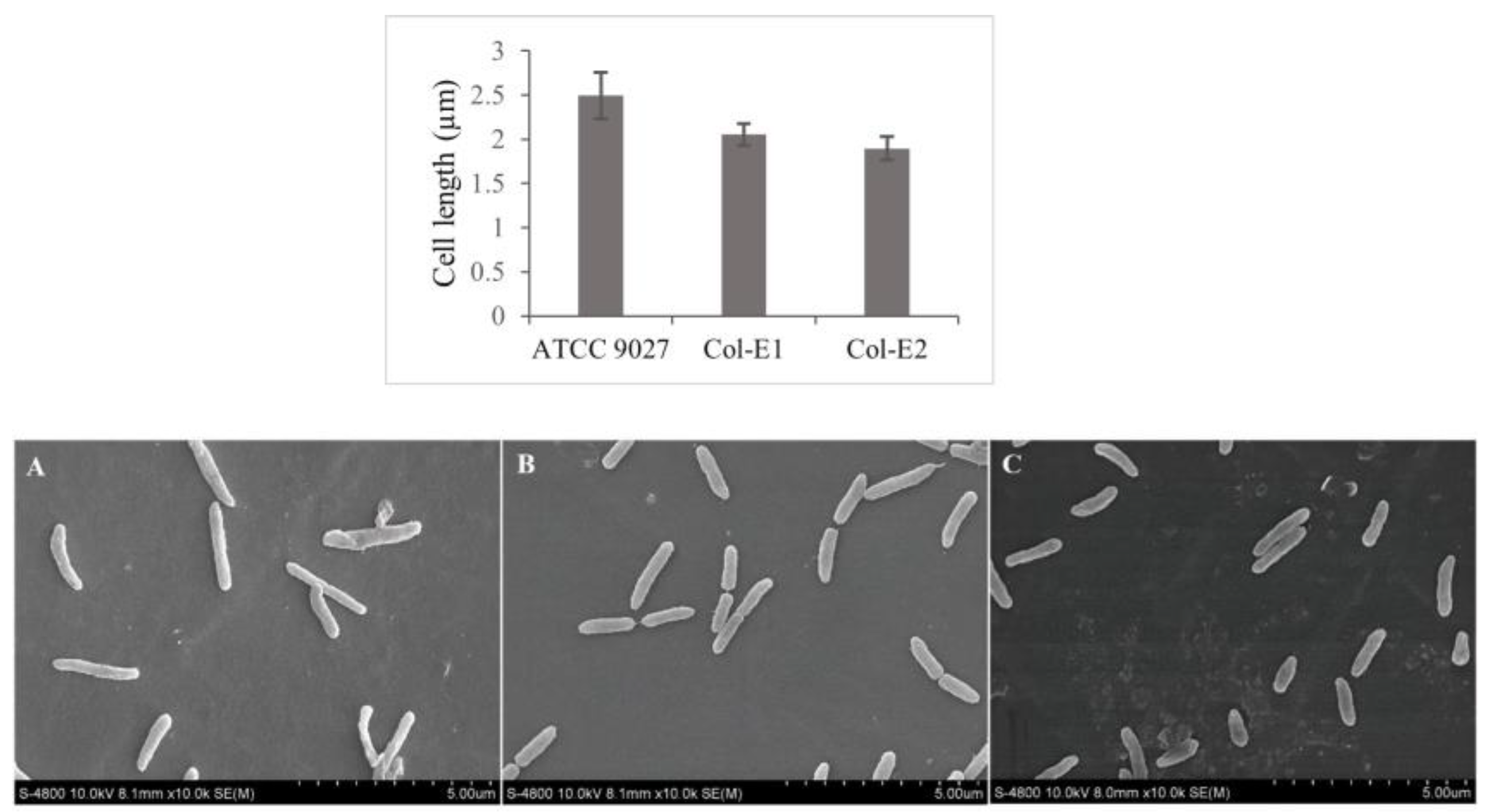
Morphology of P. aeruginosa under colistin exposure
Proteome response of P. aeruginosa to colistin
Quantitative reverse transcription PCR (RT- qPCR) analysis
Discussion
Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yero, D.; Díaz-Lobo, M.; Costenaro, L.; et al. The Pseudomonas aeruginosa substrate-binding protein Ttg2D functions as a general glycerophospholipid transporter across the periplasm. Commun Biol. 2021, 4, 448. [Google Scholar] [CrossRef]
- Sandoval-Motta, S.; Aldana, M. Adaptive resistance to antibiotics in bacteria: a systems biology perspective. WIREs Syst Biol Med. 2016, 8, 253–267. [Google Scholar] [CrossRef]
- de Andrade, J.P.L.; de Macêdo Farias, L.; Ferreira, J.F.G.; et al. Sub-inhibitory concentration of piperacillin-tazobactam may be related to virulence properties of filamentous Escherichia coli. Curr Microbiol. 2016, 72, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Gogry, F.A.; Siddiqui, M.T.; Sultan, I.; Haq, Q.M.o.h.d.R. Current update on intrinsic and acquired colistin resistance mechanisms in bacteria. Front Med (Lausanne). 2021, 8, 677720. [Google Scholar] [CrossRef] [PubMed]
- Dößelmann, B.; Willmann, M.; Steglich, M.; et al. Rapid and consistent evolution of colistin resistance in extensively drug-resistant Pseudomonas aeruginosa during morbidostat culture. Antimicrob Agents Chemother. 2017, 61, e00043–17. [Google Scholar] [CrossRef] [PubMed]
- Saleh, N.M.; Hesham, M.S.; Amin, M.A.; Samir Mohamed, R. Acquisition of colistin resistance links cell membrane thickness alteration with a point mutation in the lpxD gene in Acinetobacter baumannii. Antibiotics (Basel). 2020, 9, 164. [Google Scholar] [CrossRef]
- Tsakou, F.; Jersie-Christensen, R.; Jenssen, H.; Mojsoska, B. The role of proteomics in bacterial response to antibiotics. Pharmaceuticals (Basel). 2020, 13, 214. [Google Scholar] [CrossRef]
- Hashemi, M.M.; Holden, B.S.; Coburn, J.; et al. Proteomic analysis of resistance of Gram-negative bacteria to chlorhexidine and impacts on susceptibility to colistin, antimicrobial peptides, and ceragenins. Front Microbiol. 2019, 10, 210. [Google Scholar] [CrossRef]
- Kowalska-Krochmal, B.; Dudek-Wicher, R. The minimum inhibitory concentration of antibiotics: methods, interpretation, clinical relevance. Pathogens. 2021, 10, 165. [Google Scholar] [CrossRef]
- Sousa, A.M.; Machado, I.; Nicolau, A.; Pereira, M.O. Improvements on colony morphology identification towards bacterial profiling. J Microbiol Methods. 2013, 95, 327–335. [Google Scholar] [CrossRef]
- Nguyen, N.H.B.; Pham, T.T.V.; Huynh, T.Q.; Nguyen, T.H.; Nguyen, T.T.H. Sample preparative procedure for Pseudomonas aeruginosa observation under scanning electron microscope. Vietnam J Biotechnol. 2022, 20, 717–726. [Google Scholar] [CrossRef]
