Microbiological and Drug Resistance Patterns of Bronchoalveolar Lavage Samples Taken from Hospitalized Patients in Iran
Abstract
Introduction
Methods
Sample collection and storage
Bacterial identification using culture
Antibiotic susceptibility testing of the cultured bacteria
Identification of the fastidious bacteria using PCR
Statistical analysis
Results
Demographic and clinical information of the hospitalized patients
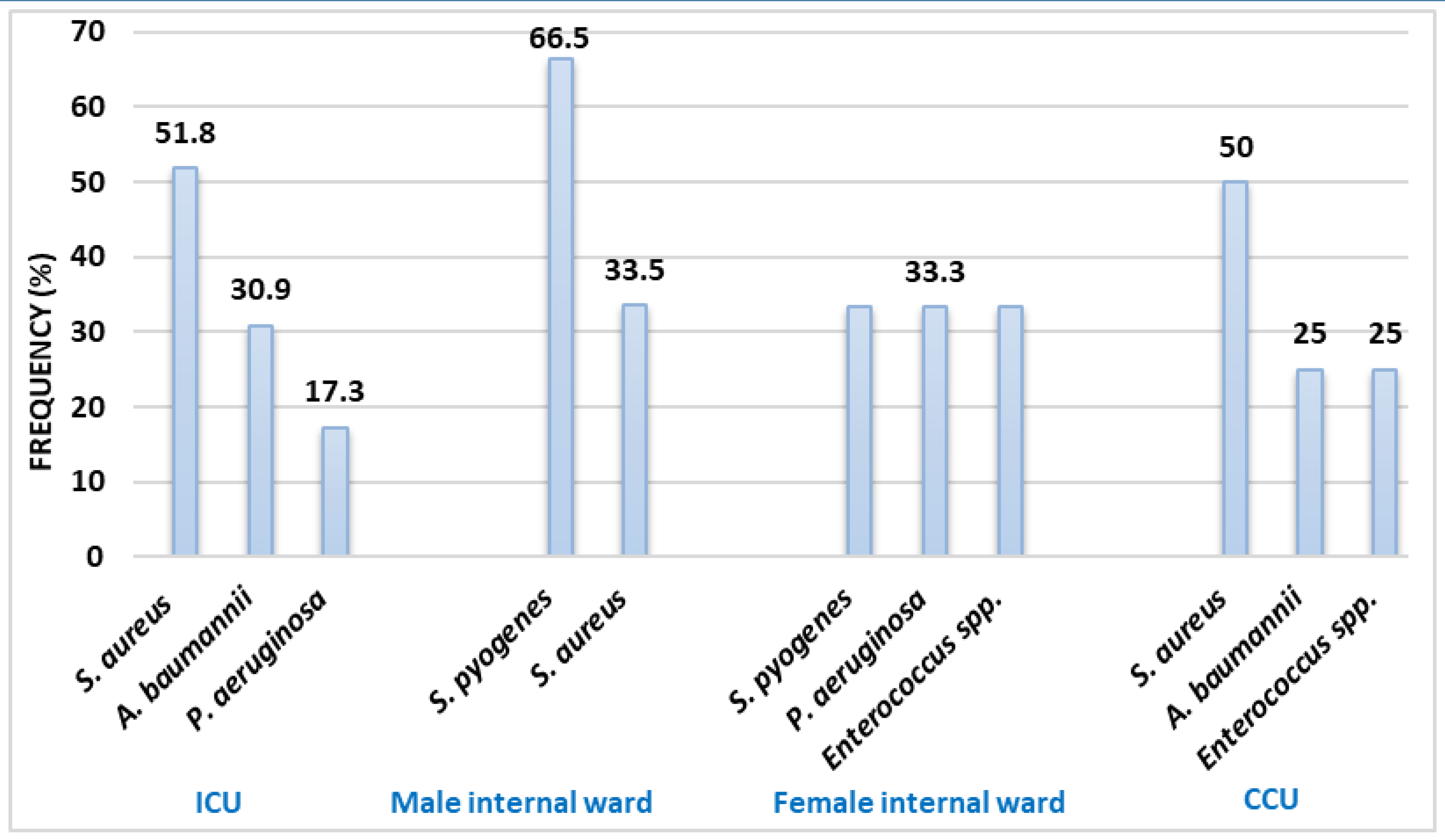
Frequency of the microorganisms isolated from BAL culture
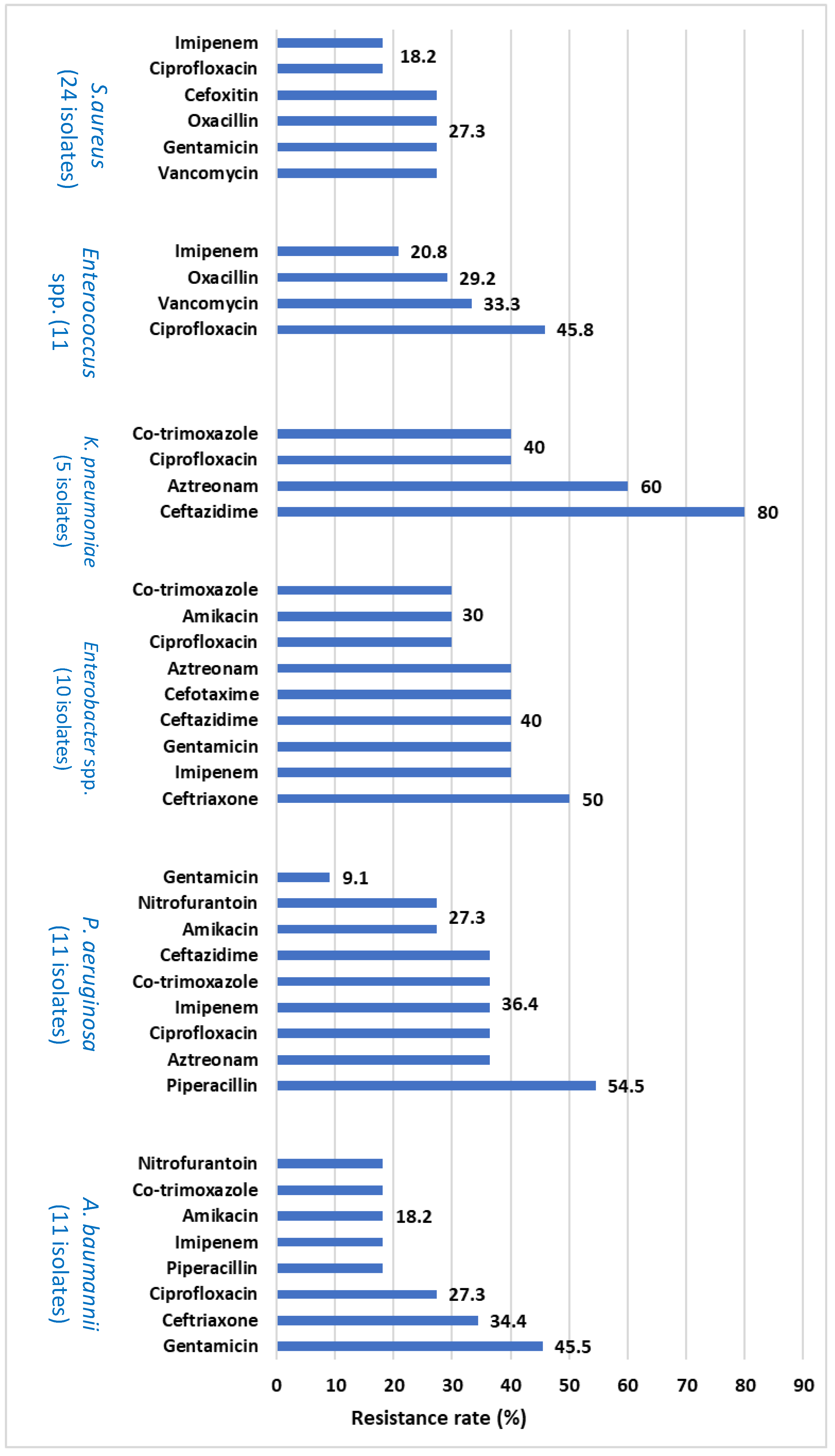
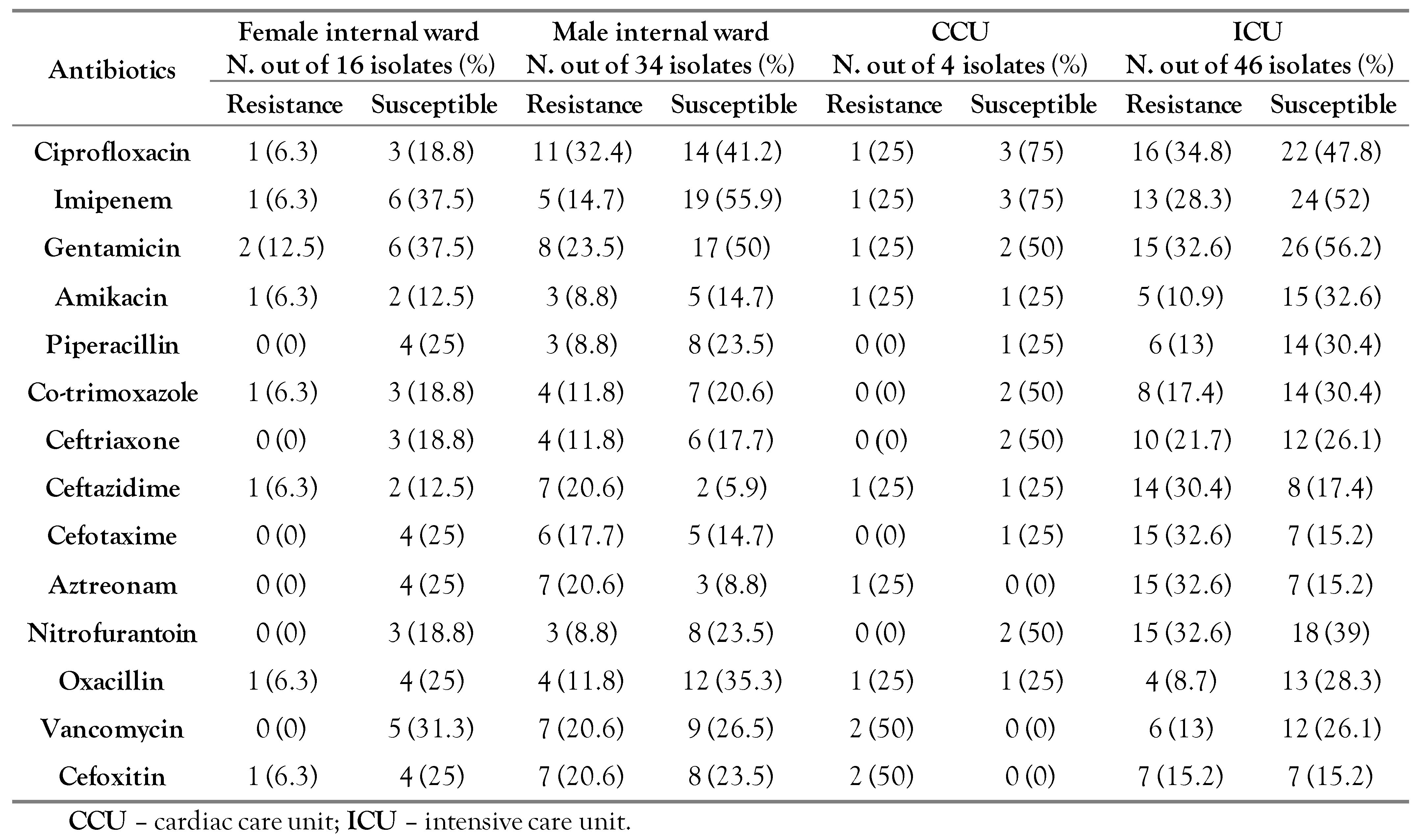
Antibiotic resistance patterns of the bacteria isolated from BAL using the DAD method
PCR detection of fastidious bacteria in the BAL sample
Discussion
Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Flanders, S.A.; Collard, H.R.; Saint, S. Nosocomial pneumonia: State of the science. Am J Infect Control. 2006, 34, 84–93. [Google Scholar] [CrossRef]
- Pezhman, B.; Fatemeh, R.; Amir, R.; Mahboobeh, R.; Mohammad, F. Nosocomial infections in an Iranian educational hospital: An evaluation study of the Iranian nosocomial infection surveillance system. BMC Infect Dis. 2021, 21, 1256. [Google Scholar] [CrossRef] [PubMed]
- Auerbach, P.S. Wilderness Medicine E-Book: Expert Consult Premium Edition-Enhanced Online Features. Elsevier Health Sciences, 2011. [Google Scholar]
- Berenholtz, S.M.; Pronovost, P.J.; Lipsett, P.A.; et al. Eliminating catheter-related bloodstream infections in the intensive care unit. Crit Care Med. 2004, 32, 2014–2020. [Google Scholar] [CrossRef] [PubMed]
