Isolation, Genomic, and Proteomic Characterization of a Novel Neotropical Strain of Bacillus thuringiensis with Mosquitocidal Activities
Abstract
1. Introduction
2. Materials and Methods
2.1. Origin and Culture of Bt TOD651 Strain
2.2. Crystal Protein Purification and SDS-PAGE Analysis
2.3. Identification of Crystal Morphology
2.4. Larvae Rearing
2.5. Bioassays
2.6. Whole-Genome Sequencing, Assembly, and Annotation
2.7. Phylogenetic Relationship
2.8. LC-MS/MS Analysis
2.9. Proteomic Data Analysis
2.10. Data Availability
3. Results
3.1. Protein Profile, Crystal Morphology, and Mosquitocidal Activity
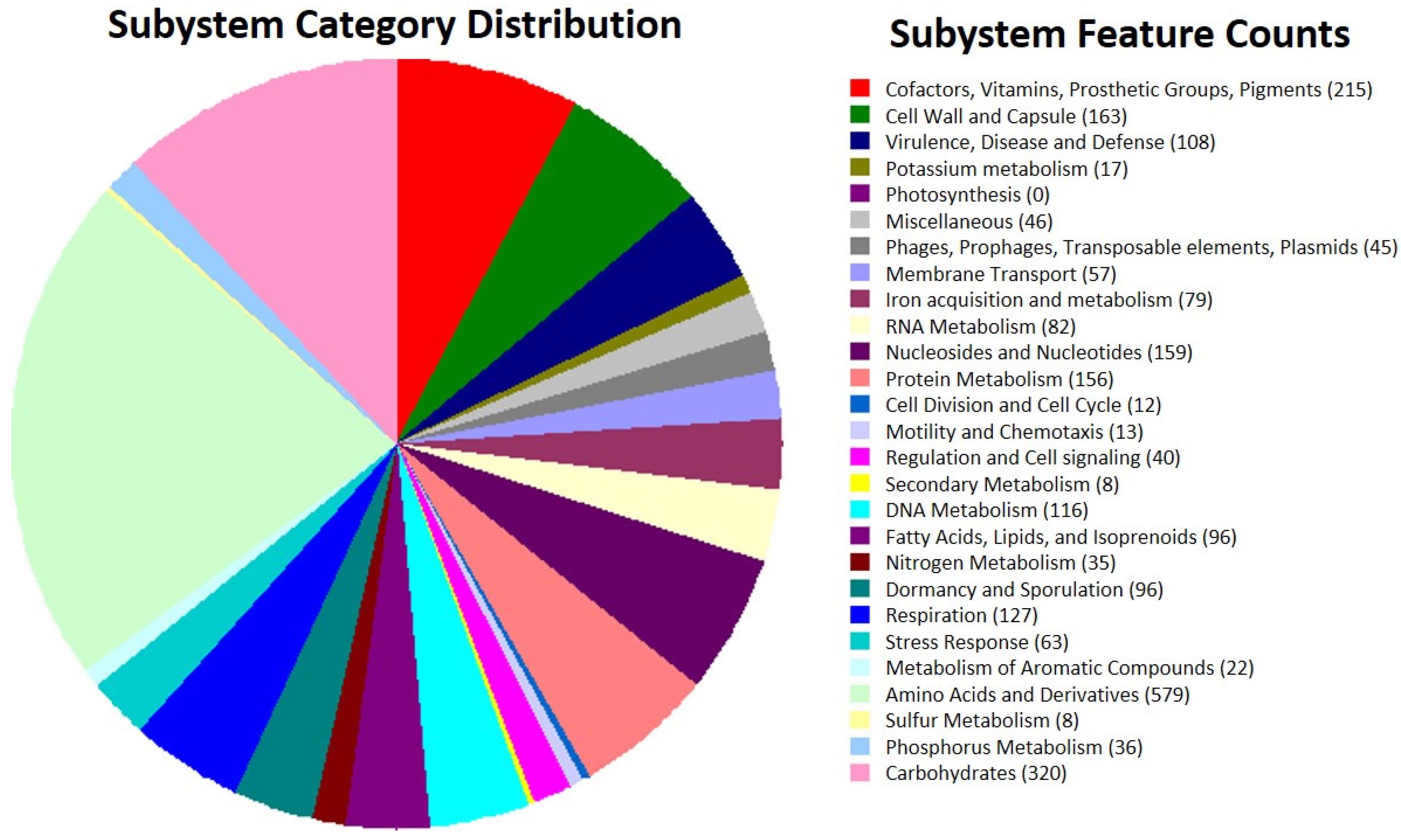
3.2. General Genomic Features
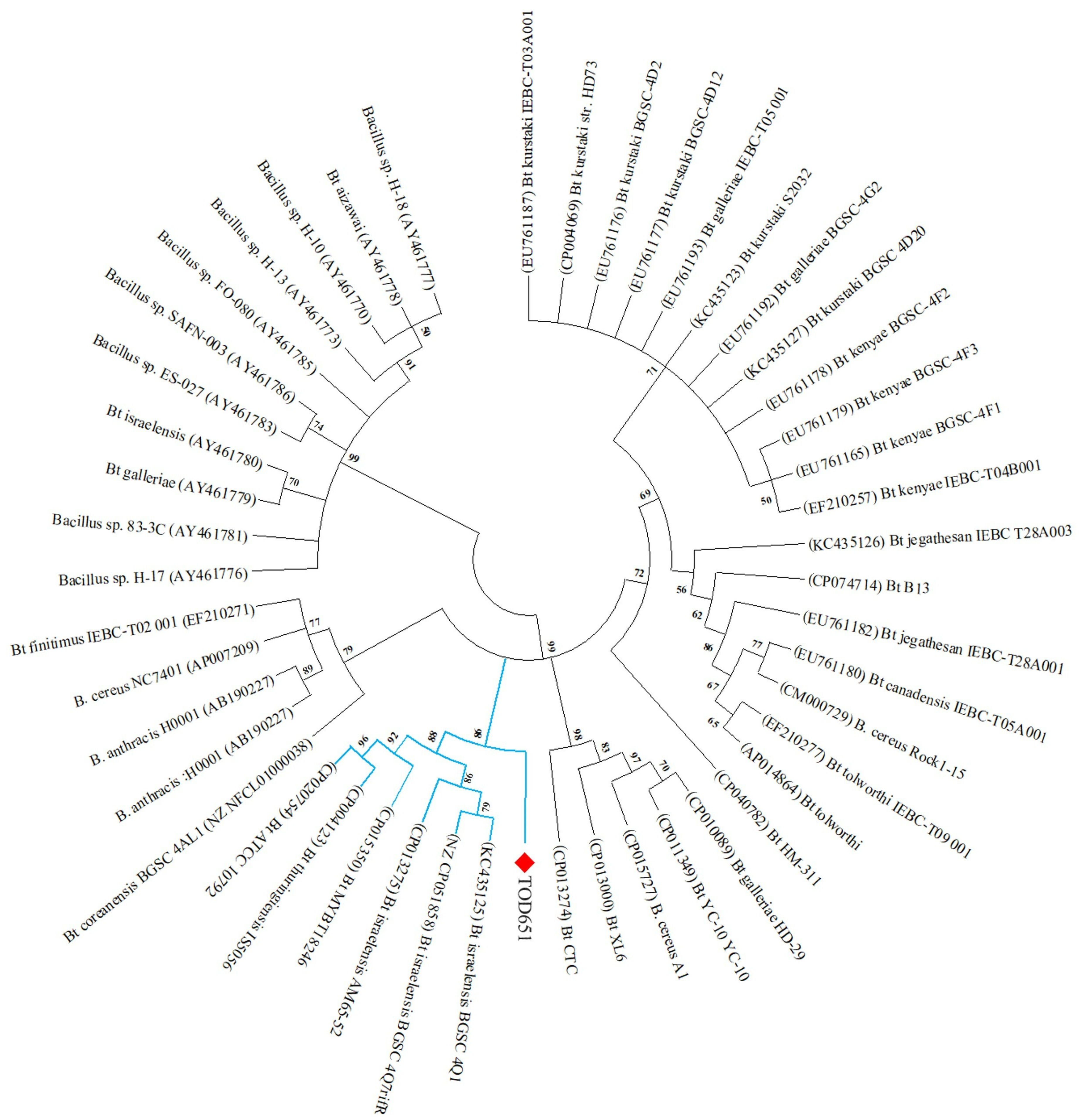
3.3. Phylogenetic Analysis
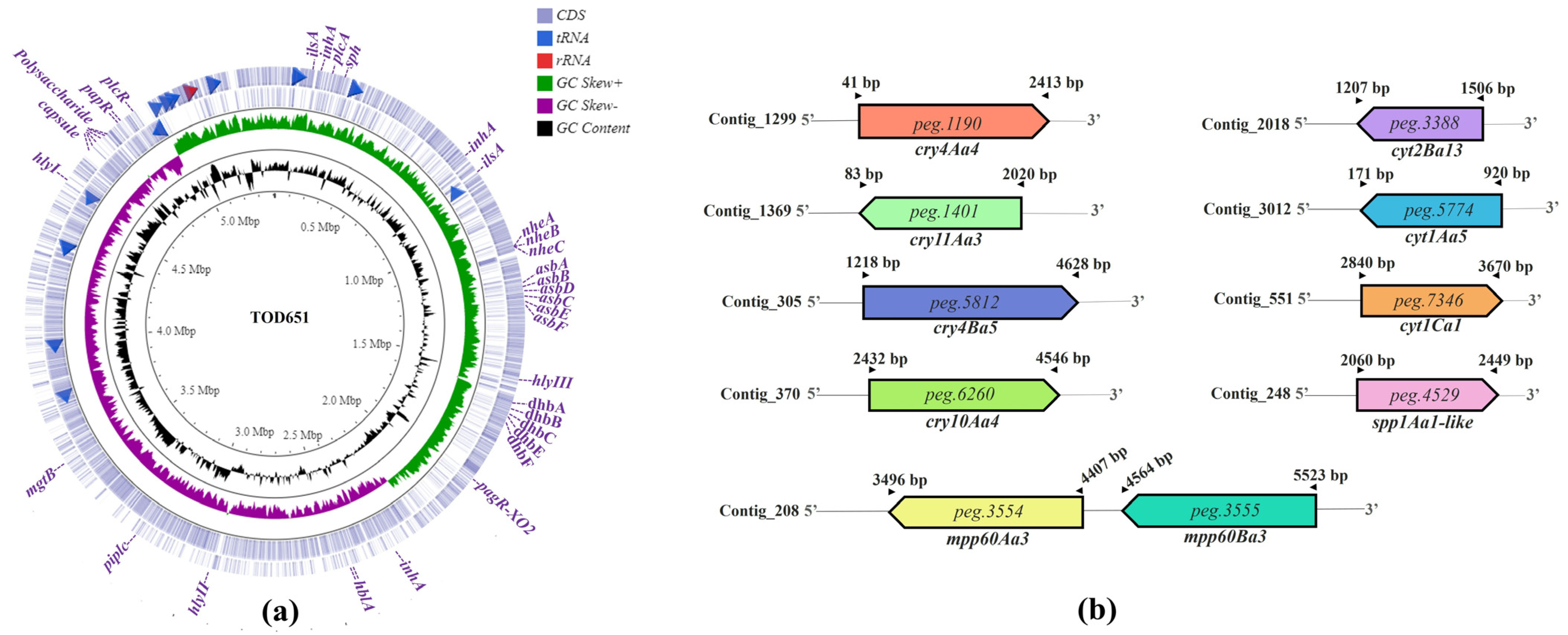
3.4. Genes Associated with Bt TOD651 Pathogenicity
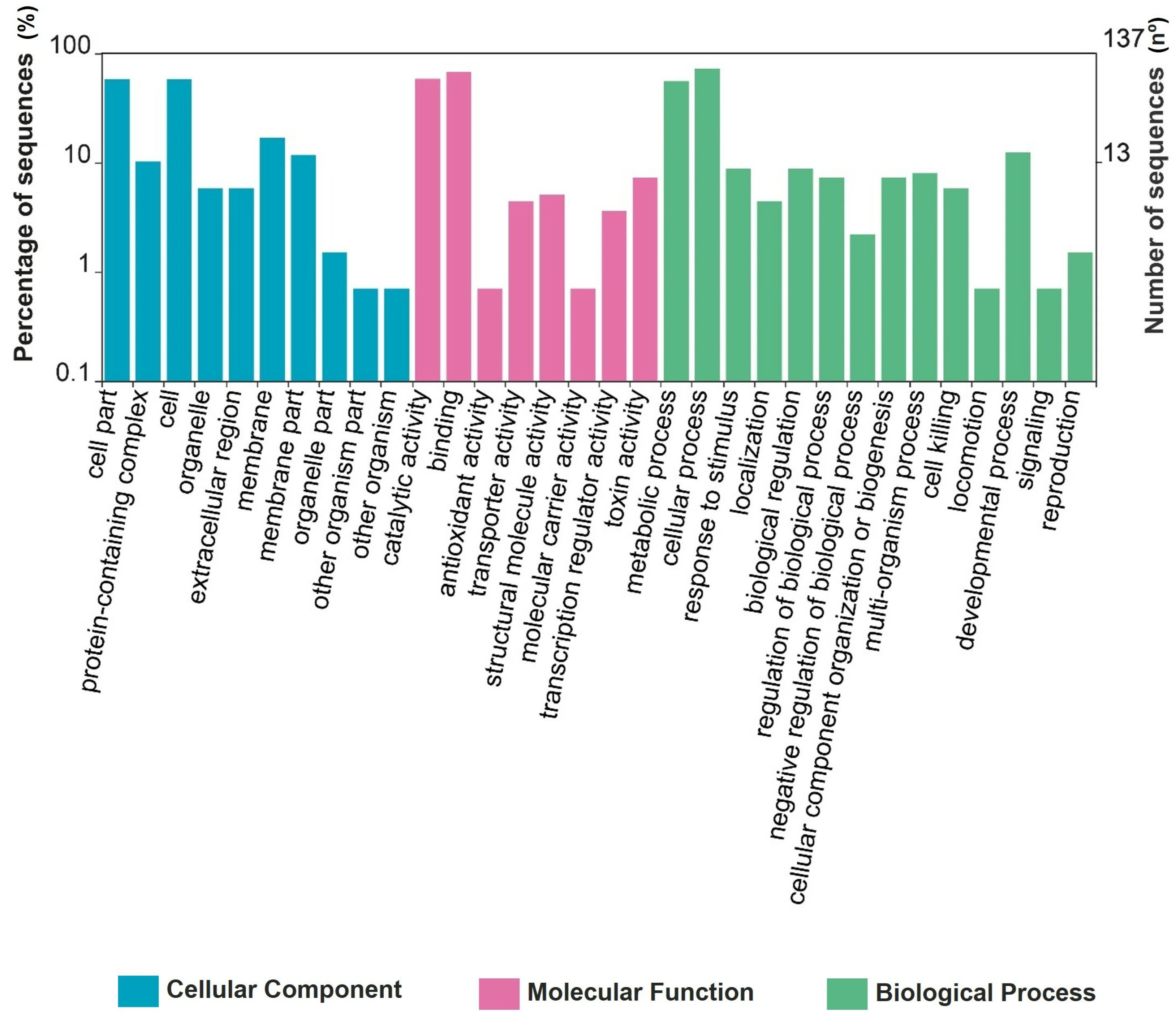
3.5. Proteomics of Spore–Crystal Mixture
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ramos-Nino, M.E.; Fitzpatrick, D.M.; Eckstrom, K.M.; Tighe, S.; Hattaway, L.M.; Hsueh, A.N.; Cheetham, S. Metagenomic analysis of Aedes aegypti and Culex quinquefasciatus mosquitoes from Grenada, West Indies. PLoS ONE 2020, 15, e0231047. [Google Scholar] [CrossRef] [PubMed]
- Boukedi, H.; Hman, M.; Khedher, S.B.; Tounsi, S.; Abdelkefi-Mesrati, L. Promising active bioinsecticides produced by Bacillus thuringiensis strain BLB427. J. Adv. Res. Rev. 2020, 8, 026–035. [Google Scholar]
- AL-FAR, I.M. Bacillus thuringiensis and its pesticidal crystal proteins. Int. J. Fauna Biol. 2020, 3, 157–162. [Google Scholar]
- Ben-Dov, E. Bacillus thuringiensis subsp. israelensis and its dipteran-specific toxins. Toxins 2014, 6, 1222–1243. [Google Scholar]
- Lutinski, J.A.; Kuczmainski, A.G.; de Quadros, S.; Busato, M.A.; Weirich, C.M.M.; Malgueiro, A.; Garcia, F.R.M. Bacillus thuringiensis var. israelensis como alternativa para o controle populacional de Aedes aegypti (Linnaeus, 1762) (Diptera: Culicidae). Ciência Nat. 2017, 39, 211–220. [Google Scholar]
- Berry, C.; O’Neil, S.; Ben-Dov, E.; Jones, A.F.; Murphy, L.; Quail, M.A.; Holden, M.T.G.; Harris, D.; Zaritsky, A.; Parkhill, J. Complete sequence and organization of pBtoxis, the toxin-coding plasmid of Bacillus thuringiensis subsp. Israelensis. Appl. Environ. Microbiol. 2002, 68, 5082–5095. [Google Scholar] [CrossRef]
- Gray, E.W.; Fusco, R. Microbial control of black flies (Diptera: Simuliidae) with Acillus thuringiensis subsp. israelensis. In Microbial Control of Insect and Mite Pests; Lacey, L.A., Ed.; Academic Press: Yakima, WA, USA, 2017; pp. 367–377. [Google Scholar]
- Valtierra-de-Luis, D.; Villanueva, M.; Berry, C.; Caballero, P. Potential for Bacillus thuringiensis and Other Bacterial Toxins as Biological Control Agents to Combat Dipteran Pests of Medical and Agronomic Importance. Toxins 2020, 12, 773. [Google Scholar] [CrossRef]
- Viana, J.L.; Soares-da-Silva, J.; Vieira-Neta, M.R.A.; Tadei, W.P.; Oliveira, C.D.; Abdalla, F.C.; Peixoto, C.A.; Pinheiro, V.C.S. Isolates of Bacillus thuringiensis from Maranhão biomes with potential insecticidal action against Aedes aegypti larvae (Diptera, Culicidae). Braz. J. Biol. 2020, 81, 114–124. [Google Scholar] [CrossRef]
