Associated Factors for Bacterial Colonization in Patients Admitted to the Intensive Care Unit of the Clinical Hospital of Infectious Diseases
Abstract
Introduction
Methods
Study population
Data collection
Microbiological data
Results
Prevalence and location of bacterial isolates
The resistance mechanisms of bacterial isolates
| Mechanism of resistance seen in bacterial colonization | (N=53), N (%) |
|---|---|
| ESBL | 20 (37.8) |
| Carbapenemase | 4 (7.6) |
| MRSA | 6 (11.3) |
| VRE | 2 (3.8) |
| ESBL/carbapenemase | 7 (13.2) |
| MRSA/VRE | 1 (1.9) |
| ESBL/carbapenemase/VRE | 6 (11.3) |
| ESBL/VRE | 3 (5.7) |
| ESBL/MRSA | 4 (7.5) |
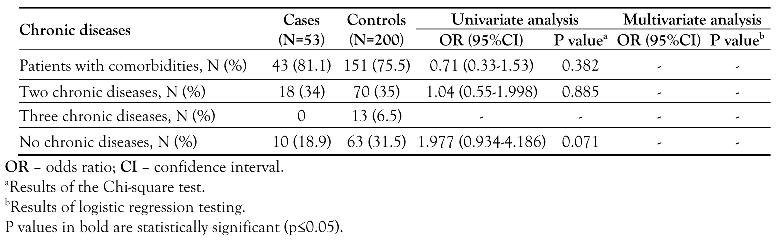
Risk factors associated with bacterial colonization


Discussion
Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Masse, J.; Elkalioubie, A.; Blazejewski, C.; et al. Colonization pressure as a risk factor of ICU-acquired multidrug resistant bacteria: A prospective observational study. Eur J Clin Microbiol Infect Dis 2017, 36, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; De Angelis, G.; Cataldo, M.A.; et al. Antibiotic usage and risk of colonization and infection with antibiotic-resistant bacteria: A hospital population-based study. Antimicrob Agents Chemother 2009, 53, 4264–4269. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; et al. Multidrug-resistant, extensively drug resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Sakai, A.M.; Iensue, T.N.A.N.; Pereira, K.O.; et al. Colonization profile and duration by multi-resistant organisms in a prospective cohort of newborns after hospital discharge. Rev Inst Med Trop Sao Paulo 2020, 62, e22. [Google Scholar] [CrossRef] [PubMed]
- Wohrley, J.D.; Bartlett, A.H. The role of the environment and colonization in healthcare-associated infections. In Healthcare-associated infections in children; McNeil, J., Campbell, J., Crews, J., Eds.; Springer: Cham, 2019. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. ECDC country visit to Romania to discuss antimicrobial resistance issues; ECDC: Stockholm, 2018. [Google Scholar]
- Slavcovici, A.; Streinu-Cercel, A.; Tatulescu, D.; et al. The role of risk factors (‘Carmeli score’) and infective endocarditis classification in the assessment of appropriate empirical therapy. Ther Pharmacol Clin Toxicol 2009, 13, 52–56. [Google Scholar]
- Tuty Kuswardhani, R.A.; Henrina, J.; Pranata, R.; Anthonius Lim, M.; Lawrensia, S.; Suastika, K. Charlson comorbidity index and a composite of poor outcomes in COVID-19 patients: A systematic review and meta-analysis. Diabetes Metab Syndr 2020, 14, 2103–2109. [Google Scholar] [CrossRef] [PubMed]
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 9.0. 2019. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_9.0_Breakpoint_Tables.pdf (accessed on 2 August 2022).
- European Centre for Disease Prevention and Control. Antimicrobial resistance in the EU/EEA (EARS-Net) - Annual Epidemiological Report 2019; ECDC: Stockholm, 2020. [Google Scholar]
- Pana, Z.D.; Zaoutis, T. Treatment of extended-spectrum β-lactamase-producing Enterobacteriaceae (ESBLs) infections: What have we learned until now? F1000Res 2018, 7, F1000 Faculty Rev-1347. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control Prevention. Healthcare facilities: information about CRE. 2019. Available online: https://www.cdc.gov/hai/organisms/cre/cre-facilities.html (accessed on 3 August 2022).
- Morrill, H.J.; Pogue, J.M.; Kaye, K.S.; LaPlante, K.L. Treatment options for carbapenem-resistant Enterobacteriaceae infections. Open Forum Infect Dis 2015, 2, ofv050. [Google Scholar] [CrossRef] [PubMed]
- Lixandru, B.E.; Cotar, A.I.; Straut, M.; et al. Carbapenemase-producing Klebsiella pneumoniae in Romania: A six-month survey. PLoS ONE 2015, 10, e0143214. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Vancomycin-resistant Enterococci (VRE) in healthcare settings. 2019. Available online: https://www.cdc.gov/hai/organisms/vre/vre.html (accessed on 23 July 2022).