- American Thoracic Society; Infectious Diseases Society of America. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am J Respir Crit Care Med. 2005, 171, 388–416. [Google Scholar] [CrossRef] [PubMed]
- Jalalpour, S.; Kermanshahi, R.K.; Nouhi, A.S.; Esfahani, H.Z. Surveying the frequency of β-lactamase enzyme and antibiotic sensitivity pattern in isolated pathogen bacteria from low and high hospital contact surfaces. Pejouhandeh. 2010, 15, 77–82. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. M100S25; CLSI: Wayne, PA, USA, 2021. [Google Scholar]
- Asadollahi, P.; Jabalameli, F.; Beigverdi, R.; Emaneini, M. Assessment of disinfectant and antibiotic susceptibility patterns and multi-locus variable number tandem repeat analysis of Staphylococcus epidermidis isolated from blood cultures. Iran J Microbiol. 2018, 10, 90–97. [Google Scholar]
- Marsh, R.L.; Kaestli, M.; Chang, A.B.; et al. The microbiota in bronchoalveolar lavage from young children with chronic lung disease includes taxa present in both the oropharynx and nasopharynx. Microbiome. 2016, 4, 37. [Google Scholar] [CrossRef]
- Kollef, M.H.; Torres, A.; Shorr, A.F.; Martin-Loeches, I.; Micek, S.T. Nosocomial infection. Crit Care Med. 2021, 49, 169–187. [Google Scholar] [CrossRef]
- de la Varga-Martínez, O.; Gómez-Sánchez, E.; Muñoz, M.F.; et al. Impact of nosocomial infections on patient mortality following cardiac surgery. J Clin Anesth. 2021, 69, 110104. [Google Scholar] [CrossRef]
- Unal, S.; Garcia-Rodriguez, J.A. Activity of meropenem and comparators against Pseudomonas aeruginosa and Acinetobacter spp. isolated in the MYSTIC Program, 2002-2004. Diagn Microbiol Infect Dis. 2005, 53, 265–271. [Google Scholar] [CrossRef]
