Abstract
A potential based on a bond dipole representation of electrostatics is reported for peptides. Different from those popular force fields using atom-centered point-charge or point-multipole to express the electrostatics, our peptide potential uses the chemical bond dipole–dipole interactions to express the electrostatic interactions. The parameters for permanent and induced bond dipoles are derived from fitting to the MP2 three-body interaction energy curves. The parameters for van der Waals are taken from AMBER99sb and further refined from fitting to the MP2 stacking interaction energy curve. The parameters for bonded terms are taken from AMBER99sb without any modification. The scale factors for intramolecular dipole–dipole interactions are determined from reproducing the highly qualified ab initio conformational energies of dipeptides and tetrapeptides. The resulting potential is validated by use to evaluate the conformational energies of polypeptides containing up to 15 amino acid residues. The calculation results show that our peptide potential produces the conformational energies much closer to the famous density functional theory M06-2X/cc-pVTZ results than the famous AMBER99sb and AMOEBAbio18 force fields. Our potential also produces accurate intermolecular interaction energies for hydrogen-bonded and stacked dimers. We anticipate the peptide potential proposed here could be helpful in computer simulations of polypeptides and proteins.
1. Introduction
Molecular simulation plays a more and more significant role in studying structures and properties of biosystems. However, its predicting ability depends upon how well the molecular interactions are described by the potential energy function.
Many popular molecular potentials have been used in the simulation of biomolecules, including fixed-charge potentials, such as CHARMM [1,2], OPLS [3,4], GROMOS [5,6], and AMBER [7,8], and polarizable ones, such as AMOEBA [9,10] and Drude [11,12,13]. Up to now, the most widely used potentials are still the fixed-charge ones. However, the disadvantage of the fixed-charge potentials limits its capability in describing the electrostatic interactions due to the weakness to capturing charge redistribution during a change in surroundings.
Electrostatic interactions, including polarization, are very important for biosystems [14,15]. In biomolecular systems, electrostatic polarization arises naturally due to the influence of other molecules, which produces significant many-body interaction energy. In order to capture the many-body polarization effects, potentials incorporating an explicit treatment of electronic polarization are needed.
Classical polarizable models with explicit polarization first appeared around the 1950s [16,17]. However, a general polarizable force field for molecular modeling has become more and more attractive only in the past decade because of the great improvement of computer technology.
Two famous polarizable potentials to deal with polarization are AMOEBA [9,10,18] and Drude [12,19]. In AMOEBA force field, the permanent electrostatic interactions are calculated using atom-centered monopoles, dipoles, and quadrupoles. For the electrostatic polarization effects, at least one atom-centered dipole is assigned to each atomic core and the induced dipoles are determined through an iterative self-consistent scheme. Recently, Liu et al. proposed an AMOEBA+ potential [20] where the effects of the charge transfer and charge penetration are considered. An AMOEBA+(CF) potential [21] is also put forward in which the charge flux effects are taken into account. Drude force field is another famous polarizable potential which was first developed around 2000 [19]. Drude potential attaches an auxiliary particle to the parent atom to mimic the electrostatic polarization explicitly. The induced dipole is expressed with the Drude charge, the electric field, and the force constant [13].
On the basis of the many-body expansion of the interaction energy, Paesani et al. have established an MB-pol potential for water [22,23], in which explicit 1B, 2B, and 3B terms are included. MB-pol model has shown its high quality in reproducing the highly qualified ab initio many-body interactions for water clusters as well as for small molecule–water systems.
Our laboratory has established a polarizable dipole–dipole interaction model for hydrogen-bonding, stacking, and T-shaped interactions [24,25,26,27,28,29]. Recently, it was further developed to compute many-body polarization effects [30,31]. In this paper, a peptide potential that contains not only the intermolecular interaction terms, but also the intramolecular terms is proposed. Different from all the existing potentials, our potential uses chemical bond dipole–dipole interactions to describe the electrostatics and polarization.
2. Theoretical Model
Our peptide potential is written as Equation (1).
In Equation (1), the first three terms are the bond stretching, angle bending, and dihedral angle, respectively. kb, b, and b0 represent the stretching force constant of a bond, actual bond length, and equilibrium bond length. kθ, θ, and θ0 represent the bending force constant of an angle, measured angle, and equilibrium angle. Vn, φ, and γn represent the Fourier coefficient, dihedral angle, and phase angle.
We use the Lennard–Jones function (the fourth term) to express the nonbonded van der Waals potential. Here, Rij represents the distance between atoms i and j. , , , . εi and Ri* are van der Waals well depth and radius parameters, respectively.
The nonbonded electrostatics of our peptide potential is written as the last term of Equation (1). Different from the traditional atom-centered partial-charge-based Coulomb electrostatic potential, our peptide potential expresses the electrostatics (including permanent and induced interactions) with chemical bond dipoles. Here, μi and μj represent two different bond dipole moments. As shown in Figure 1, rij, α, α’, and β are the geometric parameters which describe the relative geometrical relationship of two dipoles.
