Heparanase (HPSE) Associates with the Tumor Immune Microenvironment in Colorectal Cancer
Abstract
:1. Introduction
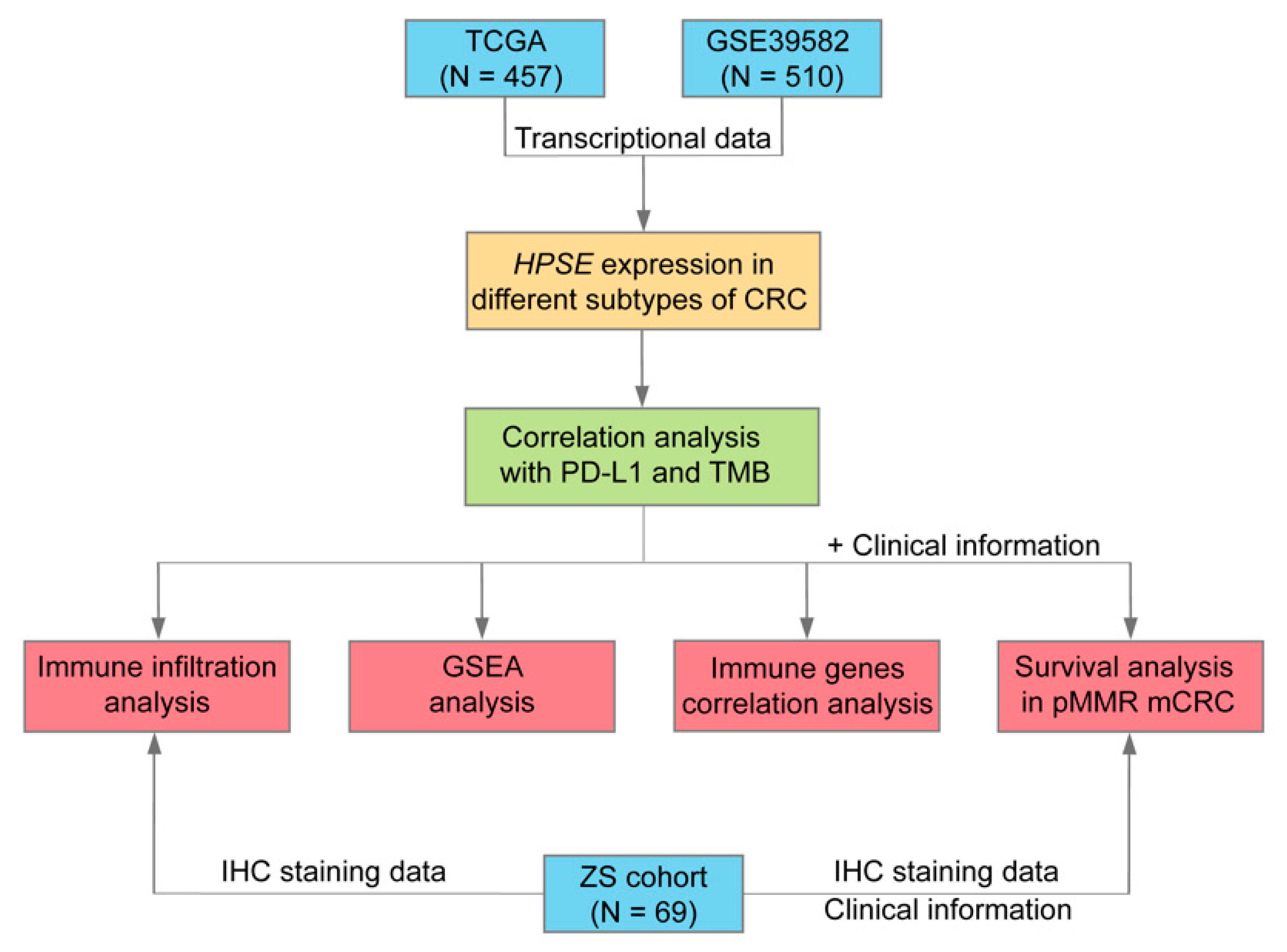
2. Materials and Methods
2.1. Public Datasets and Clinical Samples
2.2. Immune Cell Infiltration Evaluation
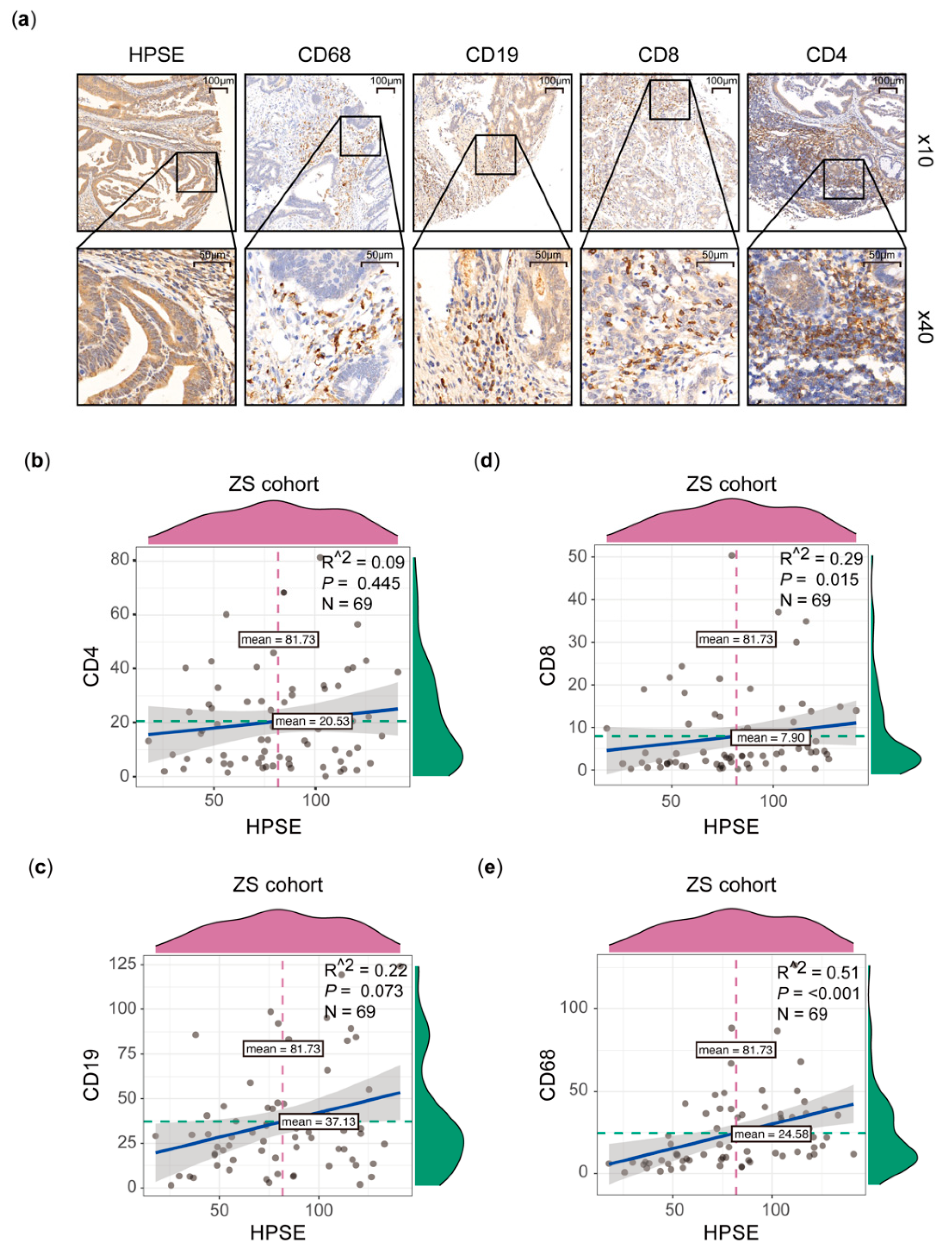
2.3. Immunohistochemical Staining (IHC)
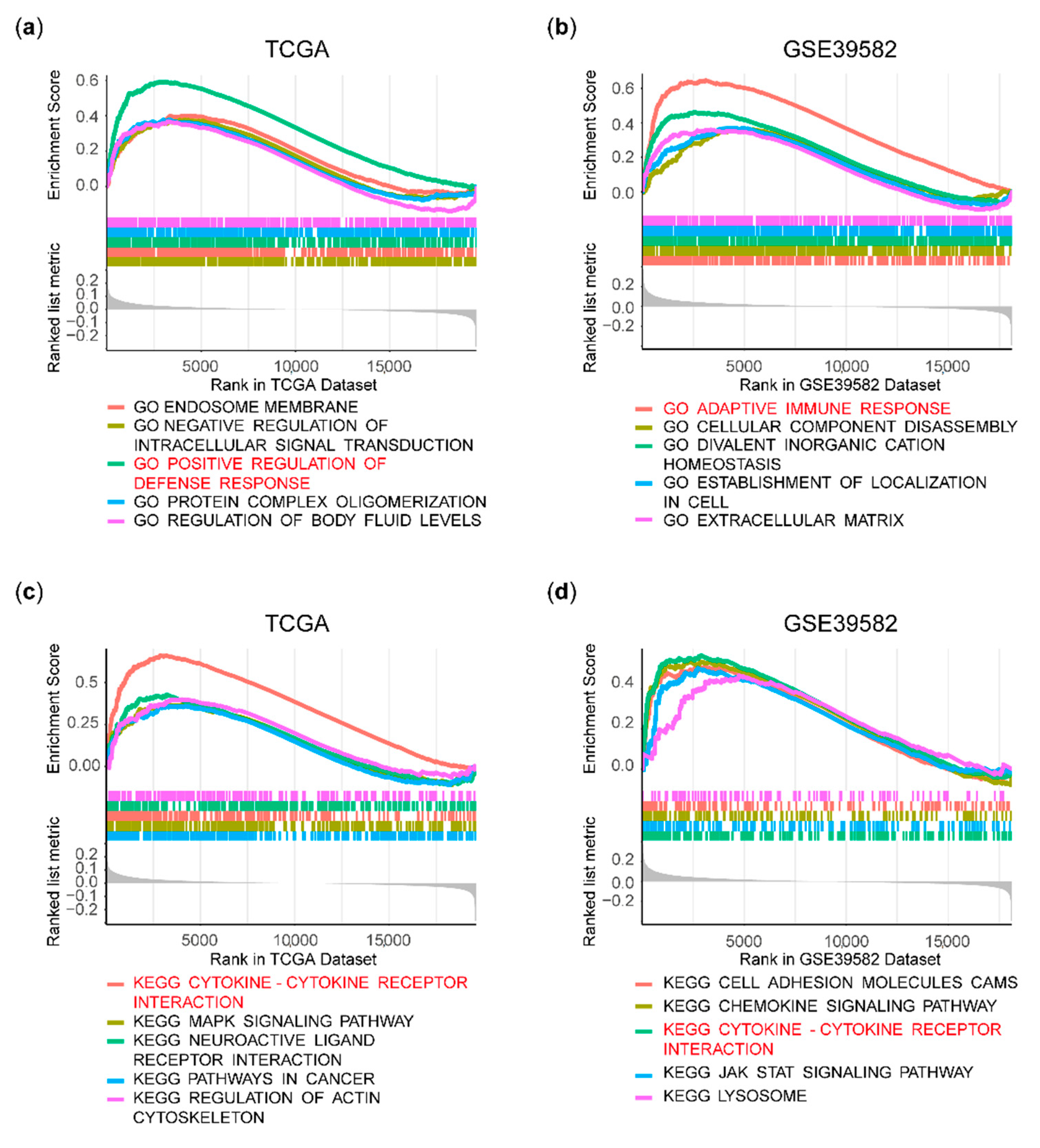
2.4. Gene Set Enrichment Analysis (GSEA)
2.5. Statistical Analysis
3. Results
3.1. HPSE Expression in Different Molecular Subtypes of CRC and Its Association with PD-L1 Expression and TMB
3.2. Higher HPSE Expression Associated with an Increased Infiltration of Immune Cells
3.3. Immune Pathways Were Enriched in the HPSE Expression-High Group
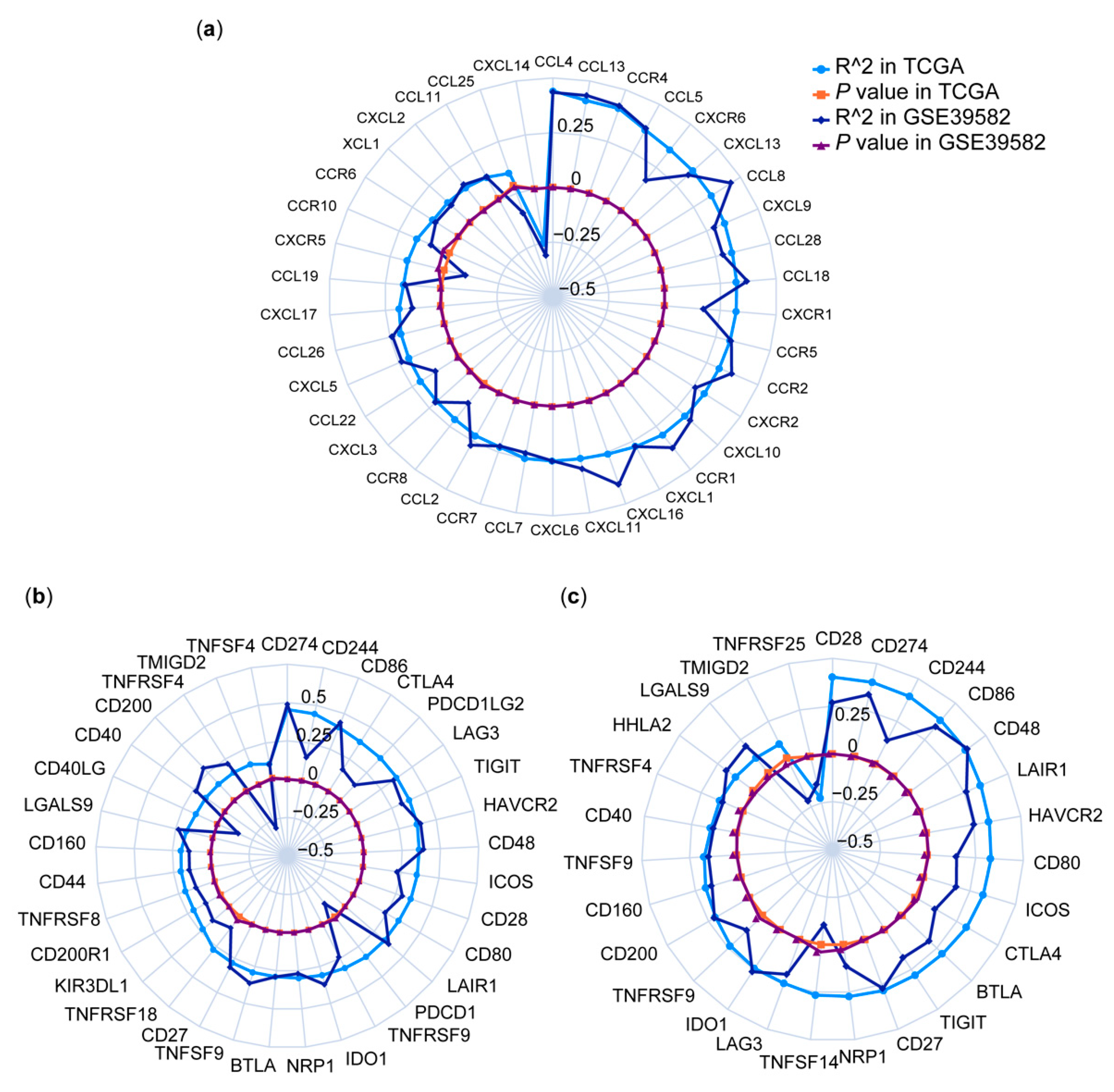
3.4. Correlations between HPSE Expression and Immune Genes
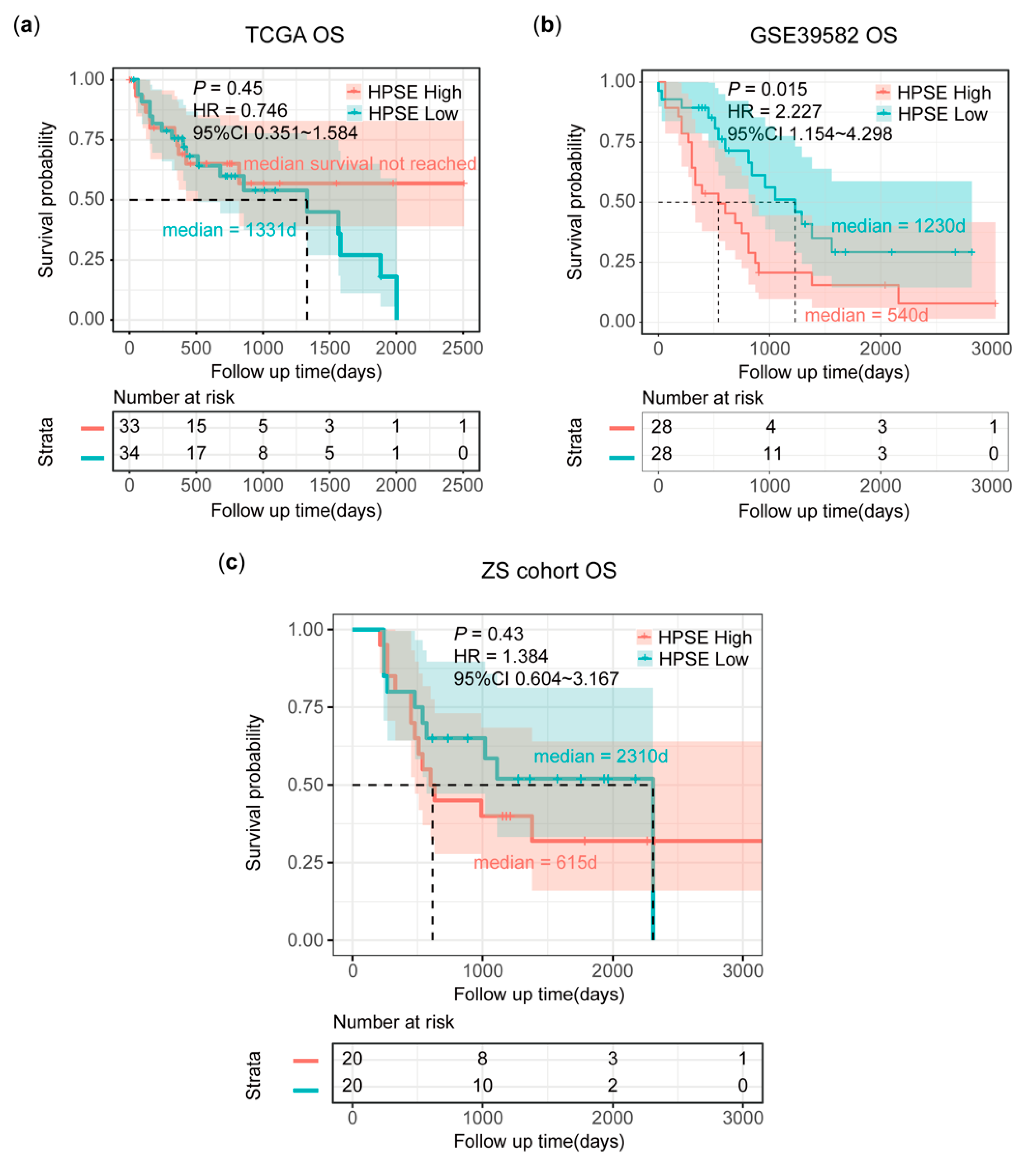
3.5. Poor Survival in pMMR mCRC Patients with High HPSE Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Dekker, E.; Tanis, P.J.; Vleugels, J.L.A.; Kasi, P.M.; Wallace, M.B. Colorectal cancer. Lancet 2019, 394, 1467–1480. [Google Scholar] [CrossRef]