- Garcia Dde, O.; Timenetsky, J.; Martinez, M.B.; Francisco, W.; Sinto, S.I.; Yanaguita, R.M. Proteases (caseinase and elastase), hemolysins, adhesion and susceptibility to antimicrobials of Stenotrophomonas maltophilia isolates obtained from clinical specimens. Braz J Microbiol. 2002, 33, 157–162. [Google Scholar] [CrossRef]
- Ahamed, F.; Devnath, P. Extraction, purification and characterization of pyocyanin produced by Pseudomonas aeruginosa and evaluation for its antimicrobial activity. Int Res J Biol Sci. 2017, 6, 1–7. [Google Scholar]
- Essar, D.W.; Eberly, L.; Hadero, A.; Crawford, I.P. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol. 1990, 172, 884–900. [Google Scholar] [CrossRef]
- Dao, K.H.T.; Hamer, K.E.; Clark, C.L.; Harshman, L.G. Pyoverdine production by Pseudomonas aeruginosa exposed to metals or an oxidative stress agent. Ecol Appl. 1999, 9, 441–448. [Google Scholar] [CrossRef]
- Banin, E.; Vasil, M.L.; Greenberg, E.P. Iron and Pseudomonas aeruginosa biofilm formation. Proc Natl Acad Sci U S A. 2005, 102, 11076–11081. [Google Scholar] [CrossRef] [PubMed]
- Thai, V.C.; Lim, T.K.; Le, K.P.U.; Lin, Q.; Nguyen, T.T.H. iTRAQ-based proteome analysis of fluoroquinolone- resistant Staphylococcus aureus. J Glob Antimicrob Resist. 2017, 8, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Chen, W.N. iTRAQ-Coupled 2-D LC-MS/MS analysis of membrane protein profile in Escherichia coli incubated with apidaecin IB. PLoS One. 2011, 6, e20442. [Google Scholar] [CrossRef] [PubMed]
- Sanz-García, F.; Hernando-Amado, S.; Martínez, J.L. Mutational evolution of Pseudomonas aeruginosa resistance to ribosome-targeting antibiotics. Front Genet. 2018, 9, 451. [Google Scholar] [CrossRef]
- Savli, H.; Karadenizli, A.; Kolayli, F.; Gundes, S.; Ozbek, U.; Vahaboglu, H. Expression stability of six housekeeping genes: a proposal for resistance gene quantification studies of Pseudomonas aeruginosa by real-time quantitative RT-PCR. J Med Microl. 2003, 52, 403–408. [Google Scholar] [CrossRef]
- Kumar, A.; Lorand, D. Robust ΔΔct estimate. Genomics. 2021, 113, 420–427. [Google Scholar] [CrossRef]
- Huynh, T.Q.; Nguyen, T.T.H. The effects of sub-MIC ciprofloxacin exposure on antibiotic susceptibility and virulence factors in Pseudomonas aeruginosa ATCC 9027. Int J Life Sci Res Arch. 2022, 3, 70–77. [Google Scholar] [CrossRef]
- Sabnis, A.; Hagart, K.L.; Klöckner, A.; et al. Colistin kills bacteria by targeting lipopolysaccharide in the cytoplasmic membrane. eLife. 2021, 10, e65836. [Google Scholar] [CrossRef] [PubMed]
