Occurrence of Cryptococcus neoformans and Other Yeast-like Fungi in Environmental Sources in Bonaire (Dutch Caribbean)
Abstract
Introduction
Methods
Sampling
Identification of isolates
Results
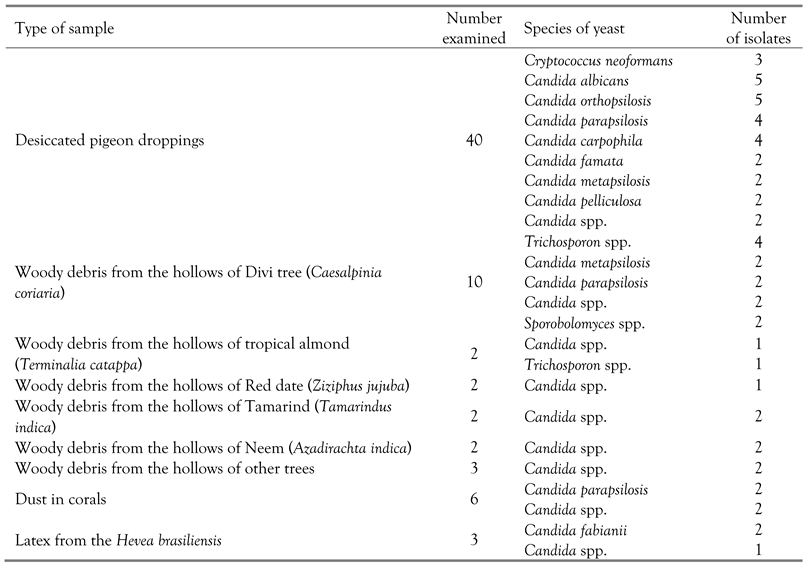
Discussion
Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hagen, F.; Khayhan, K.; Theelen, B. , et al. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet Biol. 2015, 78, 16–48. [Google Scholar] [CrossRef]
- Mitchell, T.G.; Castañeda, E.; Nielsen, K.; Wanke, B.; Lazéra, M.S. Environmental niches for Cryptococcus neoformans and Cryptococcus gattii. In: Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadevall A (eds). Cryptococcus: from human pathogen to model yeast. ASM Press: Washington, DC, USA, 2011. [Google Scholar] [CrossRef]
- Hagen, F.; Chowdhary, A.; Prakash, A.; Yntema, J.B.; Meis, J.F. Molecular characterization of Cryptococcus gattii genotype AFLP6/VGII isolated from woody debris of divi-divi (Caesalpinia coriaria), Bonaire, Dutch Caribbean. Rev Iberoam Micol. 2014, 31, 193–196. [Google Scholar] [CrossRef]
- Tendolkar, U.; Tainwala, S.; Jog, S.; Mathur, M. Use of a new medium -tobacco agar, for pigment production of Cryptococcus neoformans. Indian J Med Microbiol. 2003, 21, 277–279. [Google Scholar] [CrossRef]
- Arsic Arsenijevic, V.; Pekmezovic, M.G.; Meis, J.F.; Hagen, F. Molecular epidemiology and antifungal susceptibility of Serbian Cryptococcus neoformans isolates. Mycoses. 2014, 57, 380–387. [Google Scholar] [CrossRef]
- Loperena-Alvarez, Y.; Ren, P.; Li, X. , et al. Genotypic characterization of environmental isolates of Cryptococcus gattii from Puerto Rico. Mycopathologia. 2010, 170, 279–85. [Google Scholar] [CrossRef]
- Illnait-Zaragozi, M.T.; Martínez-Machín, G.F.; Fernández-Andreu, C.M.; Perurena-Lancha, M.R.; Hagen, F.; Meis, J.F. Cryptococcus and cryptococcosis in Cuba. A minireview. Mycoses. 2014, 57, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Vanhove, M.; Beale, M.A.; Rohdes, J. , et al Genomic epidemiology of Cryptococcus yeasts identifies adaptation to environmental niches underpinning infection across an African HIV/AIDS cohort. Mol Ecol. 2017, 26, 1991–2005. [Google Scholar] [CrossRef]
- Pfeiffer, T.J.; Ellis, D.H. Environmental isolation of Cryptococcus neoformans var. gattii from Eucalyptus terreticornis. J Med Vet Mycol. 1992, 30, 407–408. [Google Scholar] [CrossRef] [PubMed]
- Gugnani, H.C.; Mitchell, T.G.; Litvintseva, A.P.; Lengeler, K.; et al. Isolation of Cryptococcus gattii and C. neoformans var. grubii from the flowers and bark of Eucalyptus trees in India. Med Mycol. 2005, 43, 565–569. [Google Scholar] [CrossRef] [PubMed]
- Rosario Medina, I.; Román Fuentes, L.; Batista Arteaga, M. , et al. Pigeons and their droppings as reservoirs of Candida and other zoonotic yeasts. Rev Iberoam Micol. 2017, 34, 211–214. [Google Scholar] [CrossRef]
- Njoku-Obi, A.N.; Okofor, J.I.; Gugnani, H.C. Yeast-like fungi recovered from normal human skin in Nuskka (Nigeria). Antonie van Leeuwenhoek. 1976, 42, 101–105. [Google Scholar] [CrossRef]
- Asadzadeh, M.; Ahmad, S.; Al-Sweih, N.; Hagen, F.; Meis, J.F.; Khan, Z. High-resolution fingerprinting of Candida parapsilosis isolates suggests persistence and transmission of infections among neonatal intensive care unit patients in Kuwait. Sci Rep. 2019, 9, 1340. [Google Scholar] [CrossRef]
