The Emerging Role of Pseudomonas aeruginosa in Diarrhea: Where We Stand
Abstract
Introduction
Colonization by PA in human gut
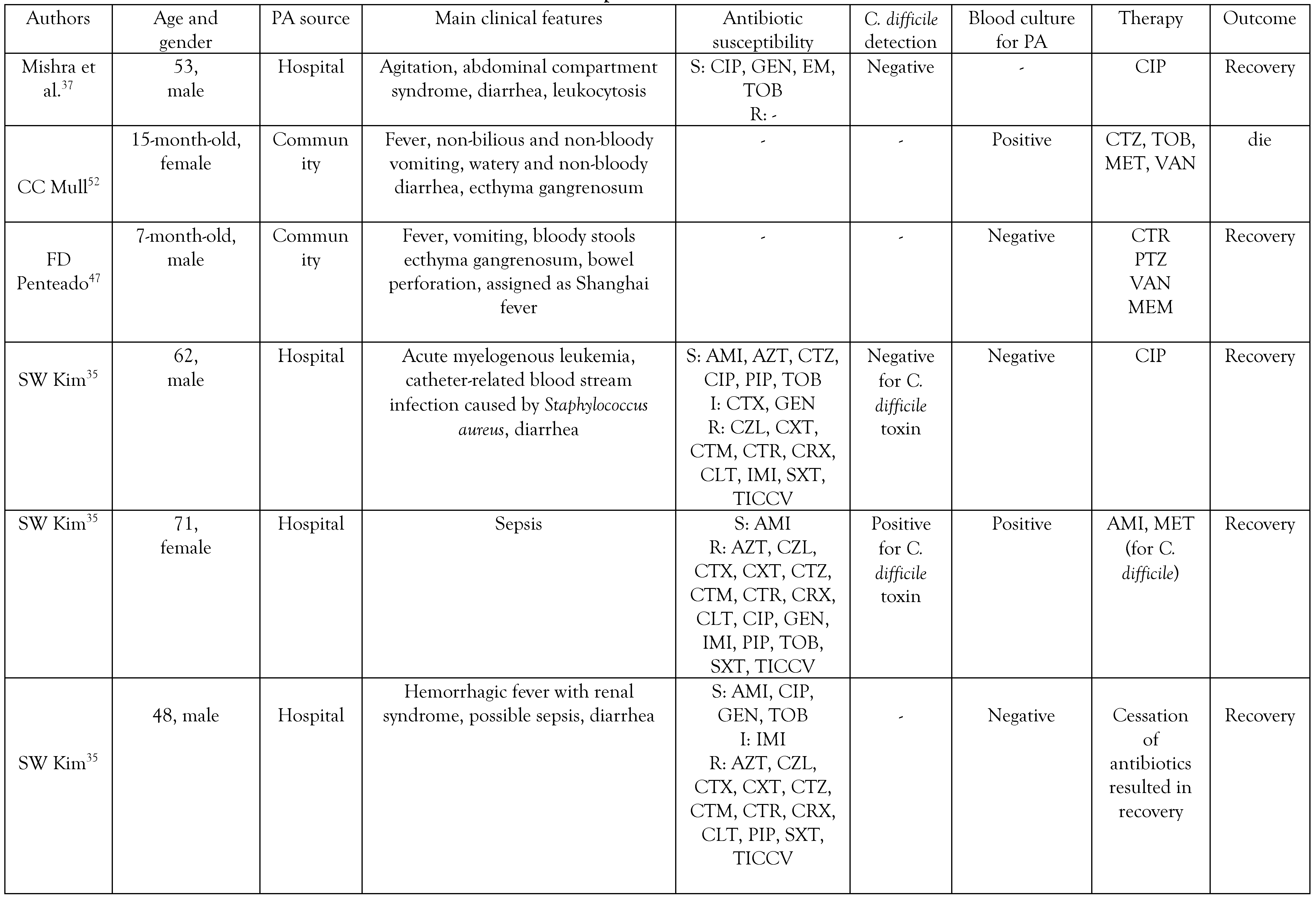
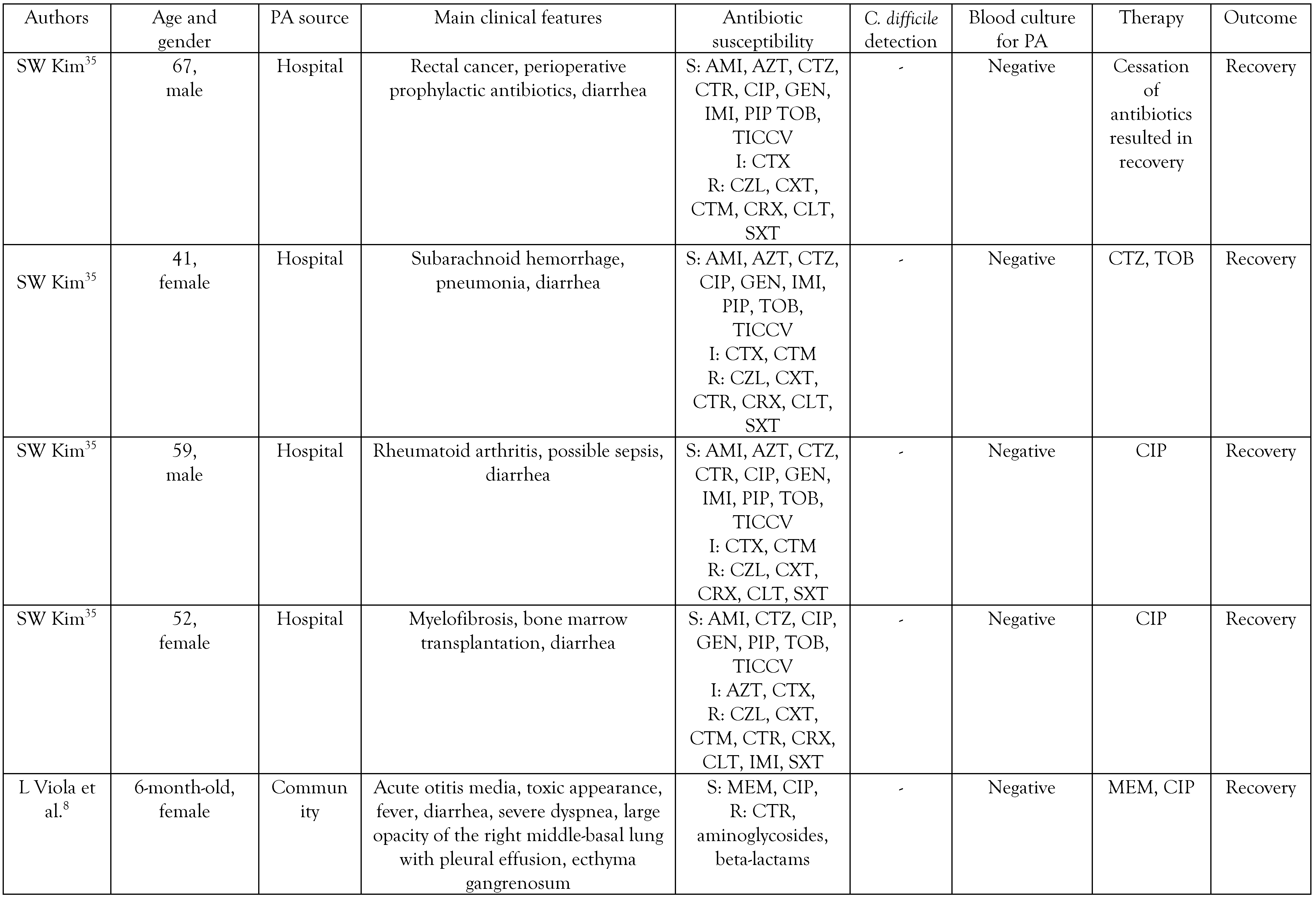
The origin and clinical manifestations of PA diarrhea
Antibiotic resistance in PA causing diarrhea
The prevention of Pseudomonas aeruginosaassociated diarrhea and conclusion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Riddle, M.S.; DuPont, H.L.; Connor, B.A. ACG clinical guideline: Diagnosis, treatment, and prevention of acute diarrheal infections in adults. Am. J. Gastroenterol. 2016, 111, 602–622. [Google Scholar] [CrossRef]
- Keusch, G.T.; et al. Chapter 19. Diarrheal diseases. In Disease control priorities in developing countries, 2nd ed.; Oxford University Press: Oxford, UK, 2006; p. 37188. [Google Scholar] [CrossRef]
- Talan, D.; Moran, G.J.; Newdow, M.; et al. Etiology of bloody diarrhea among patients presenting to United States emergency departments: Prevalence of Escherichia coli O157:H7 and other enteropathogens. Clin. Infect. Dis. 2001, 32, 573–580. [Google Scholar] [CrossRef]
- Tam, C.C.; O’Brien, S.J.; Tompkins, D.S.; et al. Changes in causes of acute gastroenteritis in the United Kingdom over 15 years: Microbiologic findings from 2 prospective, population-based studies of infectious intestinal disease. Clin. Infect. Dis. 2012, 54, 1275–1286. [Google Scholar] [CrossRef]
- Metreveli, M.; Bulia, S.; Shalamberidze, I.; et al. Campylobacteriosis, shigellosis and salmonellosis in hospitalized children with acute inflammatory diarrhea in Georgia. Pathogens 2022, 11, 232. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Zhang, J.; Li, Y.; et al. The 12 gastrointestinal pathogens spectrum of acute infectious diarrhea in a sentinel hospital, Shenzhen, China. Front. Microbiol. 2016, 7, 1926. [Google Scholar] [CrossRef] [PubMed]
- Jafari, F.; Shokrzadeh, L.; Hamidian, M.; SalmanzadehAhrabi, S.; Zali, M.R. Acute diarrhea due to enteropathogenic bacteria in patients at hospitals in Tehran. Jpn. J. Infect. Dis. 2008, 61, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Viola, L.; Langer, A.; Pulitanò, S.; Chiaretti, A.; Piastra, M.; Polidori, G. Serious Pseudomonas aeruginosa infection in healthy children: Case report and review of the literature. Pediatr. Int. 2006, 48, 330–333. [Google Scholar] [CrossRef] [PubMed]
- Marcon, A.P.; Gamba, M.A.; Vianna, L.A. Nosocomial diarrhea in the intensive care unit. Braz. J. Infect. Dis. 2006, 10, 384–389. [Google Scholar] [CrossRef]