- Pirii, L.E.; Friedrich, A.W.; Rossen, J.W.A.; et al. Extensive colonization with carbapenemase-producing microorganisms in Romanian burn patients: Infectious consequences from the Colectiv fire disaster. Eur J Clin Microbiol Infect Dis 2018, 37, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Ungureanu, A.; Zlatian, O.; Mitroi, G.; et al. Staphylococcus aureus colonisation in patients from a primary regional hospital. Mol Med Rep 2017, 16, 8771–8780. [Google Scholar] [CrossRef] [PubMed]
- Ion Nedelcu, N.; Petre Iacob, C. Predictors for MDRO carriage in adult patients of a infectious diseases clinic from Bucharest, Romania. Int J Infect Dis 2021, 101 (Suppl. 1), 25–26. [Google Scholar] [CrossRef]
- Dautzenberg, M.J.; Wekesa, A.N.; Gniadkowski, M.; et al. The association between colonization with carbapenemase-producing enterobacteriaceae and overall ICU mortality: An observational cohort study. Crit Care Med 2015, 43, 1170–1177. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, S.; Fatima, J.; Shakil, S.; Rizvi, S.M.; Kamal, M.A. Antibiotic resistance and extended spectrum beta-lactamases: Types, epidemiology and treatment. Saudi J Biol Sci 2015, 22, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Elshamy, A.A.; Aboshanab, K.M. A review on bacterial resistance to carbapenems: Epidemiology, detection and treatment options. Future Sci OA 2020, 6, FSO438. [Google Scholar] [CrossRef] [PubMed]
- Wyres, K.L.; Hawkey, J.; Mirčeta, M.; et al. Genomic surveillance of antimicrobial resistant bacterial colonisation and infection in intensive care patients. BMC Infect Dis 2021, 21, 683. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Antimicrobial resistance. National action plan. 2021. Available online: https://www.cdc.gov/drugresistance/us-activities/national-action-plan.html (accessed on 5 August 2022).
- WHO Regional Office for Europe; European Centre for Disease Prevention and Control. Antimicrobial resistance surveillance in Europe 2022 - 2020 data; WHO Regional Office for Europe: Copenhagen, 2022. [Google Scholar]
| Bacterial strains | |
|---|---|
| All microorganisms | N=53, N (%) |
| Klebsiella spp. | 12 (22.6) |
| Escherichia coli | 17 (32.1) |
| Staphylococcus aureus | 6 (11.3) |
| Enterococcus faecium | 2 (3.8) |
| Klebsiella spp. and Acinetobacter baumannii | 1 (1.9) |
| Klebsiella spp. and Enterobacter cloacae | 2 (1.9) |
| Klebsiella spp. and Staphylococcus aureus | 1 (1.9) |
| Klebsiella spp. and Enterococcus faecium | 3 (5.7) |
| Klebsiella spp. and Enterococcus faecalis | 1 (1.9) |
| Escherichia coli and Staphylococcus aureus | 1 (1.9) |
| Escherichia coli and Enterococcus faecium | 1 (1.9) |
| Staphylococcus aureus and other Enterobacteriaceae | 1 (1.9) |
| Staphylococcus aureus and Acinetobacter baumannii | 1 (1.9) |
| Staphylococcus aureus and Enterococcus faecium | 1 (1.9) |
| Klebsiella spp., Escherichia coli and Enterococcus faecium | 1 (1.9) |
| Klebsiella spp., Enterococcus faecium and Enterococcus faecalis | 2 (3.8) |
© GERMS 2023.
Share and Cite
Vlad, N.-D.; Voidăzan, S.; Căpâlnă, A.; Cernat, R.-C.; Carp, S.-D.; Mitan, R.; Dumitru, A.; Rugină, S.; Nemet, C.; Dumitru, I.M. Associated Factors for Bacterial Colonization in Patients Admitted to the Intensive Care Unit of the Clinical Hospital of Infectious Diseases. Germs 2023, 13, 10-19. https://doi.org/10.18683/germs.2023.1362
Vlad N-D, Voidăzan S, Căpâlnă A, Cernat R-C, Carp S-D, Mitan R, Dumitru A, Rugină S, Nemet C, Dumitru IM. Associated Factors for Bacterial Colonization in Patients Admitted to the Intensive Care Unit of the Clinical Hospital of Infectious Diseases. Germs. 2023; 13(1):10-19. https://doi.org/10.18683/germs.2023.1362
Chicago/Turabian StyleVlad, Nicoleta-Dorina, Septimiu Voidăzan, Andreea Căpâlnă, Roxana-Carmen Cernat, Sorina-Dalia Carp, Romelia Mitan, Andrei Dumitru, Sorin Rugină, Codruţa Nemet, and Irina Magdalena Dumitru. 2023. "Associated Factors for Bacterial Colonization in Patients Admitted to the Intensive Care Unit of the Clinical Hospital of Infectious Diseases" Germs 13, no. 1: 10-19. https://doi.org/10.18683/germs.2023.1362
APA StyleVlad, N.-D., Voidăzan, S., Căpâlnă, A., Cernat, R.-C., Carp, S.-D., Mitan, R., Dumitru, A., Rugină, S., Nemet, C., & Dumitru, I. M. (2023). Associated Factors for Bacterial Colonization in Patients Admitted to the Intensive Care Unit of the Clinical Hospital of Infectious Diseases. Germs, 13(1), 10-19. https://doi.org/10.18683/germs.2023.1362