- Santella, B.; Serretiello, E.; De Filippis, A.; et al. Lower respiratory tract pathogens and their antimicrobial susceptibility pattern: A 5-year study. Antibiotics 2021, 10, 851. [Google Scholar] [CrossRef]
- Kuo, S.C.; Liu, C.E.; Lu, P.L.; et al. Activity of ceftolozane-tazobactam against Gram-negative pathogens isolated from lower respiratory tract infections in the Asia-Pacific region: SMART 2015-2016. Int J Antimicrob Agents. 2020, 55, 105883. [Google Scholar] [CrossRef] [PubMed]
- Imani Fooladi, A.; Parvizi, E.; Soltanpour, M.; Ahmadi, A. Study of prevalence and antimicrobial susceptibility pattern of polybacterial pneumonia. Tehran Univ Med J. 2015, 73, 632–638. [Google Scholar]
- Mazloomirad, F.; Hasanzadeh, S.; Sharifi, A.; Nikbakht, G.; Roustaei, N.; Khoramrooz, S.S. Identification and detection of pathogenic bacteria from patients with hospital-acquired pneumonia in southwestern Iran; evaluation of biofilm production and molecular typing of bacterial isolates. BMC Pulm Med. 2021, 21, 408. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ren, L.; Zou, J. Risk factors and prevention strategies of nosocomial infection in geriatric patients. Can J Infect Dis Med Microbiol. 2019, 2019, 6417959. [Google Scholar] [CrossRef]
- Mohammad Shafiei, P.; Baserisalehi, M.; Mobasherizade, S. Investigating the antibiotic resistance prevalence and phenotypic and genotypic evaluation of AcrAB-OprM efflux pump in multidrug-resistant in clinical isolates of Moraxella catarrhalis in Kazerun City, Iran. Iran J Med Microbiol. 2020, 14, 388–407. [Google Scholar] [CrossRef]