- Dold, H. On pyocyaneus sepsis and intestinal infections in Shanghai due to Bacillus pyocyaneus. Chin. Med. J. 1918, 32, 435. [Google Scholar]
- Reynolds, D.; Kollef, M. The epidemiology and pathogenesis and treatment of Pseudomonas aeruginosa infections: An update. Drugs 2021, 81, 2117–2131. [Google Scholar] [CrossRef]
- Chuang, C.H.; Janapatla, R.P.; Wang, Y.H.; et al. Pseudomonas aeruginosa-associated diarrheal diseases in children. Pediatr. Infect. Dis. J. 2017, 36, 1119–1123. [Google Scholar] [CrossRef]
- Estepa, V.; Rojo-Bezares, B.; Torres, C.; Sáenz, Y. Faecal carriage of Pseudomonas aeruginosa in healthy humans: Antimicrobial susceptibility and global genetic lineages. FEMS Microbiol. Ecol. 2014, 89, 15–19. [Google Scholar] [CrossRef]
- Valenza, G.; Tuschak, C.; Nickel, S.; Krupa, E.; LehnerReindl, V.; Höller, C. Prevalence, antimicrobial susceptibility, and genetic diversity of Pseudomonas aeruginosa as intestinal colonizer in the community. Infect. Dis. 2015, 47, 654–657. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Roldán, L.; Bellés, A.; Bueno, J.; et al. Pseudomonas aeruginosa isolates from Spanish children: Occurrence in faecal samples, antimicrobial resistance, virulence, and molecular typing. Biomed. Res. Int. 2018, 2018, 8060178. [Google Scholar] [CrossRef] [PubMed]
- Gilboa-Garber, N. Psuedomonas aeruginosa lectins. Methods Enzymol. 1982, 83, 378–385. [Google Scholar] [CrossRef] [PubMed]
- Laughlin, R.S.; Musch, M.W.; Hollbrook, C.J.; Rocha, F.M.; Chang, E.B.; Alverdy, J.C. The key role of Pseudomonas aeruginosa PA-I lectin on experimental gut-derived sepsis. Ann. Surg. 2000, 232, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Okuda, J.; Hayashi, N.; Okamoto, M.; et al. Translocation of Pseudomonas aeruginosa from the intestinal tract is mediated by the binding of ExoS to an Na,K-ATPase regulator, FXYD3. Infect Immun. 2010, 78, 4511–4522. [Google Scholar] [CrossRef] [PubMed]
- Ohara, T.; Itoh, K. Significance of Pseudomonas aeruginosa colonization of the gastrointestinal tract. Intern. Med. 2003, 42, 1072–1076. [Google Scholar] [CrossRef]
- Hu, Y.; Qing, Y.; Chen, J.; et al. Prevalence, risk factors, and molecular epidemiology of intestinal carbapenemresistant Pseudomonas aeruginosa. Microbiol Spectr. 2021, 9, e0134421. [Google Scholar] [CrossRef]
- Wang, L.; He, Y.; Li, H.; Ai, Q.; Yu, J. The microbiota protects against Pseudomonas aeruginosa pneumonia via γδ T cell-neutrophil axis in mice. Microbes Infect. 2020, 22, 294–302. [Google Scholar] [CrossRef]