- Chan, T.; Yarchoan, M.; Jaffee, E.; Swanton, C.; Quezada, S.; Stenzinger, A.; Peters, S. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. Ann. Oncol. 2019, 30, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Zhao, B. Efficacy of PD-1 or PD-L1 inhibitors and PD-L1 expression status in cancer: Meta-analysis. BMJ 2018, 362, k3529. [Google Scholar] [CrossRef] [Green Version]
- Patel, S.P.; Kurzrock, R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol. Cancer Ther. 2015, 14, 847–856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibney, G.T.; Weiner, L.M.; Atkins, M.B. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. 2016, 17, e542–e551. [Google Scholar] [CrossRef] [Green Version]
- Ganesh, K.; Stadler, Z.K.; Cercek, A.; Mendelsohn, R.B.; Shia, J.; Segal, N.H.; Diaz, L.A. Immunotherapy in colorectal cancer: Rationale, challenges and potential. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 361–375. [Google Scholar] [CrossRef]
- Bellesoeur, A.; Torossian, N.; Amigorena, S.; Romano, E. Advances in theranostic biomarkers for tumor immunotherapy. Curr. Opin. Chem. Biol. 2020, 56, 79–90. [Google Scholar] [CrossRef]
- Picard, E.; Verschoor, C.P.; Ma, G.W.; Pawelec, G. Relationships Between Immune Landscapes, Genetic Subtypes and Responses to Immunotherapy in Colorectal Cancer. Front. Immunol. 2020, 11, 369. [Google Scholar] [CrossRef]