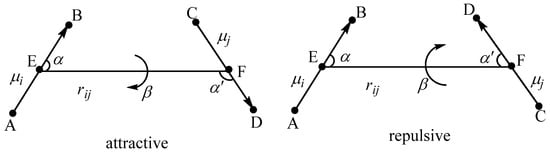
Figure 1.
The attractive and repulsive interacting dipoles. Atom A forms a polar chemical bond A-B with atom B; atom C forms another polar chemical bond C-D with atom D. The two polar bonds A-B and C-D are taken as two bond dipoles whose dipole moments are represented by μi and μj. E and F are the mid-points of the bonds A-B and C-D, respectively. rij is the distance between the two mid-points E and F. α and α’ are the angles between the line EF and the positive direction of the bond dipole. The β is the dihedral angle between the two planes BEF and EFD.
In our model, the electrostatic interactions are calculated through the dipole–dipole interactions [32] between bond dipoles in a peptide separated by no less than three chemical bonds. That means no intramolecular dipole–dipole interaction needs to be considered for small molecules, such as acetamide and water. When the geometry of a peptide molecule changes from one conformation to another, redistribution of electron density happens, which leads to polarization. The bond dipole moment changes from μ0 to μ = μ0 + δμ, where δμ represents the induced bond dipole and μ0 represents the permanent bond dipole. Our electrostatics can be further rewritten as Equation (2).
In Equation (2), the first term of the right side denotes the permanent electrostatic interaction because its value is determined only by the permanent bond dipoles, whereas the second term can be named as the induced or polarized interaction because its value is a function of induced bond dipoles. The induced bond dipole moment is expressed with a simple Formula (3).
where d represents the bond length of a chemical bond, q0 represents atomic reference charge, q represents atomic partial charge, and c is a correction factor.
3. Parameterization
Before using our potential Equation (1) to evaluate the conformational energies for polypeptides, the parameters in Equation (1) need to be first determined. The atom types used in our potential are provided in Figure 2, where the same type of atom will use the same set of parameters. For example, the symbol OH is used to represent the oxygen atom of hydroxyl group, the symbol HO is used to represent the hydrogen atom of hydroxyl group, the symbol CT is used to represent the sp3 carbon atom, the symbol HC is used to represent the hydrogen atom bonded to the sp3 carbon, etc. Then, we turn to determining the parameters needed in our potential. The details are described below:
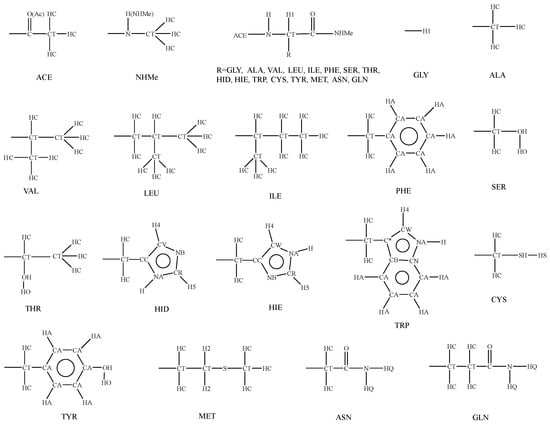
Figure 2.
Atom types.
3.1. Parameters for Electrostatics
In this work, we treat the permanent bond dipole μ0, the reference charge q0, and the correction factor c as parameters. The induced bond dipole moment δμ is calculated via Equation (3). For H-X single bond (X = N, C, O, S), q0 in Equation (3) is the H atomic partial charge of H-X bond in the extended structure of a tetrapeptide molecule. For C=O double bond, q0 in Equation (3) is the O atomic partial charges of C=O bond in the extended structure of a tetrapeptide molecule. q is the corresponding H or O atomic partial charges in any target peptide molecule. The charge q is obtained from AM1 calculations.