- Zhou, Y.; Wu, Z.; Zhang, J.; Wan, Y.; Jin, W.; Li, Y.; Fang, X. Bacillus thuringiensis novel toxin Epp is toxic to mosquitoes and Prodenia litura larvae. Braz. J. Microbiol. 2020, 51, 437–445. [Google Scholar] [CrossRef]
- Wu, J.; Wei, L.; He, J.; Fu, K.; Li, X.; Jia, L.; Wang, R.; Zhang, W. Characterization of a novel Bacillus thuringiensis toxin active against Aedes aegypti larvae. Acta Trop. 2021, 223, 106088. [Google Scholar] [CrossRef]
- Roy, M.; Chatterjee, S.; Dangar, T.K. Characterization and mosquitocidal potency of a Bacillus thuringiensis strain of rice field soil of Burdwan, West Bengal, India. Microb. Pathog. 2021, 158, 105093. [Google Scholar] [CrossRef] [PubMed]
- Day, M.; Ibrahim, M.; Dyer, D.; Bulla, L. Genome Sequence of Bacillus thuringiensis subsp. kurstaki strain HD-1. Genome Announc. 2014, 2, e00613-14. [Google Scholar] [PubMed]
- Jeong, H.; Choi, S.-K.; Park, S.-H. Genome Sequences of Bacillus thuringiensis serovar kurstaki strain BP865 and B. thuringiensis, serovar aizawai strain HD-133. Genome Announc. 2017, 5, e01544-16. [Google Scholar] [CrossRef] [PubMed]
- Sajid, M.; Geng, C.; Li, M.; Wang, Y.; Liu, H.; Zheng, J.; Sun, M. Whole-genome analysis of Bacillus thuringiensis revealing partial genes as a source of novel cry toxins. Appl. Environ. Microbiol. 2018, 84, e00277-18. [Google Scholar] [CrossRef] [PubMed]
- Lechuga, A.; Lood, C.; Salas, M.; van Noort, V.; Lavigne, R.; Redrejo-Rodríguez, M. Completed genomic sequence of Bacillus thuringiensis HER1410 reveals a Cry-containing chromosome, two megaplasmids, and an integrative plasmidial prophage. G3 Genes Genom. Genet. 2020, 10, 2927–2939. [Google Scholar]
- Susič, N.; Janežič, S.; Rupnik, M.; Stare, B.G. Whole genome sequencing and comparative genomics of two nematicidal Bacillus strains reveals a wide range of possible virulence factors. G3 Genes Genom. Genet. 2020, 10, 881–890. [Google Scholar] [CrossRef]
- Wu, D.; He, J.; Gong, Y.; Chen, D.; Zhu, X.; Qiu, N.; Sun, M.; Li, M.; Yu, Z. Proteomic analysis reveals the strategies of Bacillus thuringiensis YBT-1520 for survival under long-term heat stress. Proteomics 2011, 11, 2580–2591. [Google Scholar] [CrossRef]
- Caballero, J.; Jiménez-Moreno, N.; Orera, I.; Williams, T.; Fernández, A.B.; Villanueva, M.; Ferré, J.; Caballero, P.; Ancín-Azpilicueta, C. Unraveling the composition of insecticidal crystal proteins in Bacillus thuringiensis: A proteomics approach. Appl. Environ. Microbiol. 2020, 86, e00476-20. [Google Scholar] [CrossRef] [PubMed]
- Rang, J.; He, H.; Wang, T.; Ding, X.; Zuo, M.; Quan, M.; Sun, Y.; Yu, Z.; Hu, S.; Xia, L. Comparative analysis of genomics and proteomics in Bacillus thuringiensis 4.0718. PLoS ONE 2015, 10, e0119065. [Google Scholar] [CrossRef]
- Alves, G.B.; Melo, F.L.; Oliveira, E.E.; Haddi, K.; Costa, L.; Dias, M.L.; Campos, F.S.; Pereira, E.J.G.; Côrrea, R.F.T.; Ascêncio, S.D.; et al. Comparative genomic analysis and mosquito larvicidal activity of four Bacillus thuringiensis subsp. israelensis strains. Sci. Rep. 2020, 10, 5518. [Google Scholar] [CrossRef]