- Dienstmann, R.; Vermeulen, L.; Guinney, J.; Kopetz, S.; Tejpar, S.; Tabernero, J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat. Rev. Cancer 2017, 17, 79–92. [Google Scholar] [CrossRef]
- Baretti, M.; Le, D.T. DNA mismatch repair in cancer. Pharmacol. Ther. 2018, 189, 45–62. [Google Scholar] [CrossRef]
- Llosa, N.J.; Cruise, M.; Tam, A.; Wicks, E.C.; Hechenbleikner, E.M.; Taube, J.M.; Blosser, R.L.; Fan, H.; Wang, H.; Luber, B.S.; et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015, 5, 43–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masola, V.; Zaza, G.; Gambaro, G.; Franchi, M.; Onisto, M. Role of heparanase in tumor progression: Molecular aspects and therapeutic options. Semin. Cancer Biol. 2019, 62, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Jayatilleke, K.M.; Hulett, M.D. Heparanase and the hallmarks of cancer. J. Transl. Med. 2020, 18, 1–25. [Google Scholar] [CrossRef]
- Gutter-Kapon, L.; Alishekevitz, D.; Shaked, Y.; Li, J.P.; Aronheim, A.; Ilan, N.; Vlodavsky, I. Heparanase is required for activation and function of macrophages. Proc. Natl. Acad. Sci. USA 2016, 113, E7808–E7817. [Google Scholar] [CrossRef] [Green Version]
- Aras, S.; Zaidi, M.R. TAMeless traitors: Macrophages in cancer progression and metastasis. Br. J. Cancer 2017, 117, 1583–1591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Putz, E.M.; Mayfosh, A.; Kos, K.; Barkauskas, D.S.; Nakamura, K.; Town, L.; Goodall, K.; Yee, D.Y.; Poon, I.; Baschuk, N.; et al. NK cell heparanase controls tumor invasion and immune surveillance. J. Clin. Investig. 