First, two peptide–water trimers are devised, with each trimer containing one glycine tetrapeptide and two water molecules and the optimal structures are obtained from B3LYP/6-31+G** calculations in vacuum. The benchmark three-body interaction energies are computed using the counterpoise-corrected (CP-corrected) MP2/aug-cc-pVTZ method. Our peptide potential is also applied to these trimers to calculate the three-body interaction energies, where the initial values of parameters μ0 and q0 are from experiment bond dipole moments [33] and from the AM1 atomic partial charge of the glycine tetrapeptide extended structure, and the correction factor c is initially set to 1. The parameters μ0, q0, and c are optimized by reproducing the benchmark three-body interaction curves for these trimers repeatedly until the curves computed via our potential agree well with the benchmark MP2 results. Here, the relevant parameters for water molecules are taken directly from our previous work [30], where the three-body interaction energy of different water clusters has been calculated using dipole–dipole interaction. In Figure 3a,b, the curves produced by our potential and by the MP2 method are shown, indicating our potential yields quite similar three-body interaction curves to that of the MP2 methods. The determined parameters are collected in Table 1. The determined parameters are further tested by computing the total three-body interaction energies of two clusters (see Figure S1 of the Supporting Information for details). Cartesian co-ordinates in angstroms for two clusters are provided in Table S5. For the side chains of serine, threonine, tyrosine, cysteine, and methionine, the experiment bond dipole moments [33] of the C-O, O-H, C-S, and S-H single bonds are used as the permanent bond dipole parameters μ0 of the CT-OH, CA-OH, OH-HO, CT-S, CT-SH, and SH-HS without any modification. q0 of OH-HO and SH-HS are determined from the AM1 partial charge of the extended structure of the tetrapeptides AcGlyXGlyNHMe (X = Ser, Thr, Tyr, and Cys). The correction factor c of the CT-OH, CA-OH, CT-SH, and CT-S is equal to zero, which means no induced bond dipole is included.
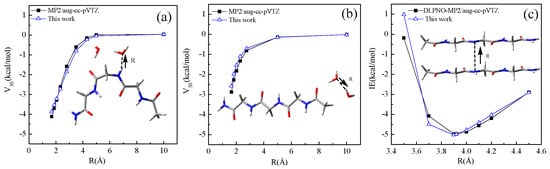
Figure 3.
Interaction energy curves. (a,b): 3−body interactions; (c): 2−body interactions.

Table 1.
Parameters for nonbonded terms.
3.2. Parameters for Van Der Waals
A stacked dimer containing two glycine tetrapeptide molecules is devised and the benchmark intermolecular stacking interaction energy curve is computed by the CP-corrected DLPNO-MP2/aug-cc-pVTZ method. Taking the van der Waals parameters of AMBER99sb as initial parameters, the stacking interaction curve is also computed using our potential with the electrostatic parameters determined above. The parameters for van der Waals are adjusted so that the stacking interaction energy curve from our potential is consistent with the corresponding MP2 one (Figure 3c). The relevant parameters determined in this way are listed in Table 1. The van der Waals parameters for other atom types are taken from AMBER99sb without any modification and also given in Table 1. The parameters different from AMBER99sb are highlighted in bold.
3.3. Parameters for Bonded Terms
In this paper, the AMBER99sb parameters are adopted for bond, angle, and dihedral terms without any modification.
3.4. Scale Factors for Intramolecular Dipole–Dipole Interactions
Scale factors are used to mask the short-range intramolecular dipole–dipole interactions. We determine the scale factors by reproducing the benchmark conformational energies of glycine and alanine peptides, including a total of 950 dipeptide structures [11] (325 structures of glycine dipeptide and 625 structures of alanine dipeptide) and 79 tetrapeptide structures [34] (28 structures of glycine tetrapeptide and 51 structures of alanine tetrapeptide). The 79 tetrapeptide structures are provided in Figure S2 of the Supporting Information. Cartesian co-ordinates in angstroms for 79 tetrapeptide structures are provided in Table S5. The C7eq conformer of alanine peptide (or the C7 conformer of glycine peptide) is chosen as zero energy reference due to the consideration that C7eq is the global minimum of the alanine dipeptide. The scale factor for interaction between dipoles is determined to be 0.75 for two dipoles separated by three bonds and 0.90 for two dipoles separated by four bonds. In Table S1, the root-mean-square errors (RMSEs) of the conformational energies produced by our potential are listed with respect to the benchmark conformational energies. Table S1 also includes the AMBER99sb and AMOEBAbio18 results for comparison. The linear correlations between the benchmark conformational energies and the results produced by our potential as well as produced by AMBER99sb and AMOEBAbio18 for all these 1029 di- and tetrapeptide structures are given in Figure S3. Both Table S1 and Figure S3 indicate that our potential is more accurate than AMBER99sb and AMOEBAbio18 in reproducing the benchmark conformational energies for these small peptide structures. The scale factors are also listed in Table 1. The parameters for bonded terms are collected in Table S2.
Within this work, the Gaussian 09 [35] or ORCA5.0.1 [36] package suite are used for the electronic structure calculations. The force field AMOEBAbio18 and AMBER99sb calculations are carried out by the Tinker 8.10.1 package [37]. Our peptide-potential-related calculations are carried out by our in-house-developed PBFF code [38].