- Dankocsik, C.; Donovan, W.P.; Jany, C.S. Activation of a cryptic crystal protein gene of Bacillus thuringiensis subspecies kurstaki by gene fusion and determination of the crystal protein insecticidal specificity. Mol. Microbiol. 1990, 4, 2087–2094. [Google Scholar] [CrossRef] [PubMed]
- Quan, M.; Xie, J.; Liu, X.; Li, Y.; Rang, J.; Zhang, T.; Zhou, F.; Xia, F.; Hu, S.; Sun, Y.; et al. Comparative analysis of genomics and proteomics in the new isolated Bacillus thuringiensis X022 revealed the metabolic regulation mechanism of carbon flux following Cu2+ treatment. Front. Microbiol. 2016, 7, 792. [Google Scholar] [CrossRef] [PubMed]
- Gomis-Cebolla, J.; Scaramal, R.A.P.; Ferré, J. A genomic and proteomic approach to identify and quantify the expressed Bacillus thuringiensis proteins in the supernatant and parasporal crystal. Toxins 2018, 10, 193. [Google Scholar] [CrossRef] [PubMed]
- Khorramnejad, A.; Gomis-Cebolla, J.; Talaei-Hassanlouei, R.; Bel, Y.; Escriche, B. Genomics and proteomics analyses revealed novel candidate pesticidal proteins in a lepidopteran-toxic Bacillus thuringiensis strain. Toxins 2020, 12, 673. [Google Scholar] [CrossRef]
- Baragamaarachchi, R.Y.; Samarasekera, J.K.R.R.; Weerasena, O.V.D.S.J.; Lamour, K.; Jurat-Fuentes, J.L. Identification of a native Bacillus thuringiensis strain from Sri Lanka active against Dipel-resistant Plutella xylostella. PeerJ 2019, 7, e7535. [Google Scholar] [CrossRef]
- Doggett, N.A.; Stubben, C.J.; Chertkov, O.; Bruce, D.C.; Detter, J.C.; Johnson, S.L.; Han, C.S. Complete genome sequence of Bacillus thuringiensis serovar israelensis strain HD-789. Genome Announc. 2013, 1, e01023-13. [Google Scholar] [CrossRef]
- Fayad, N.; Patiño-Navarrete, R.; Kambris, Z.; Antoun, M.; Osta, M.; Chopineau, J.; Mahillon, J.; El Chamy, L.; Sanchis, V.; Awad, M.K. Characterization and whole genome sequencing of AR23, a highly toxic Bacillus thuringiensis strain isolated from Lebanese soil. Curr. Microbiol. 2019, 76, 1503–1511. [Google Scholar] [CrossRef]
- Ma, W.; Chen, H.; Jiang, X.; Wang, J.; Gelbič, I.; Guan, X.; Zhang, L. Whole genome sequence analysis of the mosquitocidal Bacillus thuringiensis LLP29. Arch. Microbiol. 2020, 202, 1693–1700. [Google Scholar] [CrossRef]
- Monnerat, R.G.; Batista, A.C.; de Medeiros, P.T.; Martins, E.S.; Melatti, V.M.; Praça, L.B.; Dumas, V.F.; Morinaga, C.; Demo, C.; Gomes, A.C.M.; et al. Screening of Brazilian Bacillus thuringiensis isolates active against Spodoptera frugiperda, Plutella xylostella and Anticarsia gemmatalis. Biol. Control 2007, 41, 291–295. [Google Scholar] [CrossRef]
- Mounsef, J.R.; Salameh, D.; Kallassy, A.M.; Chamy, L.; Brandam, C.; Lteif, R. A simple method for the separation of Bacillus thuringiensis spores and crystals. J. Microbiol. Methods 2014, 107, 147–149. [Google Scholar] [CrossRef]
- Aguiar, R.W.S.; dos Santos, S.F.; Morgado, F.S.; Ascencio, S.D.; Lopes, M.M.; Viana, K.F.; Didonet, J.; Ribeiro, B.M. Insecticidal and repellent activity of Siparuna guianensis Aubl. (Negramina) against Aedes aegypti and Culex quinquefasciatus. PLoS ONE 2015, 10, e0116765. [Google Scholar]