2017, 127, 2777–2788. [Google Scholar] [CrossRef]
- Grossman, R.L.; Heath, A.P.; Ferretti, V.; Varmus, H.E.; Lowy, D.R.; Kibbe, W.A.; Staudt, L.M. Toward a Shared Vision for Cancer Genomic Data. N. Engl. J. Med. 2016, 375, 1109–1112. [Google Scholar] [CrossRef] [PubMed]
- Marisa, L.; De Reyniès, A.; Duval, A.; Selves, J.; Gaub, M.P.; Vescovo, L.; Etienne-Grimaldi, M.-C.; Schiappa, R.; Guenot, D.; Ayadi, M.; et al. Gene Expression Classification of Colon Cancer into Molecular Subtypes: Characterization, Validation, and Prognostic Value. PLoS Med. 2013, 10, e1001453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Newman, A.M.; Steen, C.B.; Liu, C.L.; Gentles, A.J.; Chaudhuri, A.A.; Scherer, F.; Khodadoust, M.S.; Esfahani, M.S.; Luca, B.A.; Steiner, D.; et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019, 37, 773–782. [Google Scholar] [CrossRef]
- Aran, D.; Hu, Z.; Butte, A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Mezzadra, R.; Schumacher, T.N. Regulation and Function of the PD-L1 Checkpoint. Immunity 2018, 48, 434–452. [Google Scholar] [CrossRef] [Green Version]
