Uncovering Quercetin’s Effects against Influenza A Virus Using Network Pharmacology and Molecular Docking
Abstract
:1. Introduction
2. Materials and Methods
2.1. Pharmacokinetic Evaluation and Therapeutic Targets Associated with Quercetin
2.2. Potential Pathological Target Genes Linked to Influenza
2.3. Construction of Compound–Disease Target (C-D) Network
2.4. Biological Function and Pathway Enrichment Analysis
2.5. Compound–Disease Target–Pathway (C-D-P) Network Construction
2.6. Construction of Protein–Protein Interaction (PPI) Network
2.7. Molecular Docking Analysis
3. Results
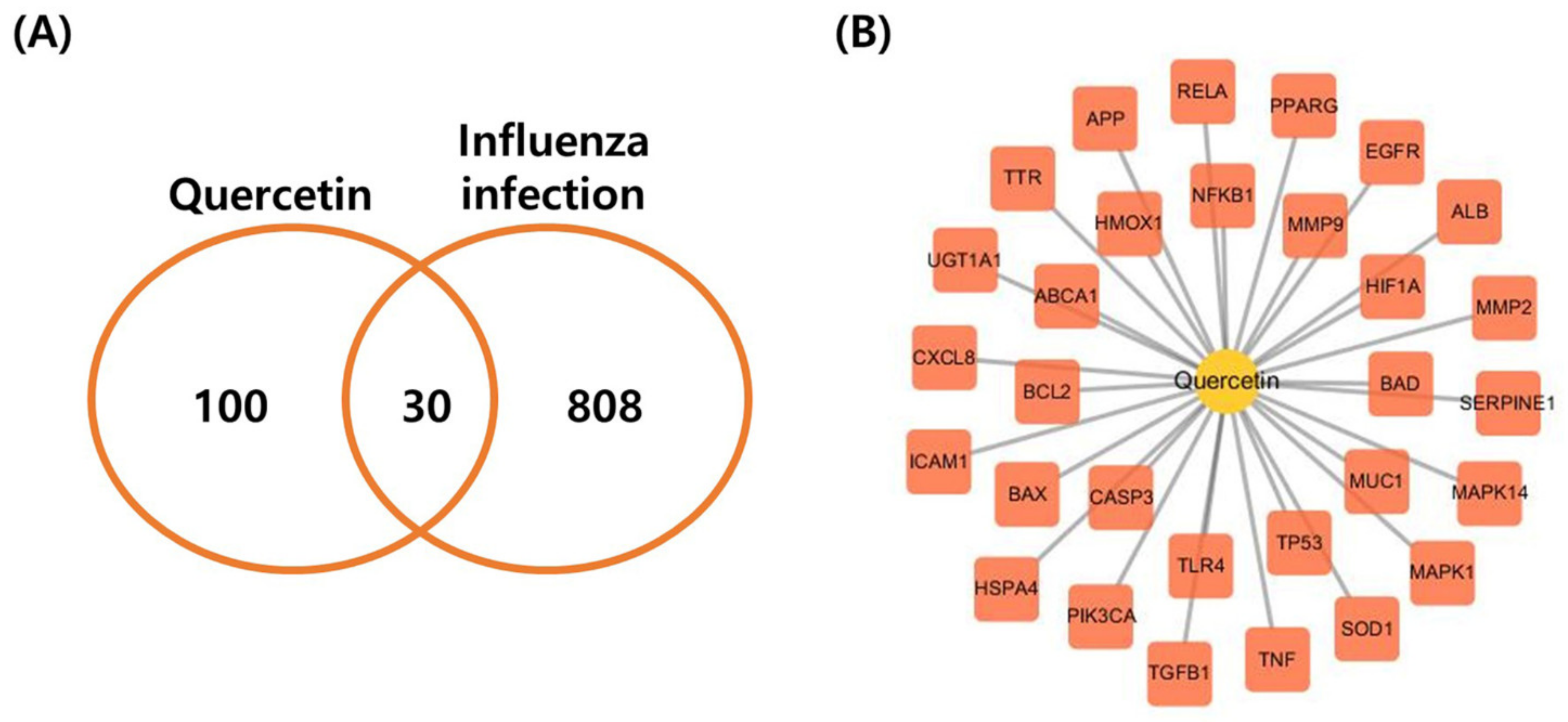
3.1. Biological Targets of Quercetin and Influenza and Compound–Disease Target Network Construction (C-D Network)
3.2. KEGG Pathway Analysis
3.3. Compound–Disease–Target–Pathway (C-D-P) Network
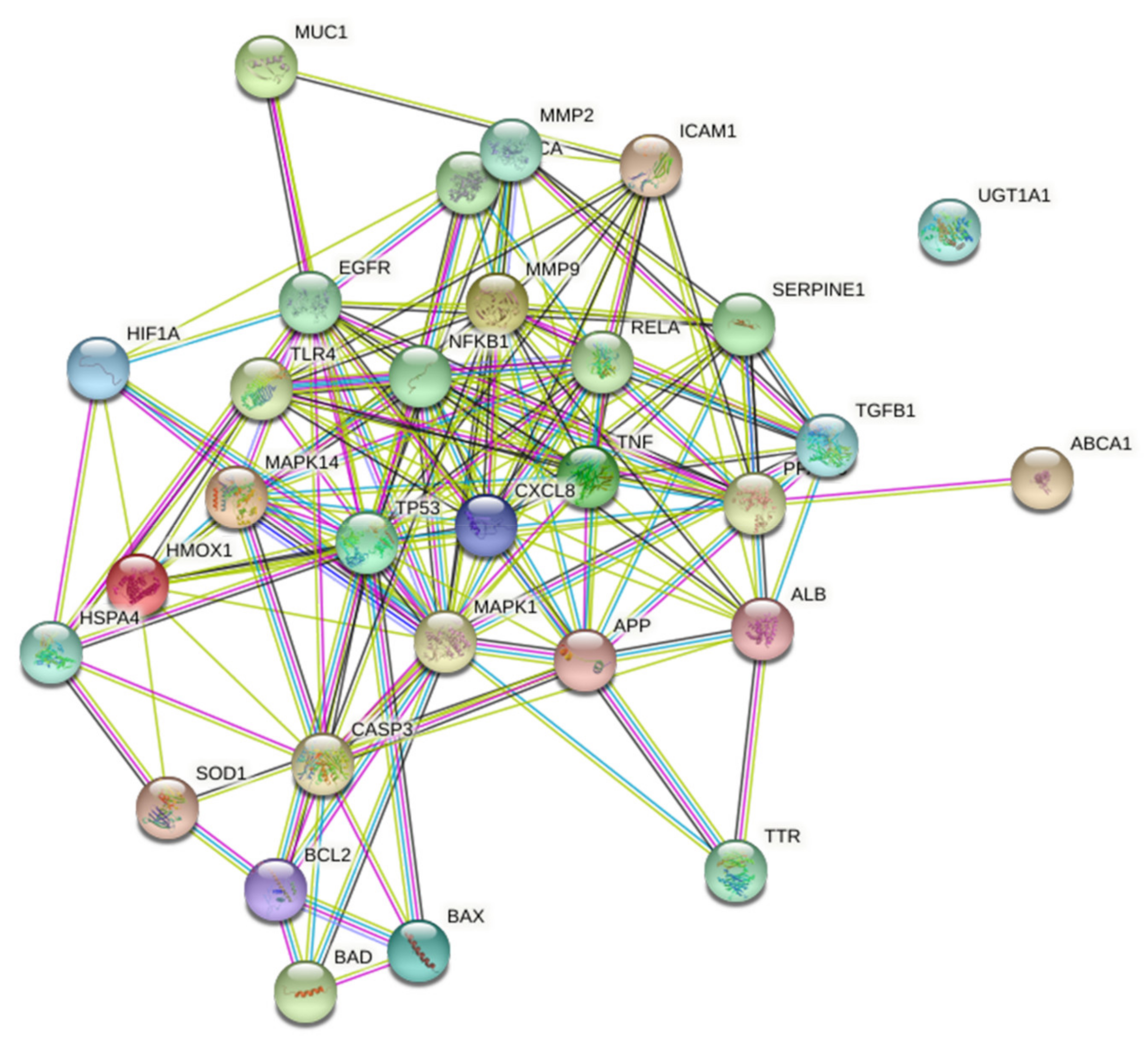
3.4. Protein–Protein Interaction of Disease Targets Associated with Quercetin
3.5. Molecular Docking Analysis
4. Discussion
Author Contributions
Funding
Informed Consent Statement
Conflicts of Interest
References
- Herold, S.; Becker, C.; Ridge, K.M.; Budinger, G.R. Influenza virus-induced lung injury: Pathogenesis and implications for treatment. Eur. Respir. J. 2015, 45, 1463–1478. [Google Scholar] [CrossRef] [Green Version]
- Krammer, F.; Smith, G.J.D.; Fouchier, R.A.M.; Peiris, M.; Kedzierska, K.; Doherty, P.C.; Palese, P.; Shaw, M.L.; Treanor, J.; Webster, R.G.; et al. Influenza. Nat. Rev. Dis. Primers 2018, 4, 3. [Google Scholar] [CrossRef]
- Wu, X.; Sun, Q.; Zhang, C.; Yang, S.; Li, L.; Jia, Z. Progress of small molecular inhibitors in the development of anti-influenza virus agents. Theranostics 2017, 7, 826–845. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.J.; Morris-Natschke, S.L.; Cheng, Y.Y.; Lee, K.H.; Li, R.T. Development of anti-influenza agents from natural products. Med. Res. Rev. 2020, 40, 2290–2338. [Google Scholar] [CrossRef]