- Valbon, W.; Andreazza, F.; Oliveira, E.E.; Liu, F.; Feng, B.; Hall, M.; Klimavicz, J.; Coats, J.R.; Dong, K. Bioallethrin activates specific olfactory sensory neurons and elicits spatial repellency in Aedes aegypti. Pest Manag. Sci. 2022, 78, 438–445. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, R.E.; Dulmage, H.T.; Alls, R.; Couch, T.L.; Dame, D.A.; Hall, I.M.; Rose, R.I.; Versoi, P.L. US standard bioassay for the potency assessment of Bacillus thuringiensis serotype H-14 against mosquito larvae. Bull. ESA 1984, 30, 26–29. [Google Scholar]
- Dulmage, H.T.; Yousten, A.A.; Singer, S.L.L.A.; Lacey, L.A. Guidelines for Production of Bacillus thuringiensis H-14 and Bacillus sphaericus; No. TDR/BCV/90.1; Unpublished; World Health Organization: Geneva, Switzerland, 1990. [Google Scholar]
- SAS Institute. SAS/STAT User’s Guide; SAS Institute: Cary, NC, USA, 2008. [Google Scholar]
- Andrews, S. FastQC: A Quality-Control Tool for High-Throughput Sequence. 2015. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 26 October 2021).
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Lischer, H.E.; Shimizu, K.K. Reference-guided de novo assembly approach improves genome reconstruction for related species. BMC Bioinform. 2017, 18, 474. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Liu, H.; Zheng, J.; Bo, D.; Yu, Y.; Ye, W.; Peng, D.; Sun, M. BtToxin_Digger: A comprehensive and high-throughput pipeline for mining toxin protein genes from Bacillus thuringiensis. Bioinformatics 2021, 38, 250–251. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Santos, M.D.; Lima, D.B.; Fischer, J.S.; Clasen, M.A.; Kurt, L.U.; Camillo-Andrade, A.C.; Monteiro, L.C.; de Aquino, P.F.; Neves-Ferreira, A.G.C.; Valente, R.H.; et al. Simple, efficient and thorough shotgun proteomic analysis with PatternLab V. Nat. Protoc. 2022, 17, 1553–1578. [Google Scholar] [CrossRef]
- Ye, J.; Zhang, Y.; Cui, H.; Liu, J.; Wu, Y.; Cheng, Y.; Xu, H.; Huang, X.; Li, S.; Zhou, A.; et al. WEGO 2.0: A web tool for analyzing and plotting GO annotations, 2018 update. Nucleic Acids Res. 2018, 46, W71–W75. [Google Scholar] [CrossRef] [PubMed]
- Elleuch, J.; Jaoua, S.; Darriet, F.; Chandre, F.; Tounsi, S.; Zghal, R.Z. Cry4Ba and Cyt1Aa proteins from Bacillus thuringiensis israelensis: Interactions and toxicity mechanism against Aedes aegypti. Toxicon 2015, 104, 83–90. [Google Scholar] [CrossRef] [PubMed]
- El-Kersh, T.A.; Ahmed, A.M.; Al-Sheikh, Y.A.; Tripet, F.; Ibrahim, M.S.; Metwalli, A.A. Isolation and characterization of native Bacillus thuringiensis strains from Saudi Arabia with enhanced larvicidal toxicity against the mosquito vector Anopheles gambiae (sl). Parasites Vectors 2016, 9, 647. [Google Scholar] [CrossRef] [PubMed]
- Nair, K.; Al-Thani, R.; Al-Thani, D.; Al-Yafei, F.; Ahmed, T.; Jaoua, S. Diversity of Bacillus thuringiensis strains from Qatar as shown by crystal morphology, δ-endotoxins and cry gene content. Front. Microbiol. 2018, 9, 708. [Google Scholar] [CrossRef]