- Nagarsheth, N.; Wicha, M.S.; Zou, W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 2017, 17, 559–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fridman, W.H.; Zitvogel, L.; Sautes-Fridman, C.; Kroemer, G. The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 2017, 14, 717–734. [Google Scholar] [CrossRef] [PubMed]
- Lortat-Jacob, H.; Grosdidier, A.; Imberty, A. Structural diversity of heparan sulfate binding domains in chemokines. Proc. Natl. Acad. Sci. USA 2002, 99, 1229–1234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, X.; Moseman, E.A.; Saito, H.; Petryniak, B.; Thiriot, A.; Hatakeyama, S.; Ito, Y.; Kawashima, H.; Yamaguchi, Y.; Lowe, J.B.; et al. Endothelial heparan sulfate controls chemokine presentation in recruitment of lymphocytes and dendritic cells to lymph nodes. Immunity 2010, 33, 817–829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodall, K.; Poon, I.; Phipps, S.; Hulett, M.D. Soluble Heparan Sulfate Fragments Generated by Heparanase Trigger the Release of Pro-Inflammatory Cytokines through TLR-4. PLoS ONE 2014, 9, e109596. [Google Scholar] [CrossRef] [PubMed]
- Massena, S.; Christoffersson, G.; Hjertström, E.; Zcharia, E.; Vlodavsky, I.; Ausmees, N.; Rolny, C.; Li, J.-P.; Phillipson, M. A chemotactic gradient sequestered on endothelial heparan sulfate induces directional intraluminal crawling of neutrophils. Blood 2010, 116, 1924–1931. [Google Scholar] [CrossRef] [Green Version]