- Mortensen, N.P.; Fowlkes, J.D.; Sullivan, C.J.; et al. Effects of colistin on surface ultrastructure and nanomechanics of Pseudomonas aeruginosa cells. Langmuir. 2009, 25, 3728–3733. [Google Scholar] [CrossRef]
- O'Driscoll, N.H.; Cushnie, T.P.T.; Matthews, K.H.; Lamb, A.J. Colistin causes profound morphological alteration but minimal cytoplasmic membrane perforation in populations of Escherichia coli and Pseudomonas aeruginosa. Arch Microbiol. 2018, 200, 793–802. [Google Scholar] [CrossRef]
- Basan, M.; Zhu, M.; Dai, X.; et al. Inflating bacterial cells by increased protein synthesis. Mol Syst Biol. 2015, 11, 836. [Google Scholar] [CrossRef]
- Schaechter, M.; MaalØe, O.; Kjeldgaard, N.O. Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella typhimurium. Microbiology. 1958, 19, 592–606. [Google Scholar] [CrossRef]
- Seiffein, N.L.; Ali, G.H. Effect of subinhibitory concentrations of selected antibiotics and propolis on pyocyanin and biofilm production among Pseudomonas aeruginosa isolates in Alexandria, Egypt. Egypt J Med Microbiol. 2021, 30, 129–137. [Google Scholar] [CrossRef]
- Jones, A.; Elphick, H.; Pettitt, E.; Everard, M.L.; Evans, G.S. Colistin stimulates the activity of neutrophil elastase and Pseudomonas aeruginosa elastase. Eur Respir J. 2002, 19, 1136–1141. [Google Scholar] [CrossRef] [PubMed]
- Carretero-Ledesma, M.; García-Quintanilla, M.; Martín- Peña, R.; Pulido, M.R.; Pachón, J.; McConnell, M.J. Phenotypic changes associated with colistin resistance due to lipopolysaccharide loss in Acinetobacter baumannii. Virulence. 2018, 9, 930–942. [Google Scholar] [CrossRef] [PubMed]
- Cianciulli Sesso, A.; Lilić, B.; Amman, F.; Wolfinger, M.T.; Sonnleitner, E.; Bläsi, U. Gene expression profiling of Pseudomonas aeruginosa upon exposure to colistin and tobramycin. Front Microbiol. 2021, 12, 937. [Google Scholar] [CrossRef] [PubMed]
- Vallet, I.; Diggle, S.P.; Stacey, R.E.; et al. Biofilm formation in Pseudomonas aeruginosa: fimbrial cup gene clusters are controlled by the transcriptional regulator MvaT. J Bacteriol. 2004, 186, 2880–2890. [Google Scholar] [CrossRef]
- Xu, H.; Lin, W.; Xia, H.; et al. Influence of ptsP gene on pyocyanin production in Pseudomonas aeruginosa. FEMS Microbiol Lett. 2005, 253, 103–109. [Google Scholar] [CrossRef]
- Kozak, M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983, 47, 1–45. [Google Scholar] [CrossRef]
- Lipowska, J.; Miks, C.D.; Kwon, K.; et al. Pyrimidine biosynthesis in pathogens - structures and analysis of dihydroorotases from Yersinia pestis and Vibrio cholerae. Int J Biol Macromol. 2019, 136, 1176–1187. [Google Scholar] [CrossRef]
- UniProt. 2023. grpE - Protein GrpE - Pseudomonas aeruginosa. UniProtKB. Available online: https://www.uniprot.org/uniprotkb/A0A069Q2K4/entry (accessed on 18 July 2023).