- Hollensteiner, J.; Poehlein, A.; Spröer, C.; Bunk, B.; Sheppard, A.E.; Rosentstiel, P.; Schulenburg, H.; Liesegang, H. Complete genome sequence of the nematicidal Bacillus thuringiensis MYBT18246. Stand. Genom. Sci. 2017, 12, 48. [Google Scholar] [CrossRef]
- Swiecicka, I.; Bideshi, D.K.; Federici, B.A. Novel isolate of Bacillus thuringiensis subsp. thuringiensis that produces a Quasicuboidal crystal of Cry1Ab21 toxic to larvae of Trichoplusia ni. Appl. Environ. Microbiol. 2008, 74, 923–930. [Google Scholar]
- Vilas-Bôas, G.T.; Alvarez, R.C.; dos Santos, C.A.; Vilas-Boas, L.A. Fatores de virulência de Bacillus thuringiensis: O que existe além das proteínas Cry. EntomoBrasilis 2012, 5, 1–10. [Google Scholar] [CrossRef]
- Guillemet, E.; Cadot, C.; Tran, S.L.; Guinebretiere, M.H.; Lereclus, D.; Ramarao, N. The InhA metalloproteases of Bacillus cereus contribute concomitantly to virulence. J. Bacteriol. 2010, 192, 286–294. [Google Scholar] [CrossRef]
- Pohare, M.B.; Wagh, S.G.; Udayasuriyan, V. Bacillus thuringiensis as potential biocontrol agent for sustainable agriculture. In Current Trends in Microbial Biotechnology for Sustainable Agriculture, 1st ed.; Yadav, A.N., Singh, J., Singh, C., Yadav, N., Eds.; Springer: Singapore, 2020; pp. 439–468. [Google Scholar]
- Stein, C.; Jones, G.W.; Chalmers, T.; Berry, C. Transcriptional analysis of the toxin-coding plasmid pBtoxis from Bacillus thuringiensis subsp. israelensis. Appl. Environ. Microbiol. 2006, 72, 1771–1776. [Google Scholar] [CrossRef]
- Cohen, S.; Albeck, S.; Ben-Dov, E.; Cahan, R.; Firer, M.; Zaritsky, A.; Dym, O. Cyt1Aa toxin: Crystal structure reveals implications for its membrane-perforating function. J. Mol. Biol. 2011, 413, 804–814. [Google Scholar] [CrossRef]
- Otieno-Ayayo, Z.N.; Zaritsky, A.; Wirth, M.C.; Manasherob, R.; Khasdan, V.; Cahan, R.; Ben-Dov, E. Variations in the mosquito larvicidal activities of toxins from Bacillus thuringiensis ssp. israelensis. Environ. Microbiol. 2008, 10, 2191–2199. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, M.A.F.; Alzate, O.; Mohammad, M.; McNall, R.J.; Adang, M.J.; Dean, D.H. Introduction of Culex toxicity into Bacillus thuringiensis Cry4Ba by protein engineering. Appl. Environ. Microbiol. 2003, 69, 5343–5353. [Google Scholar] [CrossRef]
- Sun, Y.; Zhao, Q.; Xia, L.; Ding, X.; Hu, Q.; Federici, B.A.; Park, H.W. Identification and characterization of three previously undescribed crystal proteins from Bacillus thuringiensis subsp. jegathesan. Appl. Environ. Microbiol. 2013, 79, 3364–3370. [Google Scholar] [CrossRef] [PubMed]
- Juárez-Pérez, V.; Guerchicoff, A.; Rubinstein, C.; Delecluse, A. Characterization of Cyt2Bc toxin from Bacillus thuringiensis subsp. medellin. Appl. Environ. Microbiol. 2002, 68, 1228–1231. [Google Scholar] [CrossRef]
- Torres-Quintero, M.C.; Gómez, I.; Pacheco, S.; Sánchez, J.; Flores, H.; Osuna, J.; Mendoza, G.; Soberón, M.; Bravo, A. Engineering Bacillus thuringiensis Cyt1Aa toxin specificity from dipteran to lepidopteran toxicity. Sci. Rep. 2018, 8, 4989. [Google Scholar] [CrossRef]
- Cantón, P.E.; Reyes, E.Z.; Ruiz De Escudero, I.; Bravo, A.; Soberon, M. Binding of Bacillus thuringiensis subsp. israelensis Cry4Ba to Cyt1Aa has an important role in synergism. Peptides 2011, 32, 595–600. [Google Scholar] [PubMed]