- Cassetta, L.; Pollard, J.W. Targeting macrophages: Therapeutic approaches in cancer. Nat. Rev. Drug Discov. 2018, 17, 887–904. [Google Scholar] [CrossRef]
- van der Leun, A.M.; Thommen, D.S.; Schumacher, T.N. CD8(+) T cell states in human cancer: Insights from single-cell analysis. Nat. Rev. Cancer 2020, 20, 218–232. [Google Scholar] [CrossRef]
- Caruana, I.; Savoldo, B.; Hoyos, V.; Weber, G.; Liu, H.; Kim, E.; Ittmann, M.M.; Marchetti, D.; Dotti, G. Heparanase promotes tumor infiltration and antitumor activity of CAR-redirected T lymphocytes. Nat. Med. 2015, 21, 524–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasaki, S.; Baba, T.; Nishimura, T.; Hayakawa, Y.; Hashimoto, S.-I.; Gotoh, N.; Mukaida, N. Essential roles of the interaction between cancer cell-derived chemokine, CCL4, and intra-bone CCR5-expressing fibroblasts in breast cancer bone metastasis. Cancer Lett. 2016, 378, 23–32. [Google Scholar] [CrossRef] [Green Version]
- Lien, M.Y.; Tsai, H.C.; Chang, A.C.; Tsai, M.H.; Hua, C.H.; Wang, S.W.; Tang, C.H. Chemokine CCL4 Induces Vascular Endothelial Growth Factor C Expression and Lymphangiogenesis by miR-195-3p in Oral Squamous Cell Carcinoma. Front. Immunol. 