4. Applications
4.1. Conformational Energy
We first check our peptide potential in the reproduction of peptides’ conformational energies of quantum mechanical calculations. The neutral polypeptide molecules Ac(Ala)4X(Ala)4NHMe, where X = Val, Leu, Ile, Phe, Ser, Thr, Hid, Hie, Trp, Cys, Tyr, Met, Asn, and Gln, are chosen as testing systems. For all these testing peptides except X = Hid, three important conformation structures [39], helices, β-strand, and 27-ribbon, are obtained from B3LYP/6-31G* calculations in vacuum (6-31G* is used for geometry optimization due to the efficiency consideration). For the polypeptide with X = Hid, we succeeded in optimizing the helix and 27-ribbon structures but failed in optimizing the β-strand structure. In addition, this is another reason why we use the 27-ribbon (C7eq) conformer not the β-strand (C5) conformer as zero energy reference. Some important geometrical parameters, such as backbone dihedral angles and hydrogen bond lengths of these testing systems are collected in Table S3. The Cartesian co-ordinates of these testing systems are collected in Table S5.
Our potential as well as the parameters listed in Table 1 and Table S2 are employed to rapidly calculate the conformational energies for these testing peptides. The M06-2X/cc-pVTZ conformation energies are taken as benchmark because the MP2 method is too expensive for these large peptides. In Table 2, the conformational energies produced by our potential, by the M06-2X method, and by the force field AMBER99sb and AMOEBAbio18 methods are collected. Table 2 shows that the famous M06-2X method yields negative conformational energies for all helices and positive conformational energies for all β-strands for all these testing peptides, suggesting that the helix conformer is more stable and the β-strand conformer is more unstable than the 27-ribbon conformer. Both our potential and the AMBER99sb force field produce the conformational energies qualitatively consistent with the M06-2X results for all the testing peptides, whereas the AMOEBAbio18 force field produces negative conformational energies for β-strand conformers, which are in conflict with the M06-2X results.

Table 2.
Conformational energies (kcal/mol) relative to the C7eq conformer for AcAla4XAla4NHMe.
As listed in Table 2, for all the 14 testing peptides, our potential reproduces the benchmark M06-2X conformational energies with an RMSE of 2.04 kcal/mol, whereas AMBER99sb and AMOEBAbio18 had RMSEs of 5.22 and 14.08 kcal/mol, respectively. The maximum absolute error of our potential is 4.71 kcal/mol, found for the helix conformer of X = Hie. The maximum absolute error of AMBER99sb is 9.42 kcal/mol, found for the helix conformer of X = Hid, and that of AMOEBAbio18 is 21.79 kcal/mol, found for the β-strand conformer of X = Trp. These comparisons suggest that our potential is more accurate in predicting the conformational energies of peptides than AMBER99sb and AMOEBAbio18.
Two larger peptides, AcAla13NH2 and Ac(Ala2GlnAla2)3NH2, are further taken as testing systems. The conformational energies of the helix and β-strand conformer, relative to the 27-ribbon conformer, produced by our potential as well as by M06-2X/cc-pVTZ, AMBER99sb, and AMOEBAbio18, are collected in Table 3. For the peptide AcAla13NH2, all methods suggest a negative conformational energy for the helix conformer; the deviation from the M06-2X result is 2.00 kcal/mol for our potential, −11.84 kcal/mol for AMBER99sb, and −13.32 kcal/mol for AMOEBAbio18. M06-2X, AMBER99sb and our potential suggest a positive conformational energy for the β-strand conformer, whereas AMOEBAbio18 suggests a negative value for the β-strand. Similar results can be found for the peptide Ac(Ala2GlnAla2)3NH2. These comparisons further indicate our potential has a good ability to predict the conformation stability.

Table 3.
Conformational energies (kcal/mol) relative to the C7eq conformer for larger peptides.
In Figure 4, the linear correlations between the benchmark conformational energies and the force field ones are given for all testing systems of Table 2 and Table 3. Both the correlation coefficients and the RMSEs in Figure 4 further demonstrate that the peptide potential proposed in this work can reproduce the benchmark conformational energies reasonably and is more accurate than the famous AMBER99sb and AMOEBAbio18 force fields. It is found that the AMOEBAbio18 force field was not good at predicting the conformational energy in vacuum, which may be because it is designed to best reproduce energies in water, not energies in vacuum.
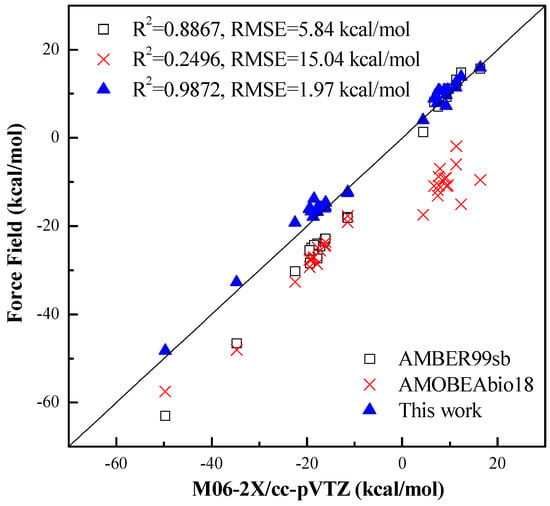
Figure 4.