- Fu, Z.; Sun, Y.; Xia, L.; Ding, X.; Mo, X.; Li, X.; Huang, K.; Zhang, Y. Assessment of protoxin composition of Bacillus thuringiensis strains by use of polyacrylamide gel block and mass spectrometry. Appl. Environ. Microbiol. 2008, 79, 875–880. [Google Scholar] [CrossRef]
- Valtierra-de-Luis, D.; Villanueva, M.; Lai, L.; Williams, T.; Caballero, P. Potential of Cry10Aa and Cyt2Ba, two minority δ-endotoxins produced by Bacillus thuringiensis ser. israelensis, for the control of Aedes aegypti larvae. Toxins 2020, 12, 355. [Google Scholar]
- Wirth, M.C.; Park, H.W.; Walton, W.E.; Federici, B.A. Cyt1A of Bacillus thuringiensis delays evolution of resistance to Cry11A in the mosquito Culex quinquefasciatus. Appl. Environ. Microbiol. 2005, 71, 185–189. [Google Scholar] [CrossRef]
- Sauka, D.H.; Peralta, C.; Pérez, M.P.; Onco, M.I.; Fiodor, A.; Caballero, J.; Caballero, P.; Berry, C.; Del Valle, E.E.; Palma, L. Bacillus toyonensis biovar thuringiensis: A novel entomopathogen with insecticidal activity against lepidopteran and coleopteran pests. Biol. Control 2022, 167, 104838. [Google Scholar] [CrossRef]
- Banik, A.; Chattopadhyay, A.; Ganguly, S.; Mukhopadhyay, S.K. Characterization of a tea pest specific Bacillus thuringiensis and identification of its toxin by MALDI-TOF mass spectrometry. Ind. Crops Prod. 2019, 137, 549–556. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, H.; Xie, T.; Huang, Q.; Xiong, X.; Liu, Q.; Wang, G. The YmdB protein regulates biofilm formation dependent on the repressor SinR in Bacillus cereus 0–9. World J. Microbiol. Biotechnol. 2020, 36, 165. [Google Scholar] [CrossRef]
- Chung, M.C.; Popova, T.G.; Millis, B.A.; Mukherjee, D.V.; Zhou, W.; Liotta, L.A.; Petricoin, E.F.; Chandhoke, V.; Bailey, C.; Popov, S.G. Secreted neutral metalloproteases of Bacillus anthracis as candidate pathogenic factors. J. Biol. Chem. 2006, 281, 31408–31418. [Google Scholar] [CrossRef]
- Johnson, D.E.; Oppert, B.; McGaughey, W.H. Spore coat protein synergizes Bacillus thuringiensis crystal toxicity for the Indianmeal moth (Plodia interpunctella). Curr. Microbiol. 1998, 36, 278–282. [Google Scholar] [CrossRef]
- Ding, X.; Huang, J.; Xia, L.; Li, X.; Yuan, C.; Dan, S. A proteomic analysis approach to study insecticidal crystal proteins from different strains of Bacillus thuringiensis. Biocontrol Sci. Technol. 2009, 19, 289–299. [Google Scholar] [CrossRef]
- Xie, J.; Peng, J.; Yi, Z.; Zhao, X.; Li, S.; Zhang, T.; Quan, M.; Yang, S.; Lu, J.; Zhou, P.; et al. Role of hsp20 in the Production of Spores and Insecticidal Crystal Proteins in Bacillus thuringiensis. Front. Microbiol. 2019, 10, 2059. [Google Scholar] [CrossRef]

| Strain | A. aegypti | C. quinquefasciatus | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| LC50 (µg/mL) | CL95 (µg/mL) | SLOPE | χ2 | p | LC50 (µg/mL) | LC95 (µg/mL) | SLOPE | χ 2 | p | |
| TOD651 | 0.011 | 0.030 | 3.726 | 5.62 | 0.05 | 0.023 | 0.055 | 4.311 | 6.68 | 0.27 |
| AM65-52 | 0.013 | 0.037 | 3.725 | 4.33 | 0.36 | 0.028 | 0.069 | 4.467 | 6.49 | 0.16 |
| General Features | Value |