- Horcajada, J.P.; Montero, M.; Oliver, A.; et al. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin Microbiol Rev. 2019, 32, e00031-19. [Google Scholar] [CrossRef]
- Pettigrew, M.M.; Gent, J.F.; Kong, Y.; et al. Gastrointestinal microbiota disruption and risk of colonization with carbapenem-resistant Pseudomonas aeruginosa in intensive care unit patients. Clin. Infect. Dis. 2019, 69, 604–613. [Google Scholar] [CrossRef] [PubMed]
- Markou, P.; Apidianakis, Y. Pathogenesis of intestinal Pseudomonas aeruginosa infection in patients with cancer. Front. Cell Infect. Microbiol. 2014, 3, 115. [Google Scholar] [CrossRef] [PubMed]
- Wheatley, R.M.; Caballero, J.D.; van der Schalk, T.E.; et al. Gut to lung translocation and antibiotic mediated selection shape the dynamics of Pseudomonas aeruginosa in an ICU patient. Nat. Commun. 2022, 13, 6523. [Google Scholar] [CrossRef]
- Cohen, R.; Babushkin, F.; Cohen, S.; et al. A prospective survey of Pseudomonas aeruginosa colonization and infection in the intensive care unit. Antimicrob. Resist. Infect. Control. 2017, 6, 7. [Google Scholar] [CrossRef]
- Gómez-Zorrilla, S.; Camoez, M.; Tubau, F.; et al. Antibiotic pressure is a major risk factor for rectal colonization by multidrug-resistant Pseudomonas aeruginosa in critically ill patients. Antimicrob. Agents Chemother. 2014, 58, 586370. [Google Scholar] [CrossRef]
- Polage, C.R.; Solnick, J.V.; Cohen, S.H. Nosocomial diarrhea: Evaluation and treatment of causes other than Clostridium difficile. Clin. Infect. Dis. 2012, 55, 982–989. [Google Scholar] [CrossRef] [PubMed]
- Ford-Jones, E.L.; Mindorff, C.M.; Gold, R.; Petric, M. The incidence of viral-associated diarrhea after admission to a pediatric hospital. Am. J. Epidemiol. 1990, 131, 711–718. [Google Scholar] [CrossRef]
- McFarland, L.V. Diarrhea acquired in the hospital. Gastroenterol. Clin. North. Am. 1993, 22, 563–577. [Google Scholar] [CrossRef]
- Chuang, C.H.; Janapatla, R.P.; Wang, Y.H.; Chang, H.J.; Chen, C.L.; Chiu, C.H. Association between histo-blood group antigens and Pseudomonas aeruginosa-associated diarrheal diseases. J. Microbiol. Immunol. Infect. 2023, 56, 367–372. [Google Scholar] [CrossRef]
- Steinbrückner, B.; Fehrenbach, J.; Philippczik, G.; Kist, M.; Bauer, T.M. Clinical significance of pure or predominant growth of Pseudomonas aeruginosa in faecal specimens of medical patients. J. Hosp. Infect. 1999, 43, 164–165. [Google Scholar] [CrossRef]