The linear correlation between the M06−2X conformational energies and the force field ones.
4.2. Intermolecular Interaction Energy
Hydrogen bonding is ubiquitous in biosystems [40,41]. We further assess our potential by calculating the intermolecular interaction energies of several hydrogen-bonded dimers rapidly. For these testing dimers, the structures (Figure S4) are taken from references [27], and the Cartesian co-ordinates are included in Table S5. In Table 4 the interaction energies predicted by our potential and by AMBER99sb and AMOEBAbio18 force fields are collected. The quantum mechanical interaction energies taken from references [27] are chosen as benchmark and also given in Table 4. For the hydrogen-bonded dimers, relative to the benchmark interaction energies, the RMSE of our potential is 1.02 kcal/mol and that of AMBER99sb and AMOEBAbio18 is 1.70 and 2.21 kcal/mol, respectively. Our peptide potential can also yield the interaction energies of hydrogen bonding and stacking dimers accurately. The electrostatic contributions in parentheses are also listed in Table 4 (see Table S4 of the Supporting Information for details). For these hydrogen-bonded dimers, the electrostatic contribution ranges from 81.0% to 88.7% predicted by our model, 81.0% to 96.5% predicted by AMBER99sb, and 95.8% to 113.9% predicted by AMOEBAbio18. AMOEBAbio18 produces much higher electrostatic contributions due to its repulsive vdW interaction.

Table 4.
Interaction energies (kcal/mol) for hydrogen-bonded dimers.
4.3. Structures
Our peptide potential is further examined in the optimization of peptide structures based on the numerical first derivatives evaluated by the three-point forward difference formula. The optimal structures produced by our potential are compared with those from B3LYP/6-31G*, AMBER99sb, and AMOEBAbio18 calculations. Several selected optimal structures are given in Figure 5 as examples. The numbers in parentheses are the RMSEs of the force field optimal structures relative to the B3LYP optimal structures. It can be seen from Figure 5 that the RMSEs of our potential are 0.47, 0.66, and 0.63 Å for the helix, β-strands, and 27-ribbon, respectively. The RMSEs of AMBER99sb are 0.65, 0.76, and 1.50 Å for the helix, β-strands, and 27-ribbon, respectively. The RMSEs of AMOEBAbio18 are 0.46, 0.24, and 1.16 Å for the helix, β-strands, and 27-ribbon, respectively. These comparisons show that our potential, AMBER99sb, and AMOEBAbio18 exhibit similar accuracy in reproducing the B3LYP structures, indicating our peptide potential’s ability in producing reasonable secondary structures for polypeptide molecules.
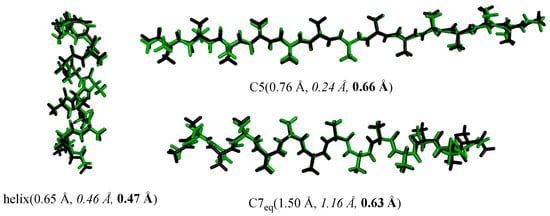
Figure 5.
Superposition of the B3LYP/6-31G(d) structures (black) and the force field ones (green) of AcAla13NH2. The values in parentheses are RMSEs of the force field structures relative to the B3LYP structure of the heavy atoms. AMBER99sb: normal; AMOEBAbio18: italic; our potential: bold.
4.4. Efficiency
Efficiency of our peptide potential is evaluated by calculating the single-point energy of the helix conformer of the peptide Ac(Ala2GlnAla2)3NH2 on a workstation with Intel(R) Xeon(R) CPU E5-2696 v4 @ 2.20 GHz. For the helix structure of Ac(Ala2GlnAla2)3NH2, it takes about 48 h and 11 min CPU time to finish an M06-2X/cc-pVTZ single-point calculation. The CPU time consumed by AMBER99sb, AMOEBAbio18, and our model is only 2.676, 4.174, and 2.049 s, respectively, indicating the high efficiency of our potential.
5. Conclusions
A peptide potential is proposed based on a bond dipole representation of electrostatic interaction. The parameters are derived from fitting to the high qualified ab initio results. Comparisons with the conformational energies yielded by the M06-2X, AMBER99sb, and AMOEBAbio18 and with the interaction energies produced by MP2, AMBER99sb, and AMOEBAbio18 show that the peptide potential reported here works much better than the famous AMBER99sb and AMOEBAbio18 force fields.