|---|---|
| Mean depth coverage | 95.9× |
| Chromosome size (bp) | 5,409,948 |
| Gapped sites (%) | 7.2 |
| GC content (%) | 35.9 |
| No. of CDS | 5130 |
| No. of rRNA | 1 |
| No. of tRNA | 71 |
| Sequence | Predicted CDS | Length (aa) | Homologous Protein | Coverage (%) | Pairwise Identity (%) | E-Value |
|---|---|---|---|---|---|---|
| Contig_1299 | peg.1190 | 698 | Cry4Aa4 | 59.07 1/99.90 2 | 99.43 1,2 | 0.0 |
| Contig_1369 | peg.1401 | 645 | Cry11Aa3 | 100.00 1,2 | 100.00 1,2 | 0.0 |
| Contig_305 | peg.5812 | 1161 | Cry4Ba5 | 100.00 1,2 | 100.00 1,2 | 0.0 |
| Contig_370 | peg.6260 | 674 | Cry10Aa4 | 97.19 1/100.00 2 | 100.00 1,2 | 0.0 |
| Contig_2018 | peg.3388 | 99 | Cyt2Ba13 | 40.24 1/100.00 2 | 100.00 1,2 | 0.0 |
| Contig_3012 | peg.5774 | 262 | Cyt1Aa5 | 100.00 1 | 100.00 1 | 0.0 |
| Contig_551 | peg.7346 | 291 | Cyt1Ca1 | 51.43 1/97.80 2 | 98.90 1,2 | 0.0 |
| Contig_208 | peg.3554 | 323 | Mpp60Aa3 | 100.00 1,2 | 100.00 1,2 | 0.0 |
| Contig_208 | peg.3552 | 319 | Mpp60Ba3 | 100.00 1,2 | 100.00 1,2 | 0.0 |
| Contig_248 | peg.4529 | 323 | Spp1Aa1 | 58.70 1 | 80.81 1 | 0.0 |
| CDS ID | Description 1 | Length (bp) | Peptide Sequence (No.) | Unique Peptide (No.) 4 | Coverage 5 | Protein Score 6 | NSAF 7 |
|---|---|---|---|---|---|---|---|
| peg.5812 | Cry4Ba5 2 | 1136 | 62 | 60 | 0.5599 | 211.396 | 0.0492475 |
| peg.1190 | Cry4Aa4 2 | 791 | 51 | 47 | 0.6587 | 184.808 | 0.0732234 |
| peg.1401 | Cry11Aa3 2 | 645 | 46 | 46 | 0.6171 | 161.116 | 0.1816369 |
| peg.3553 | Mpp60Ba3 2 | 303 | 29 | 29 | 0.8119 | 97.915 | 0.0868883 |
| peg.6260 | Cry10Aa4 2 | 705 | 31 | 29 | 0.4057 | 104.199 | 0.031742 |
| peg.3039 | Metallophosphoesterase (Mppe) 3 | 471 | 25 | 25 | 0.7113 | 91.843 | 0.0440183 |
| peg.5774 | Cyt1Aa5 2 | 249 | 13 | 13 | 0.6345 | 50.49 | 0.1850302 |
| peg.1543 | Heat-shock protein (GroEL) | 544 | 9 | 9 | 0.1397 | 27.31 | 0.0060494 |
| peg.5502 | L-alanyl-gamma-D-glutamyl-L-diamino acid endopeptidase | 325 | 9 | 9 | 0.4215 | 33.37 | 0.0131636 |
| peg.3700 | Spore coat protein (CotB) | 174 | 8 | 8 | 0.546 | 28.416 | 0.0264785 |
| peg.3439 | Aminopeptidase | 466 | 8 | 8 | 0.2833 | 27.635 | 0.0063558 |
| peg.4239 | Elongation factor Tu | 320 | 8 | 8 | 0.3906 | 31.32 | 0.0133693 |
| peg.8774 | Enolase | 431 | 7 | 7 | 0.2877 | 27.999 | 0.0068719 |
| peg.2515 | Spore coat protein (CotG) | 186 | 7 | 7 | 0.1828 | 22.946 | 0.0212316 |
| peg.3388 | Cyt2Ba13 2 | 99 | 7 | 7 | 0.6768 | 24.895 | 0.0465379 |
| peg.7343 | Cyt1Ca1 2 | 85 | 5 | 5 | 0.2588 | 14.095 | 0.0309731 |
| peg.7029 | Dihydrolipoamide dehydrogenase of pyruvate dehydrogenase complex | 470 | 4 | 4 | 0.0723 | 11.744 | 0.003501 |
| peg.8531 | Chaperone protein (DnaK) | 611 | 4 | 4 | 0.0917 | 12.256 | 0.0021544 |
| peg.5180 | Spore coat protein (GerQ) | 139 | 4 | 4 | 0.295 | 12.928 | 0.0165728 |
| peg.5234 | LSU ribosomal protein L7p/L12p(P1/P2) | 119 | 4 | 4 | 0.5462 | 13.206 | 0.0110618 |
| peg.7670 | NAD-dependent glyceraldehyde-3-phosphate dehydrogenase | 334 | 4 | 4 | 0.2395 | 16.849 | 0.0049265 |
| peg.7030 | Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex | 227 | 4 | 4 | 0.1674 | 8.968 | 0.0072487 |