- Voorhies, A.A.; Lorenzi, H.A. The challenge of maintaining a healthy microbiome during long-duration space missions. Front. Astron. Space Sci. 2016, 3, 23. [Google Scholar] [CrossRef]
- Larcombe, S.; Hutton, M.L.; Riley, T.V.; Abud, H.E.; Lyras, D. Diverse bacterial species contribute to antibioticassociated diarrhoea and gastrointestinal damage. J. Infect. 2018, 77, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.W.; Peck, K.R.; Jung, S.I.; et al. Pseudomonas aeruginosa as a potential cause of antibiotic-associated diarrhea. J. Korean Med. Sci. 2001, 16, 742–744. [Google Scholar] [CrossRef][Green Version]
- Hoff, R.T.; Patel, A.; Shapiro, A. Pseudomonas aeruginosa: An uncommon cause of antibiotic-associated diarrhea in an immunocompetent ambulatory adult. Case Rep. Gastrointest. Med. 2020, 2020, 6261748. [Google Scholar] [CrossRef]
- Mishra, S.; Mann, B.; Besmanos, C.; Raza, N.; Heidari, A. A unique case of Pseudomonas aeruginosa-associated diarrhea in a long-term hospitalized adult patient. Cureus 2023, 15, e39978. [Google Scholar] [CrossRef] [PubMed]
- Bresee, J.S.; Marcus, R.; Venezia, R.A.; et al. The etiology of severe acute gastroenteritis among adults visiting emergency departments in the United States. J. Infect. Dis. 2012, 205, 1374–1381. [Google Scholar] [CrossRef]
- Brad, G.F.; Sabau, I.; Simedrea, I.; Belei, O. Pseudomonas aeruginosa and antibiotic-associated diarrhea in children. Timis. Med. J. 2011, 61, 237–242. [Google Scholar]
- Albert, M.J.; Faruque, A.S.; Faruque, S.M.; Sack, R.B.; Mahalanabis, D. Case-control study of enteropathogens associated with childhood diarrhea in Dhaka, Bangladesh. J. Clin. Microbiol. 1999, 37, 3458–3464. [Google Scholar] [CrossRef]
- Lee, A. Adverse drug reactions; Pharmaceutical press, 2006. [Google Scholar] [CrossRef]
- Fang, L.C.; Peng, C.C.; Chi, H.; Lee, K.S.; Chiu, N.C. Pseudomonas aeruginosa sepsis with ecthyma gangrenosum and pseudomembranous pharyngolaryngitis in a 5month-old boy. J. Microbiol. Immunol. Infect. 2014, 47, 158–161. [Google Scholar] [CrossRef]
- De, A.; Mathurkar, H.; Baveja, S.; Manglani, M.V. Pseudomonas diarrhea in a child suffering from acute lymphatic leukemia. Indian. J. Med. Paediatr. Oncol. 2009, 30, 147–148. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stringer, A.M.; Al-Dasooqi, N.; Bowen, J.M.; et al. Biomarkers of chemotherapy-induced diarrhoea: A clinical study of intestinal microbiome alterations, inflammation and circulating matrix metalloproteinases. Support. Care Cancer. 2013, 21, 1843–1852. [Google Scholar] [CrossRef]