Compared with the famous atom-centered point-charge-based AMBER force filed and the famous atom-centered point-multipole-based AMOEBA force field, our bond-dipole-based peptide potential has much less electrostatic terms. For example, the peptide Ac(Ala2GlnAla2)3NH2 possesses 180 atoms, which corresponds to 180 point charges and, thus, results in a total of 15,613 charge–charge interaction terms according to the AMBER99sb force field. The AMOEBA force field assigns each atom one atom-centered partial charge, one or two nonzero atom-centered dipoles, and no less than three nonzero atom-centered quadrupoles. These result in 15,613 charge–charge interaction terms, many times of 15,613 dipole–dipole interaction terms, and many charge–dipole, charge–quadrupole, dipole–quadrupole, quadrupole–quadrupole interaction terms for the peptide Ac(Ala2GlnAla2)3NH2. Our model regards the chemical bonds C=O, N-H, and C-H of the Ac(Ala2GlnAla2)3NH2 molecule as bond dipoles. Here, we like to stress that, in our potential, the electrostatics are expressed as a function of the chemical bond dipoles, which is quite distinct from the atom dipoles used in the AMOEBA potential. According to our peptide potential model, there are only 57 bond dipole moments in the peptide Ac(Ala2GlnAla2)3NH2, which results in only 1493 dipole–dipole interaction terms. Despite this evident simplification in electrostatics, the performance of our potential is encouraging. We hope this will lead to a new path for polarizable force field development.
Further applications of our peptide potential to more complicated biomolecular systems, such as the peptides containing charged side chains, as well as the extensive sampling of Monte–Carlo simulations and molecular dynamic simulations, are not examined at our current stage, which should be a focus of our future work to validate and parameterize our model. Research on these directions is now being undertaken.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pr11041291/s1, Two clusters which contain 1 glycine tetrapeptide and 30 water molecules are provided in Figure S1. The 79 conformations used to derive the scale factors for intramolecular dipole–dipole interactions are provided in Figure S2. RMSEs of different methods with respect to the benchmark QM conformational energies are provided in Table S1. The linear correlations between the QM conformational energies and the force field ones for all the structures included in Table S1 are provided in Figure S3. Bond stretching, angle bending, and dihedral angle parameters used in our potential are provided in Table S2. The backbone dihedral angles and hydrogen bond lengths for polypeptides AcAla4XAla4NHMe (X = Val, Ile, Leu, Asn, Gln, Ser, Thr, Phe, Cys, Met, Hid, Hie, Trp, and Tyr), AcAla13NH2, and Ac(Ala2GlnAla2)3NH2 are provided in Table S3. The hydrogen-bonded dimers are provided in Figure S4. Physical components of the interaction energy (IE) for eight hydrogen-bonded dimers are provided in Table S4. Cartesian co-ordinates in angstroms for Cluster A and B, 28 glycine tetrapeptide conformers, 51 alanine tetrapeptide conformers, AcAla4XAla4NHMe (X = Val, Ile, Leu, Asn, Gln, Ser, Thr, Phe, Cys, Met, Hid, Hie, Trp, and Tyr), AcAla13NH2, Ac(Ala2GlnAla2)3NH2, and hydrogen-bonded dimers are provided in Table S5.
Author Contributions
Conceptualization, C.-S.W.; methodology, C.-S.W., Y.-M.L. and X.-H.Z.; software, Q.H.; validation, Y.-M.L., X.-H.Z., C.-M.L. and Q.L.; formal analysis, C.-S.W., Y.-M.L., X.-H.Z., L.W. and Q.H.; investigation, Y.-M.L. and X.-H.Z.; data curation, Y.-M.L. and X.-H.Z.; writing—original draft preparation, Y.-M.L., X.-H.Z., C.-M.L. and Q.L.; writing—review and editing, Y.-M.L., X.-H.Z., L.W., Q.H. and C.-S.W.; funding acquisition, C.-S.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China, grant number 21773102 and Department of Education of Liaoning Province, grant number LJKMZ20221411, LQ2020024.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
We gratefully acknowledge the financial support from National Natural Science Foundation of China (21773102) and Department of Education of Liaoning Province (LJKMZ20221411, LQ2020024).
Conflicts of Interest
The authors declare no competing financial interest.