2018, 9, 412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korbecki, J.; Kojder, K.; Simińska, D.; Bohatyrewicz, R.; Gutowska, I.; Chlubek, D.; Baranowska-Bosiacka, I. CC Chemokines in a Tumor: A Review of Pro-Cancer and Anti-Cancer Properties of the Ligands of Receptors CCR1, CCR2, CCR3, and CCR4. Int. J. Mol. Sci. 2020, 21, 8412. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Nozawa, K.; Fujishiro, M.; Kawasaki, M.; Suzuki, F.; Takamori, K.; Ogawa, H.; Takasaki, Y.; Sekigawa, I. CC motif chemokine ligand 13 is associated with rheumatoid arthritis pathogenesis. Mod. Rheumatol. 2013, 23, 856–863. [Google Scholar] [CrossRef] [PubMed]
- Hui, E.; Cheung, J.; Zhu, J.; Su, X.; Taylor, M.J.; Wallweber, H.A.; Sasmal, D.K.; Huang, J.; Kim, J.M.; Mellman, I.; et al. T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science 2017, 355, 1428–1433. [Google Scholar] [CrossRef] [PubMed]


| Number of Patients | ||||
|---|---|---|---|---|
| TCGA | GSE39582 | ZS Cohort | ||
| Total | 457 | 510 | 69 | |
| Gender | Female | 213 | 233 | 23 |
| Male | 241 | 277 | 46 | |
| Age | Mean (SD) | 66.4 (12.7) | 66.9 (13.1) | 60.1 (10.25) |
| Site | Left | 223 | 303 | 46 |
| Right | 141 | 207 | 23 | |
| CMS | CMS1 | 45 | 86 | - |
| CMS2 | 175 | 204 | - | |