- Chua, L.L.; Rajasuriar, R.; Lim, Y.A.L.; Woo, Y.L.; Loke, P.; Ariffin, H. Temporal changes in gut microbiota profile in children with acute lymphoblastic leukemia prior to commencement-, during-, and post-cessation of chemotherapy. BMC Cancer 2020, 20, 151. [Google Scholar] [CrossRef]
- Stein, A.; Voigt, W.; Jordan, K. Chemotherapy-induced diarrhea: Pathophysiology, frequency and guidelinebased management. Ther. Adv. Med. Oncol. 2010, 2, 51–63. [Google Scholar] [CrossRef]
- Penteado, F.D.; Bain, V.; Durigon, G.S.; Litvinov, N.; Pereira, M.F.B.; de Sousa Marques, H.H. Shanghai fever in a healthy infant: First report in South America. Pediatr. Infect. Dis. J. 2018, 37, e278–e279. [Google Scholar] [CrossRef]
- Chuang, C.H.; Wang, Y.H.; Chang, H.J.; et al. Shanghai fever: A distinct Pseudomonas aeruginosa enteric disease. Gut 2014, 63, 736–743. [Google Scholar] [CrossRef]
- Huang, Y.C.; Lin, T.Y.; Wang, C.H. Community-acquired Pseudomonas aeruginosa sepsis in previously healthy infants and children: Analysis of forty-three episodes. Pediatr. Infect. Dis. J. 2002, 21, 1049–1052. [Google Scholar] [CrossRef] [PubMed]
- Chuang, C.H. Case 48. A 4-month-old infant with fever, diarrhea, followed by abdominal distention and an ulcerlike skin lesion: Community-acquired Pseudomonas aeruginosa sepsis (Shanghai fever). In Paediatric Infectious Diseases; Huang, Y.C., Lee, P.I., Chen, P.Y., Eds.; Springer: Singapore, 2023. [Google Scholar] [CrossRef]
- Ortí, A.; Escrig, R.; Pérez-Tamarit, D.; Otero Mdel, C.; Diosdado, N.; Muro, M.D.; Asensi, F. Pseudomonas aeruginosa infection in a previously healthy infant. Clin. Pediatr. 2002, 41, 525–528. [Google Scholar] [CrossRef]
- Mull, C.C.; Scarfone, R.J.; Conway, D. Ecthyma gangrenosum as a manifestation of Pseudomonas sepsis in a previously healthy child. Ann. Emerg. Med. 2000, 36, 383–387. [Google Scholar] [CrossRef] [PubMed]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Matamoros, S.; Gras-Leguen, C.; Le Vacon, F.; Potel, G.; de La Cochetiere, M.F. Development of intestinal microbiota in infants and its impact on health. Trends Microbiol. 2013, 21, 167–173. [Google Scholar] [CrossRef]
- Fan, Q.; Yi, M.; Liu, H.; et al. The impact of age and pathogens type on the gut microbiota in infants with diarrhea in Dalian, China. Can. J. Infect. Dis. Med. Microbiol. 2020, 2020, 8837156. [Google Scholar] [CrossRef] [PubMed]