- Kim, C.U.; Lew, W.; Williams, M.A.; Liu, H.; Zhang, L.; Swaminathan, S.; Bischofberger, N.; Chen, M.S.; Mendel, D.B.; Tai, C.Y.; et al. Influenza neuraminidase inhibitors possessing a novel hydrophobic interaction in the enzyme active site: Design, synthesis, and structural analysis of carbocyclic sialic acid analogues with potent anti-influenza activity. J. Am. Chem. Soc. 1997, 119, 681–690. [Google Scholar] [CrossRef]
- Choi, H.J.; Song, J.H.; Park, K.S.; Kwon, D.H. Inhibitory effects of quercetin 3-rhamnoside on influenza A virus replication. Eur. J. Pharm. Sci. Off. J. Eur. Fed. Pharm. Sci. 2009, 37, 329–333. [Google Scholar] [CrossRef]
- Wu, W.; Li, R.; Li, X.; He, J.; Jiang, S.; Liu, S.; Yang, J. Quercetin as an Antiviral Agent Inhibits Influenza A Virus (IAV) Entry. Viruses 2015, 8, 6. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, A.L. Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 2008, 4, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006, 34, D668–D672. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1–30. [Google Scholar] [CrossRef]
- NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018, 46, D8–D13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piñero, J.; Ramírez-Anguita, J.M.; Saüch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2019, 48, D845–D855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Assenov, Y.; Ramírez, F.; Schelhorn, S.-E.; Lengauer, T.; Albrecht, M. Computing topological parameters of biological networks. Bioinformatics 2007, 24, 282–284. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Zhu, X.; Bai, H.; Ning, K. Network Pharmacology Databases for Traditional Chinese Medicine: Review and Assessment. Front. Pharmacol. 2019, 10, 123. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Li, M.; Shen, X.; Liu, S. Influenza A virus entry inhibitors targeting the hemagglutinin. Viruses 2013, 5, 352–373. [Google Scholar] [CrossRef] [Green Version]