- Poole, R.K. Flavohaemoglobin: the pre-eminent nitric oxide-detoxifying machine of microorganisms. F1000Res. 2020, 9, 7. [Google Scholar] [CrossRef]
- Damron, F.H.; Napper, J.; Teter, M.A.; Yu, H.D. Lipotoxin F of Pseudomonas aeruginosa is an AlgU-dependent and alginate-independent outer membrane protein involved in resistance to oxidative stress and adhesion to A549 human lung epithelia. Microbiology (Reading). 2009, 155, 1028–1038. [Google Scholar] [CrossRef]
- Macfarlane, E.L.; Kwasnicka, A.; Ochs, M.M.; Hancock, R.E. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol Microbiol. 1999, 34, 305–316. [Google Scholar] [CrossRef]
- Lin, Y.M.; Wu, S.J.; Chang, T.W.; et al. Outer membrane protein I of Pseudomonas aeruginosa is a target of cationic antimicrobial peptide/protein. J Biol Chem. 2010, 285, 8985–8994. [Google Scholar] [CrossRef] [PubMed]
- Han, M.L.; Velkov, T.; Zhu, Y.; et al. Polymyxin-induced lipid A deacylation in Pseudomonas aeruginosa perturbs polymyxin penetration and confers high-level resistance. ACS Chem Biol. 2018, 13, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Luo, Y.F.; Williams, B.J.; Blackwell, T.S.; Xie, C.M. Structure and function of OprD protein in Pseudomonas aeruginosa: from antibiotic resistance to novel therapies. Int J Med Microbiol. 2012, 302, 63–68. [Google Scholar] [CrossRef]
- Muller, C.; Plésiat, P.; Jeannot, K. A two-component regulatory system interconnects resistance to polymyxins, aminoglycosides, fluoroquinolones, and β-lactams in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2011, 55, 1211–1221. [Google Scholar] [CrossRef]
- Ghosh, D.; Veeraraghavan, B.; Elangovan, R.; Vivekanandan, P. Antibiotic resistance and epigenetics: more to it than meets the eye. Antimicrob Agents Chemother. 2020, 64, e02225–19. [Google Scholar] [CrossRef]
- Riber, L.; Hansen, L.H. Epigenetic memories: the hidden drivers of bacterial persistence? Trends Microbiol. 2021, 29, 190–194. [Google Scholar] [CrossRef]
- Adam, M.; Murali, B.; Glenn, N.O.; Potter, S.S. Epigenetic inheritance based evolution of antibiotic resistance in bacteria. BMC Evol Biol. 2008, 8, 52. [Google Scholar] [CrossRef] [PubMed]
- Ontong, J.C.; Ozioma, N.F.; Voravuthikunchai, S.P.; Chusri, S. Synergistic antibacterial effects of colistin in combination with aminoglycoside, carbapenems, cephalosporins, fluoroquinolones, tetracyclines, fosfomycin, and piperacillin on multidrug resistant Klebsiella pneumoniae isolates. PLoS One. 2021, 16, e0244673. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.; Daikos, G.L.; Durante-Mangoni, E.; et al. Colistin alone versus colistin plus meropenem for treatment of severe infections caused by carbapenem-resistant Gram- negative bacteria: an open-label, randomised controlled trial. Lancet Infect Dis. 2018, 18, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Kaye, K.S.; Marchaim, D.; Thamlikitkul, V.; et al. Colistin monotherapy versus combination therapy for carbapenem-resistant organisms. NEJM Evid. 2022, 2, EVIDoa2200131. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Chen, I.; Tang, T. Colistin Monotherapy versus colistin plus meropenem combination therapy for the treatment of multidrug-resistant Acinetobacter baumannii infection: a meta-analysis. J Clin Med. 2022, 11, 3239. [Google Scholar] [CrossRef] [PubMed]