References
- MacKerell, A.D.; Wiórkiewicz-Kuczera, J.; Karplus, M. An All-Atom Empirical Energy Function for the Simulation of Nucleic Acids. J. Am. Chem. Soc. 1995, 117, 11946–11975. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grübmuller, H.; MacKerell, A.D., Jr. CHARMM36M: An Improved Force Field for Folded and Intrinsically Disordered Proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Maxwell, D.S.; Tirado-Rives, J. Development and Testing of the OPLS All-Atom Force Field on Conformational Energetics and Properties of Organic Liquids. J. Am. Chem. Soc. 1996, 118, 11225–11236. [Google Scholar] [CrossRef]
- Robertson, M.J.; Tirado-Rives, J.; Jorgensen, W.L. Improved Peptide and Protein Torsional Energetics with the OPLS-AA Force Field. J. Chem. Theory Comput. 2015, 11, 3499–3509. [Google Scholar] [CrossRef]
- Oostenbrink, C.; Villa, A.; Mark, A.E.; van Gunsteren, W.F. A Biomolecular Force Field Based on the Free Enthalpy of Hydration and Solvation: The GROMOS Force-Field Parameter Sets 53A5 and 53A6. J. Comput. Chem. 2004, 25, 1656–1676. [Google Scholar] [CrossRef]
- Schmid, N.; Eichenberger, A.P.; Choutko, A.; Riniker, S.; Winger, M.; Mark, A.E.; van Gunsteren, W.F. Definition and Testing of the GROMOS Force-Field Versions 54A7 and 54B7. Eur. Biophys. J. 2011, 40, 843–856. [Google Scholar] [CrossRef]
- Cornell, W.D.; Cieplak, P.; Bayly, C.I.; Gould, I.R.; Merz, K.M.; Ferguson, D.M.; Spellmeyer, D.C.; Fox, T.; Caldwell, J.W.; Kollman, P.A. A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. J. Am. Chem. Soc. 1995, 117, 5179–5197. [Google Scholar] [CrossRef]
- Tian, C.; Kasavajhala, K.; Belfon, K.A.A.; Raguette, L.; Huang, H.; Migues, A.N.; Bickel, J.; Wang, Y.; Pincay, J.; Wu, Q.; et al. ff19SB: Amino-Acid-Specific Protein Backbone Parameters Trained against Quantum Mechanics Energy Surfaces in Solution. J. Chem. Theory Comput. 2020, 16, 528–552. [Google Scholar] [CrossRef]
- Shi, Y.; Xia, Z.; Zhang, J.; Best, R.; Wu, C.; Ponder, J.W.; Ren, P. Polarizable Atomic Multipole-Based AMOEBA Force Field for Proteins. J. Chem. Theory Comput. 2013, 9, 4046–4063. [Google Scholar] [CrossRef]
- Zhang, C.; Lu, C.; Jing, Z.; Wu, C.; Piquemal, J.P.; Ponder, J.W.; Ren, P. AMOEBA Polarizable Atomic Multipole Force Field for Nucleic Acids. J. Chem. Theory Comput. 2018, 14, 2084–2108. [Google Scholar] [CrossRef]
- Lopes, P.E.M.; Huang, J.; Shim, J.; Luo, Y.; Li, H.; Roux, B.; MacKerell, A.D. Polarizable Force Field for Peptides and Proteins Based on the Classical Drude Oscillator. J. Chem. Theory Comput. 2013, 9, 5430–5449. [Google Scholar] [CrossRef]
- Kognole, A.A.; Aytenfisu, A.H.; MacKerell, A.D. Extension of the CHARMM Classical Drude Polarizable Force Field to N- and O-Linked Glycopeptides and Glycoproteins. J. Phys. Chem. B 2022, 126, 6642–6653. [Google Scholar] [CrossRef]
- Lemkul, J.A.; Huang, J.; Roux, B.; MacKerell, A.D., Jr. An Empirical Polarizable Force Field Based on the Classical Drude Oscillator Model: Development History and Recent Applications. Chem. Rev. 2016, 116, 4983–5013. [Google Scholar] [CrossRef]
- Marshall, G.R. Limiting Assumptions in Molecular Modeling: Electrostatics. J. Comput. Aided Mol. Des. 2013, 27, 107–114. [Google Scholar] [CrossRef]
- Cisneros, G.A.; Karttunen, M.; Ren, P.; Sagui, C. Classical Electrostatics for Biomolecular Simulations. Chem. Rev. 2014, 114, 779–814. [Google Scholar] [CrossRef]
- Barker, J.A. Statistical Mechanics of Interacting Dipoles. Proc. R. Soc. A 1953, 219, 367–372. [Google Scholar]
- Buckingham, A.D.; Pople, J.A. The Statistical Mechanics of Imperfect Polar Gases. Part 1.—Second Virial Coefficients. Trans. Faraday Soc. 1955, 51, 1173–1179. [Google Scholar] [CrossRef]
- Ren, P.; Ponder, J.W. Polarizable Atomic Multipole Water Model for Molecular Mechanics Simulation. J. Phys. Chem. B 2003, 107, 5933–5947. [Google Scholar] [CrossRef]
- Lamoureux, G.; MacKerell, A.D.; Roux, B.T. A Simple Polarizable Model of Water Based on Classical Drude Oscillators. J. Chem. Phys. 2003, 119, 5185–5197. [Google Scholar] [CrossRef]