- Martini, C.; Torelli, R.; de Groot, T. , et al. Prevalence and clonal distribution of azole-resistant Candida parapsilosis isolates causing bloodstream infections in a large Italian hospital. Front Cell Infect Microbiol. 2020, 10, 232. [Google Scholar] [CrossRef]
- Trofa, D.; Gácser, A.; Nosanchuk, J.D. Candida parapsilosis, an emerging fungal pathogen. Clin Microbial Rev. 2008, 21, 606–625. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, I.M.; Gupta, A.; Gould, K.; Clark, S.C. A fatal fungus. Ann Thorac Surg. 2005, 80, 723–724. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Mi, H.; Wroe, C.; Jaques, B.; Talbot, D. Fatal Candida famata peritonitis complicating sclerosing peritonitis in a peritoneal dialysis patient. Nephrol Dial Transplant. 2006, 21, 2036–2037. [Google Scholar] [CrossRef][Green Version]
- Pisa, D.; Ramos, M.; García, P. , et al. Fungal infection in patients with serpiginous choroiditis or acute zonal occult outer retinopathy. J Clin Microbiol. 2008, 46, 130–5. [Google Scholar] [CrossRef] [PubMed]
- Gácser, A.; Schäfer, W.; Nosanchuk, J.S.; Salomon, S.; Nosanchuk, J.D. Virulence of Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis in reconstituted human tissue models. Fungal Genet Biol. 2007, 44, 1336–1341. [Google Scholar] [CrossRef]
- de Paula Menezes, R.; Silva, F.F.; Melo, S.G.O. , et al. Characterization of Candida species isolated from the hands of the healthcare workers in the neonatal intensive care unit. Med Mycol. 2019, 57, 588–594. [Google Scholar] [CrossRef]
- Maganti, H.B. Species and genotype diversities of yeasts in the clinical and natural environments in Hamilton. A Thesis submitted to the School of Graduate Studies in partial fulfillment of the requirements for the degree Master of Science, McMaster University, 2011.
- Azcón, R.; Perálvarez Mdel, C.; Roldán, A.; Barea, J.M. Arbuscular mycorrhizal fungi, Bacillus cereus, and Candida parapsilosis from a multi contaminated soil alleviate metal toxicity in plants. Microb Ecol. 2010, 59, 668–677. [Google Scholar] [CrossRef]
- Denning, D.W.; Gugnani, H.C. Burden of serious fungal infections in Trinidad and Tobago. Mycoses 2015, 58 Suppl. 5, 80–84. [Google Scholar] [CrossRef] [PubMed]
© GERMS 2020.
Share and Cite
Gugnani, H.C.; Hagen, F.; Meis, J.F.; Chakrabarti, A. Occurrence of Cryptococcus neoformans and Other Yeast-like Fungi in Environmental Sources in Bonaire (Dutch Caribbean). Germs 2020, 10, 195-200. https://doi.org/10.18683/germs.2020.1205
Gugnani HC, Hagen F, Meis JF, Chakrabarti A. Occurrence of Cryptococcus neoformans and Other Yeast-like Fungi in Environmental Sources in Bonaire (Dutch Caribbean). Germs. 2020; 10(3):195-200. https://doi.org/10.18683/germs.2020.1205
Chicago/Turabian StyleGugnani, Harish C, Ferry Hagen, Jacques F Meis, and Arunaloke Chakrabarti. 2020. "Occurrence of Cryptococcus neoformans and Other Yeast-like Fungi in Environmental Sources in Bonaire (Dutch Caribbean)" Germs 10, no. 3: 195-200. https://doi.org/10.18683/germs.2020.1205
APA StyleGugnani, H. C., Hagen, F., Meis, J. F., & Chakrabarti, A. (2020). Occurrence of Cryptococcus neoformans and Other Yeast-like Fungi in Environmental Sources in Bonaire (Dutch Caribbean). Germs, 10(3), 195-200. https://doi.org/10.18683/germs.2020.1205