- Jiang, C.; Chen, Q.; Xie, M. Smoking increases the risk of infectious diseases: A narrative review. Tob Induc Dis. 2020, 18, 60. [Google Scholar] [CrossRef]
- Keivanfar, M.; Zibanejad, N.; Rahimi, H.; Babaei, S.; Emadoleslami, M.S.; Reisi, M. Treatment protocol of ventilator-associated pneumonia based on microbial susceptibility in pediatric intensive care unit, Isfahan, Iran. Int J Pediatr. 2020, 8, 12039–12051. [Google Scholar]
- Broekema, N.M.; Van, T.T.; Monson, T.A.; Marshall, S.A.; Warshauer, D.M. Comparison of cefoxitin and oxacillin disk diffusion methods for detection of mecA-mediated resistance in Staphylococcus aureus in a large-scale study. J Clin Microbiol. 2009, 47, 217–219. [Google Scholar] [CrossRef]
- Uddin, M.; Ahn, J. Associations between resistance phenotype and gene expression in response to serial exposure to oxacillin and ciprofloxacin in Staphylococcus aureus. Lett Appl Microbiol. 2017, 65, 462–468. [Google Scholar] [CrossRef]
- Naccache, S.N.; Callan, K.; Burnham, C.A.; et al. Evaluation of oxacillin and cefoxitin disk diffusion and microbroth dilution methods for detecting mecA-mediated β-lactam resistance in contemporary Staphylococcus epidermidis isolates. J Clin Microbiol. 2019, 57, e00961–19. [Google Scholar] [CrossRef]
- Peacock, S.J.; Paterson, G.K. Mechanisms of methicillin resistance in Staphylococcus aureus. Annu Rev Biochem. 2015, 84, 577–601. [Google Scholar] [CrossRef]
- Tang, M.; Wang, D.; Tong, X.; et al. Comparison of different detection methods for Mycoplasma pneumoniae infection in children with community-acquired pneumonia. BMC Pediatr. 2021, 21, 90. [Google Scholar] [CrossRef]