- Mizumura, K.; Hashimoto, S.; Maruoka, S.; Gon, Y.; Kitamura, N.; Matsumoto, K.; Hayashi, S.; Shimizu, K.; Horie, T. Role of mitogen-activated protein kinases in influenza virus induction of prostaglandin E2 from arachidonic acid in bronchial epithelial cells. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2003, 33, 1244–1251. [Google Scholar] [CrossRef] [PubMed]
- Marjuki, H.; Alam, M.I.; Ehrhardt, C.; Wagner, R.; Planz, O.; Klenk, H.D.; Ludwig, S.; Pleschka, S. Membrane accumulation of influenza A virus hemagglutinin triggers nuclear export of the viral genome via protein kinase Calpha-mediated activation of ERK signaling. J. Biol. Chem. 2006, 281, 16707–16715. [Google Scholar] [CrossRef] [Green Version]
- Cho, S.Y.; Park, S.J.; Kwon, M.J.; Jeong, T.S.; Bok, S.H.; Choi, W.Y.; Jeong, W.I.; Ryu, S.Y.; Do, S.H.; Lee, C.S.; et al. Quercetin suppresses proinflammatory cytokines production through MAP kinases andNF-kappaB pathway in lipopolysaccharide-stimulated macrophage. Mol. Cell Biochem. 2003, 243, 153–160. [Google Scholar] [CrossRef]
- Nanua, S.; Zick, S.M.; Andrade, J.E.; Sajjan, U.S.; Burgess, J.R.; Lukacs, N.W.; Hershenson, M.B. Quercetin blocks airway epithelial cell chemokine expression. Am. J. Respir. Cell Mol. Biol. 2006, 35, 602–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casciuc, I.; Horvath, D.; Gryniukova, A.; Tolmachova, K.A.; Vasylchenko, O.V.; Borysko, P.; Moroz, Y.S.; Bajorath, J.; Varnek, A. Pros and cons of virtual screening based on public “Big Data”: In silico mining for new bromodomain inhibitors. Eur. J. Med. Chem. 2019, 165, 258–272. [Google Scholar] [CrossRef] [PubMed]