- Liu, C.; Piquemal, J.P.; Ren, P. AMOEBA+ Classical Potential for Modeling Molecular Interactions. J. Chem. Theory Comput. 2019, 15, 4122–4139. [Google Scholar] [CrossRef]
- Liu, C.; Piquemal, J.P.; Ren, P. Implementation of Geometry-Dependent Charge Flux into the Polarizable AMOEBA+ Potential. J. Phys. Chem. Lett. 2020, 11, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Medders, G.R.; Babin, V.; Paesani, F. A Critical Assessment of Two-Body and Three-Body Interactions in Water. J. Chem. Theory Comput. 2013, 9, 1103–1114. [Google Scholar] [CrossRef] [PubMed]
- Bull-Vulpe, E.F.; Riera, M.; Götz, A.W.; Paesani, F. MB-Fit: Software Infrastructure for Data-Driven Many-Body Potential Energy Functions. J. Chem. Phys. 2021, 155, 124801–124817. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.-L.; Jiang, X.-N.; Wang, C.-S. An Analytic Potential Energy Function for the Amide-Amide and Amide-Water Intermolecular Hydrogen Bonds in Peptides. J. Comput. Chem. 2009, 30, 2567–2575. [Google Scholar] [CrossRef]
- Jiang, X.-N.; Sun, C.-L.; Wang, C.-S. A Scheme for Rapid Prediction of Cooperativity in Hydrogen Bond Chains of Formamides, Acetamides, and N-Methylformamides. J. Comput. Chem. 2010, 31, 1410–1420. [Google Scholar] [CrossRef]
- Li, Y.; Jiang, X.-N.; Wang, C.-S. Rapid Evaluation of the Binding Energies in Hydrogen-Bonded Amide-Thymine and Amide-Uracil Dimers in Gas Phase. J. Comput. Chem. 2011, 32, 953–966. [Google Scholar] [CrossRef]
- Li, S.-S.; Huang, C.-Y.; Hao, J.-J.; Wang, C.-S. A Polarizable Dipole-Dipole Interaction Model for Evaluation of the Interaction Energies for N-H···O=C and C-H···O=C Hydrogen-Bonded Complexes. J. Comput. Chem. 2014, 35, 415–426. [Google Scholar] [CrossRef]
- Gao, X.-C.; Hao, Q.; Wang, C.-S. Improved Polarizable Dipole-Dipole Interaction Model for Hydrogen Bonding, Stacking, T-Shaped, and X-H···π Interactions. J. Chem. Theory Comput. 2017, 13, 2730–2741. [Google Scholar] [CrossRef]
- Huang, C.; Hao, Q.; Wang, C. Rapid Prediction of Interaction Energies for Nucleoside-Containing Hydrogen-Bonded Complexes: Lone-Pair Dipole Moment Treatment for Adenine, Cytosine and Guanine. Chem. Res. Chin. Univ. 2017, 33, 94–99. [Google Scholar] [CrossRef]
- Li, X.-L.; Li, C.-M.; Zhu, J.-Y.; Zhou, Z.; Hao, Q.; Wang, C.-S. A Scheme for Rapid Evaluation of the Intermolecular Three-body Polarization Effect in Water Clusters. J. Comput. Chem. 2023, 44, 677–686. [Google Scholar] [CrossRef]
- Li, X.-L.; Sun, Y.-J.; Ying, T.; Wang, C.-S. Rapid and Accurate Calculation of the Three⁃body Interaction Strength in the Hydrogen-bonded Complexes of Alcohols or Deoxyribose with Water. Chem. J. Chin. Univ. 2021, 42, 3664–3671. [Google Scholar]
- Buckingham, A.D. Molecular Quadrupole Moments. Q. Rev. Chem. Soc. 1959, 13, 183–214. [Google Scholar] [CrossRef]
- Speight, J.G. Lange’s Handbook of Chemistry, 16th ed.; McGraw-Hill: New York, NY, USA, 2005. [Google Scholar]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of Multiple Amber Force Fields and Development of Improved Protein Backbone Parameters. Proteins Struct. Funct. Bioinf. 2006, 65, 712–725. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision D.01; Gaussian, Inc.: Wallingford, CT, USA, 2013. [Google Scholar]
- Neese, F. Software Update: The ORCA Program System—Version 5.0. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2022, 12, e1606. [Google Scholar] [CrossRef]
- Ponder, J.W. TINKER Molecular Modeling Package, V8.10.1; Washington University Medical School: St. Louis, MO, USA, 2021. [Google Scholar]
- PBFF Is an In-House Developed Fortran Code Which Is Publicly. Available online: http://cswang.home.lnnu.edu.cn (accessed on 20 October 2022).
- Wu, Y.-D.; Zhao, Y.-L. A Theoretical Study on the Origin of Cooperativity in the Formation of 310- and α-helices. J. Am. Chem. Soc. 2001, 123, 5313–5319. [Google Scholar] [CrossRef]
- Dill, K.A.; MacCallum, J.L. The Protein-Folding Problem, 50 Years On. Science 2012, 338, 1042–1046. [Google Scholar] [CrossRef]
- Gromiha, M.M.; Siebers, J.G.; Selvaraj, S.; Kono, H.; Sarai, A. Role of Inter and Intramolecular Interactions in Protein-DNA Recognition. Gene 2005, 364, 108–113. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).