- Ieven, M.; Ursi, D.; Van Bever, H.; Quint, W.; Niesters, H.; Goossens, H. Detection of Mycoplasma pneumoniae by two polymerase chain reactions and role of M. pneumoniaein acute respiratory tract infections in pediatric patients. J Infect Dis. 1996, 173, 1445–1452. [Google Scholar] [CrossRef]
 |
 |
 |
© GERMS 2022.
Share and Cite
Tahmasebi, Z.; Asadollahi, P.; Sadeghifard, N.; Ghafourian, S.; Kalani, B.S.; Kalaei, E.G.P.; Pakzad, I. Microbiological and Drug Resistance Patterns of Bronchoalveolar Lavage Samples Taken from Hospitalized Patients in Iran. GERMS 2022, 12, 333-343. https://doi.org/10.18683/germs.2022.1337
Tahmasebi Z, Asadollahi P, Sadeghifard N, Ghafourian S, Kalani BS, Kalaei EGP, Pakzad I. Microbiological and Drug Resistance Patterns of Bronchoalveolar Lavage Samples Taken from Hospitalized Patients in Iran. GERMS. 2022; 12(3):333-343. https://doi.org/10.18683/germs.2022.1337
Chicago/Turabian StyleTahmasebi, Zahra, Parisa Asadollahi, Nourkhoda Sadeghifard, Sobhan Ghafourian, Behrooz Sadeghi Kalani, Esmail Ghasemi Pasha Kalaei, and Iraj Pakzad. 2022. "Microbiological and Drug Resistance Patterns of Bronchoalveolar Lavage Samples Taken from Hospitalized Patients in Iran" GERMS 12, no. 3: 333-343. https://doi.org/10.18683/germs.2022.1337
APA StyleTahmasebi, Z., Asadollahi, P., Sadeghifard, N., Ghafourian, S., Kalani, B. S., Kalaei, E. G. P., & Pakzad, I. (2022). Microbiological and Drug Resistance Patterns of Bronchoalveolar Lavage Samples Taken from Hospitalized Patients in Iran. GERMS, 12(3), 333-343. https://doi.org/10.18683/germs.2022.1337