- Qian, T.; Zhu, S.; Hoshida, Y. Use of big data in drug development for precision medicine: An update. Expert Rev. Precis. Med. Drug Dev. 2019, 4, 189–200. [Google Scholar] [CrossRef]


| Pathway and Disease (KEGG) | Gene Counts |
|---|---|
| hsa05200: Pathways in cancer | 16 |
| hsa05161: Hepatitis B | 14 |
| hsa05205: Proteoglycans in cancer | 12 |
| hsa05152: Tuberculosis | 11 |
| hsa04066: HIF-1 signaling pathway | 10 |
| hsa05142: Chagas disease (American trypanosomiasis) | 10 |
| hsa05145: Toxoplasmosis | 10 |
| hsa05160: Hepatitis C | 10 |
| hsa04210: Apoptosis | 9 |
| hsa04668: TNF signaling pathway | 9 |
| hsa04722: Neurotrophin signaling pathway | 9 |
| hsa04071: Sphingolipid signaling pathway | 9 |
| hsa05164: Influenza A | 9 |
| hsa04010: MAPK signaling pathway | 9 |
| hsa04151: PI3K-Akt signaling pathway | 9 |
| Ligand-Target | Binding Affinity (kcal/moL) |
|---|---|
| QT-MAPK1 | −8.3 |
| QT-NFKB1 | −7.3 |
| QT-RELA | −7.1 |
| QT-TP53 | −6.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.; Kim, Y.B. Uncovering Quercetin’s Effects against Influenza A Virus Using Network Pharmacology and Molecular Docking. Processes 2021, 9, 1627. https://doi.org/10.3390/pr9091627
Kim M, Kim YB. Uncovering Quercetin’s Effects against Influenza A Virus Using Network Pharmacology and Molecular Docking. Processes. 2021; 9(9):1627. https://doi.org/10.3390/pr9091627
Chicago/Turabian StyleKim, Minjee, and Young Bong Kim. 2021. "Uncovering Quercetin’s Effects against Influenza A Virus Using Network Pharmacology and Molecular Docking" Processes 9, no. 9: 1627. https://doi.org/10.3390/pr9091627
APA StyleKim, M., & Kim, Y. B. (2021). Uncovering Quercetin’s Effects against Influenza A Virus Using Network Pharmacology and Molecular Docking. Processes, 9(9), 1627. https://doi.org/10.3390/pr9091627