| peg.7476 | Hypothetical protein | 250 | 3 | 3 | 0.156 | 10.916 | 0.0052654 |
| peg.2818 | Fructose-bisphosphate aldolase class II | 267 | 3 | 3 | 0.1723 | 9.314 | 0.0049302 |
| peg.1753 | N-acetylmuramoyl-L-alanine amidase | 271 | 3 | 3 | 0.1144 | 9.547 | 0.0048574 |
| peg.8176 | Uncharacterized protein YmfJ | 82 | 3 | 3 | 0.4634 | 8.95 | 0.0120399 |
| peg.4909 | N-acetylmuramoyl-L-alanine amidase | 327 | 3 | 3 | 0.1437 | 8.267 | 0.0030192 |
| peg.3986 | Extracellular neutral protease B (NprB) | 426 | 3 | 3 | 0.1244 | 11.317 | 0.0038626 |
| peg.9127 | SSU ribosomal protein S2p (SAe) | 233 | 2 | 2 | 0.1159 | 5.189 | 0.0028248 |
| peg.7282 | Hypotetical protein | 143 | 2 | 2 | 0.1678 | 4.502 | 0.0046026 |
| peg.4363 | Superoxide dismutase | 203 | 2 | 2 | 0.1823 | 6.977 | 0.0048634 |
| peg.782 | Hypothetical protein | 129 | 2 | 2 | 0.186 | 5.041 | 0.0051022 |
| peg.4419 | Cell division trigger factor | 404 | 2 | 2 | 0.0965 | 7.55 | 0.0016292 |
| peg.3911 | DNA-binding protein (Hbsu) | 90 | 2 | 2 | 0.3667 | 7.282 | 0.0073131 |
| peg.7358 | Spore coat protein (CotE) | 180 | 2 | 2 | 0.2111 | 6.641 | 0.0036565 |
| peg.969 | Phage tail fiber protein | 431 | 2 | 2 | 0.051 | 4.816 | 0.0015271 |
| peg.3496 | Hypotetical protein | 108 | 2 | 2 | 0.1481 | 4.385 | 0.0060942 |
| peg.9081 | Uncharacterized protein BA5373 | 68 | 2 | 2 | 0.5 | 6.717 | 0.0096791 |
| peg.8583 | RNA-binding protein (Hfq) | 74 | 2 | 2 | 0.3243 | 6.323 | 0.0088943 |
| peg.3589 | Spore coat protein of CotY/CotZ family | 155 | 2 | 2 | 0.2 | 6.649 | 0.0063695 |
| peg.1544 | Heat-shock protein (GroES) | 94 | 2 | 2 | 0.2447 | 5.286 | 0.0070019 |
| peg.5565 | Tricarboxylate transport sensor protein (TctE) | 92 | 2 | 2 | 0.3261 | 3.967 | 0.0071541 |
| peg.8113 | Exosporium protein K | 118 | 2 | 2 | 0.1102 | 5.905 | 0.0083667 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alves, G.B.; Dias, M.L.; Oliveira, E.E.d.; Santos, G.R.d.; Ribeiro, B.M.; Aguiar, R.W.d.S. Isolation, Genomic, and Proteomic Characterization of a Novel Neotropical Strain of Bacillus thuringiensis with Mosquitocidal Activities. Processes 2023, 11, 1455. https://doi.org/10.3390/pr11051455
Alves GB, Dias ML, Oliveira EEd, Santos GRd, Ribeiro BM, Aguiar RWdS. Isolation, Genomic, and Proteomic Characterization of a Novel Neotropical Strain of Bacillus thuringiensis with Mosquitocidal Activities. Processes. 2023; 11(5):1455. https://doi.org/10.3390/pr11051455
Chicago/Turabian StyleAlves, Giselly Batista, Marcelo Leite Dias, Eugenio Eduardo de Oliveira, Gil Rodrigues dos Santos, Bergmann Morais Ribeiro, and Raimundo Wagner de Souza Aguiar. 2023. "Isolation, Genomic, and Proteomic Characterization of a Novel Neotropical Strain of Bacillus thuringiensis with Mosquitocidal Activities" Processes 11, no. 5: 1455. https://doi.org/10.3390/pr11051455
APA StyleAlves, G. B., Dias, M. L., Oliveira, E. E. d., Santos, G. R. d., Ribeiro, B. M., & Aguiar, R. W. d. S. (2023). Isolation, Genomic, and Proteomic Characterization of a Novel Neotropical Strain of Bacillus thuringiensis with Mosquitocidal Activities. Processes, 11(5), 1455. https://doi.org/10.3390/pr11051455