- Fakhkhari, P.; Tajeddin, E.; Azimirad, M.; et al. Involvement of Pseudomonas aeruginosa in the occurrence of community and hospital acquired diarrhea, and its virulence diversity among the stool and the environmental samples. Int. J. Environ. Health Res. 2022, 32, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, G.; Das, B.; Kumar, S.; et al. Genomic insights into extensively drug-resistant Pseudomonas aeruginosa isolated from a diarrhea case in Kolkata, India. Future Microbiol. 2023, 18, 173–186. [Google Scholar] [CrossRef]
- Adlard, P.A.; Kirov, S.M.; Sanderson, K.; Cox, G.E. Pseudomonas aeruginosa as a cause of infectious diarrhoea. Epidemiol. Infect. 1998, 121, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Tumbarello, M.; Raffaelli, F.; Peghin, M.; et al. Characterisation risk factor profiling of Pseudomonas aeruginosa urinary tract infections: Pinpointing those likely to be caused by multidrug-resistant strains. Int. J. Antimicrob. Agents. 2020, 55, 105900. [Google Scholar] [CrossRef]
- Viasus, D.; Puerta-Alcalde, P.; Cardozo, C.; et al. Predictors of multidrug-resistant Pseudomonas aeruginosa in neutropenic patients with bloodstream infection. Clin. Microbiol. Infect. 2020, 26, 345–350. [Google Scholar] [CrossRef]
- Farooq, L.; Memon, Z.; Ismail, M.O.; Sadiq, S. Frequency and antibiogram of multi-drug resistant Pseudomonas aeruginosa in a tertiary care hospital of Pakistan. Pak. J. Med. Sci. 2019, 35, 1622–1626. [Google Scholar] [CrossRef]
- Procop, G.W.; Church, D.L.; Hall, G.S.; Janda, W.M.; Koneman, E.W.; Schreckenberger, P.C. Koneman’s color atlas and textbook of diagnostic microbiology; Jones & Bartlett Learning: Burlington, MA, USA, 2020. [Google Scholar]
   |
© GERMS 2025.
Share and Cite
Khaledi, M.; Saghabashi, A.; Ghahramanpour, H. The Emerging Role of Pseudomonas aeruginosa in Diarrhea: Where We Stand. Germs 2024, 14, 179-188. https://doi.org/10.18683/germs.2024.1429
Khaledi M, Saghabashi A, Ghahramanpour H. The Emerging Role of Pseudomonas aeruginosa in Diarrhea: Where We Stand. Germs. 2024; 14(2):179-188. https://doi.org/10.18683/germs.2024.1429
Chicago/Turabian StyleKhaledi, Mansoor, Ahdiyeh Saghabashi, and Hossein Ghahramanpour. 2024. "The Emerging Role of Pseudomonas aeruginosa in Diarrhea: Where We Stand" Germs 14, no. 2: 179-188. https://doi.org/10.18683/germs.2024.1429
APA StyleKhaledi, M., Saghabashi, A., & Ghahramanpour, H. (2024). The Emerging Role of Pseudomonas aeruginosa in Diarrhea: Where We Stand. Germs, 14(2), 179-188. https://doi.org/10.18683/germs.2024.1429