| CMS3 | 47 | 65 | - | |
| CMS4 | 101 | 113 | - | |
| MMR | dMMR | 52 | 72 | 0 |
| pMMR | 403 | 391 | 69 | |
| BRAF | Mutant | 31 | 51 | 3 |
| Wildtype | 426 | 459 | 66 | |
| KRAS | Mutant | 123 | 204 | 37 |
| Wildtype | 334 | 306 | 32 | |
| Metastasis | M0 | 365 | 446 | 29 |
| M1 | 70 | 60 | 40 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, M.; Liu, Q.; Yuan, Y.; Li, S.; Dong, Y.; Liang, L.; Zou, Z.; Liu, T. Heparanase (HPSE) Associates with the Tumor Immune Microenvironment in Colorectal Cancer. Processes 2021, 9, 1605. https://doi.org/10.3390/pr9091605
Liu M, Liu Q, Yuan Y, Li S, Dong Y, Liang L, Zou Z, Liu T. Heparanase (HPSE) Associates with the Tumor Immune Microenvironment in Colorectal Cancer. Processes. 2021; 9(9):1605. https://doi.org/10.3390/pr9091605
Chicago/Turabian StyleLiu, Mengling, Qing Liu, Yitao Yuan, Suyao Li, Yu Dong, Li Liang, Zhiguo Zou, and Tianshu Liu. 2021. "Heparanase (HPSE) Associates with the Tumor Immune Microenvironment in Colorectal Cancer" Processes 9, no. 9: 1605. https://doi.org/10.3390/pr9091605
APA StyleLiu, M., Liu, Q., Yuan, Y., Li, S., Dong, Y., Liang, L., Zou, Z., & Liu, T. (2021). Heparanase (HPSE) Associates with the Tumor Immune Microenvironment in Colorectal Cancer. Processes, 9(9), 1605. https://doi.org/10.3390/pr9091605