- Sanz-García, F.; Hernando-Amado, S.; Martínez, J.L. Mutational evolution of Pseudomonas aeruginosa resistance to ribosome-targeting antibiotics. Front Genet. 2018, 9, 451. [Google Scholar] [CrossRef]
- Savli, H.; Karadenizli, A.; Kolayli, F.; et al. Expression stability of six housekeeping genes: a proposal for resistance gene quantification studies of Pseudomonas aeruginosa by real-time quantitative RT-PCR. J Med Microbiol. 2003, 52, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Tomás, M.; Doumith, M.; Warner, M.; et al. Efflux pumps, OprD porin, AmpC beta-lactamase, and multiresistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob Agents Chemother. 2010, 54, 2219–2224. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Lorand, D. Robust ΔΔct estimate. Genomics 2021, 113, 420–427. [Google Scholar] [CrossRef] [PubMed]



| Virulence factors | P. aeruginosa ATCC 9027 | Col-E1 | Col-E2 |
|---|---|---|---|
| Elastase (mm) | 3.58 ± 0.63 | 3.50 ± 0.50 | 3.67 ± 1.61 |
| Protease (mm) | 3.17 ± 0.14 | 2.75 ± 0.25 | 2.66 ± 0.38 |
| Rhamnolipid (mm) | 2.92 ± 0.76 | 2.44 ± 0.64 | 2.58 ± 0.40 |
| Pyocyanin (µg/mL)* | 1.26 ± 0.13 | 0.40 ± 0.24 | 1.60 ± 0.42 |
| Pyoverdine (excitation: 405 nm, emission: 450 nm) | 175.46 ± 19.97 | 169.92 ± 6.37 | 210.39 ± 3.71 |
| Biofilm (OD550nm) | 5.95 ± 1.34 | 2.42 ± 2.00 | 7.96 ± 3.85 |
© GERMS 2024.
Share and Cite
Tran, N.B.V.; Huynh, T.Q.; Ngo, H.L.; Nguyen, N.H.B.; Nguyen, T.H.; Tong, T.H.; Trinh, T.T.L.; Nguyen, V.D.; Pham, L.N.M.; Das, P.P.; et al. Comparative Phenotypic and Proteomic Analysis of Colistin-Exposed Pseudomonas aeruginosa. GERMS 2024, 14, 246-266. https://doi.org/10.18683/germs.2024.1436
Tran NBV, Huynh TQ, Ngo HL, Nguyen NHB, Nguyen TH, Tong TH, Trinh TTL, Nguyen VD, Pham LNM, Das PP, et al. Comparative Phenotypic and Proteomic Analysis of Colistin-Exposed Pseudomonas aeruginosa. GERMS. 2024; 14(3):246-266. https://doi.org/10.18683/germs.2024.1436
Chicago/Turabian StyleTran, Nguyen Bao Vy, Thuc Quyen Huynh, Hong Loan Ngo, Ngoc Hoa Binh Nguyen, Thi Hiep Nguyen, Thi Hang Tong, Thi Truc Ly Trinh, Van Dung Nguyen, Le Nhat Minh Pham, Prem Prakash Das, and et al. 2024. "Comparative Phenotypic and Proteomic Analysis of Colistin-Exposed Pseudomonas aeruginosa" GERMS 14, no. 3: 246-266. https://doi.org/10.18683/germs.2024.1436
APA StyleTran, N. B. V., Huynh, T. Q., Ngo, H. L., Nguyen, N. H. B., Nguyen, T. H., Tong, T. H., Trinh, T. T. L., Nguyen, V. D., Pham, L. N. M., Das, P. P., Lim, T. K., Lin, Q., & Nguyen, T. T. H. (2024). Comparative Phenotypic and Proteomic Analysis of Colistin-Exposed Pseudomonas aeruginosa. GERMS, 14(3), 246-266. https://doi.org/10.18683/germs.2024.1436




