Apolipoprotein Signature of HDL and LDL from Atherosclerotic Patients in Relation with Carotid Plaque Typology: A Preliminary Report
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Lipoproteins Purification
2.3. SDS-PAGE Analysis
2.4. Shotgun Proteomics
2.5. Protein–Protein Interaction (PPI) Network and Gene Ontology (GO) Analysis
3. Results
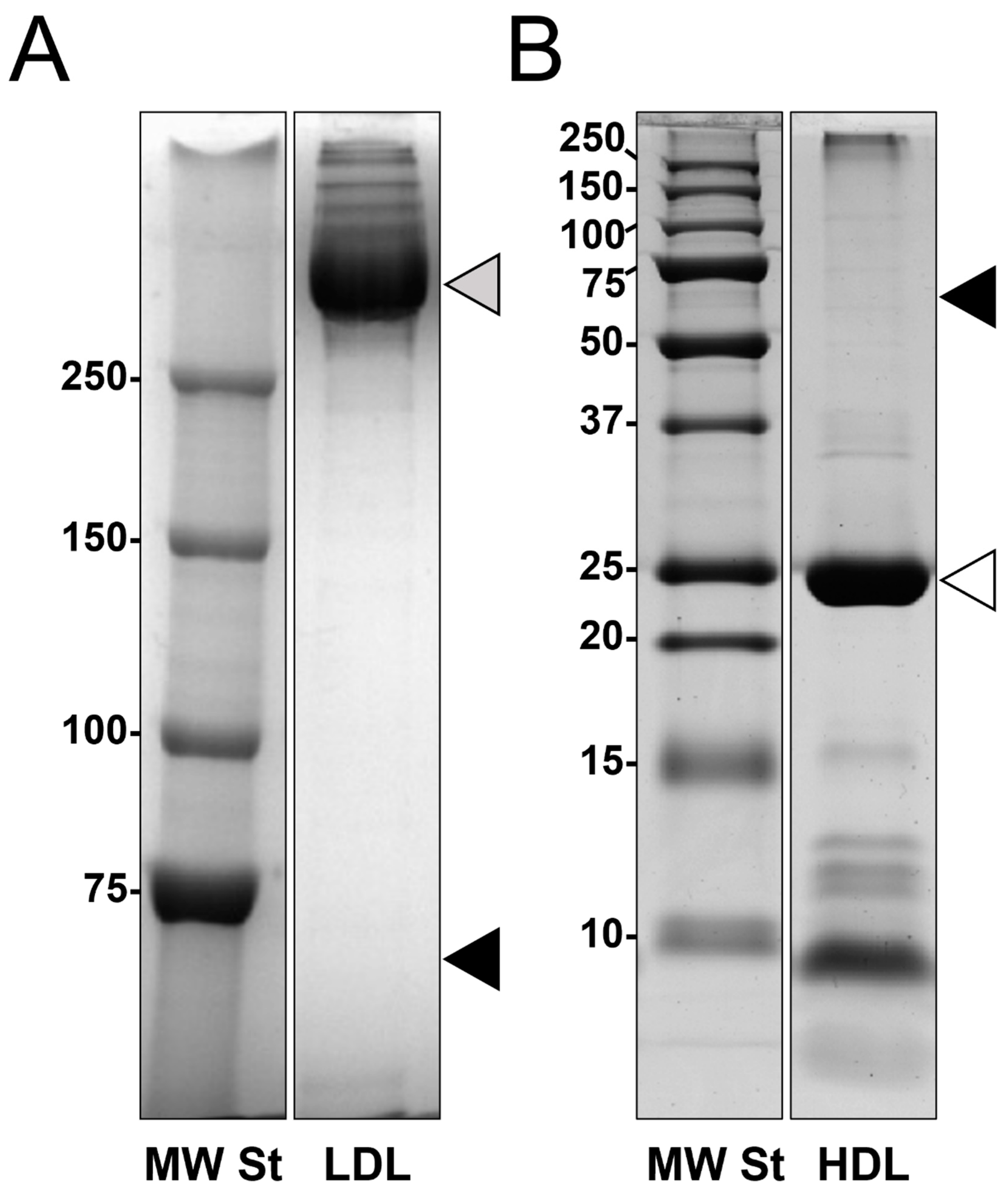
3.1. Lipoproteins Purification
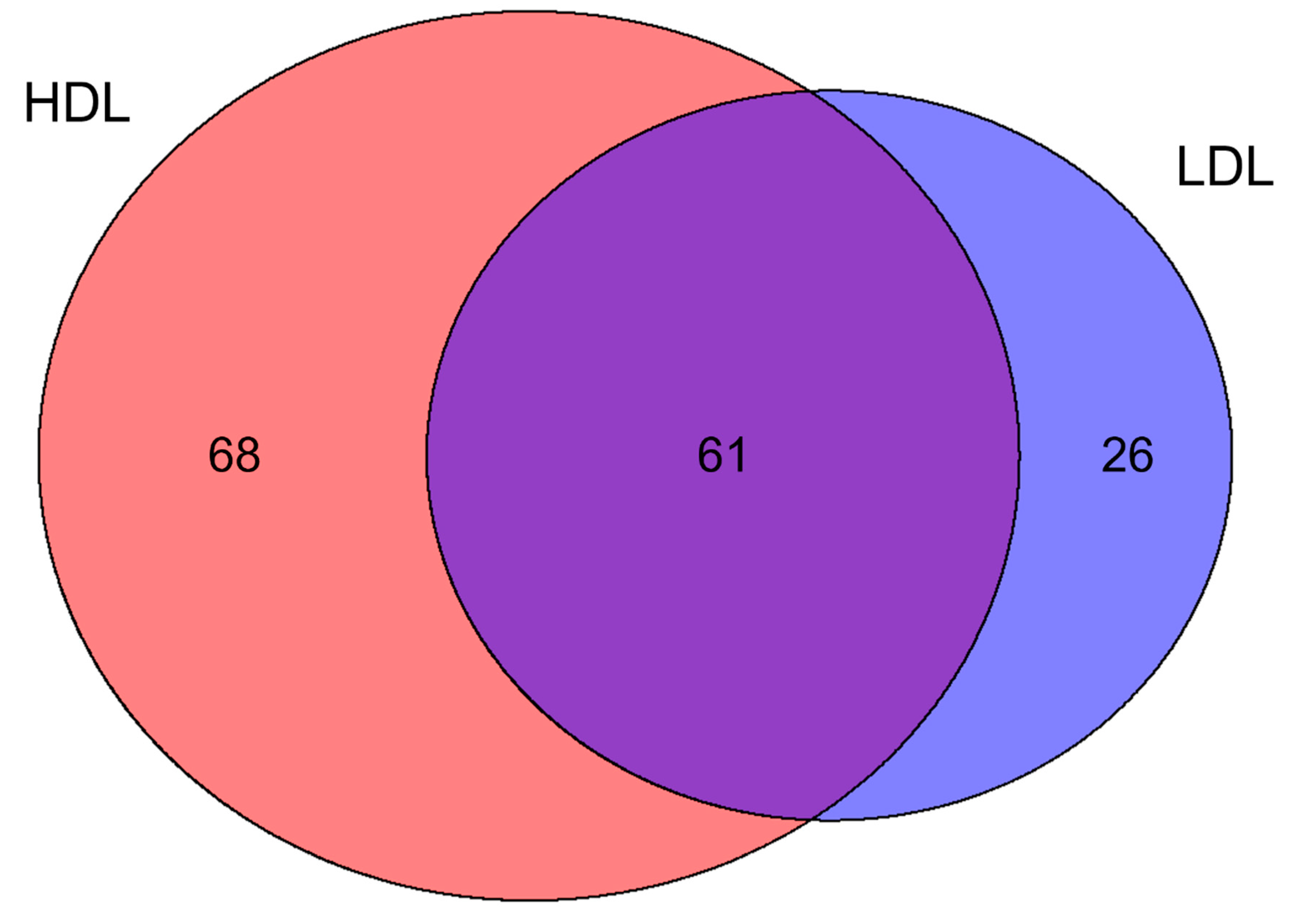
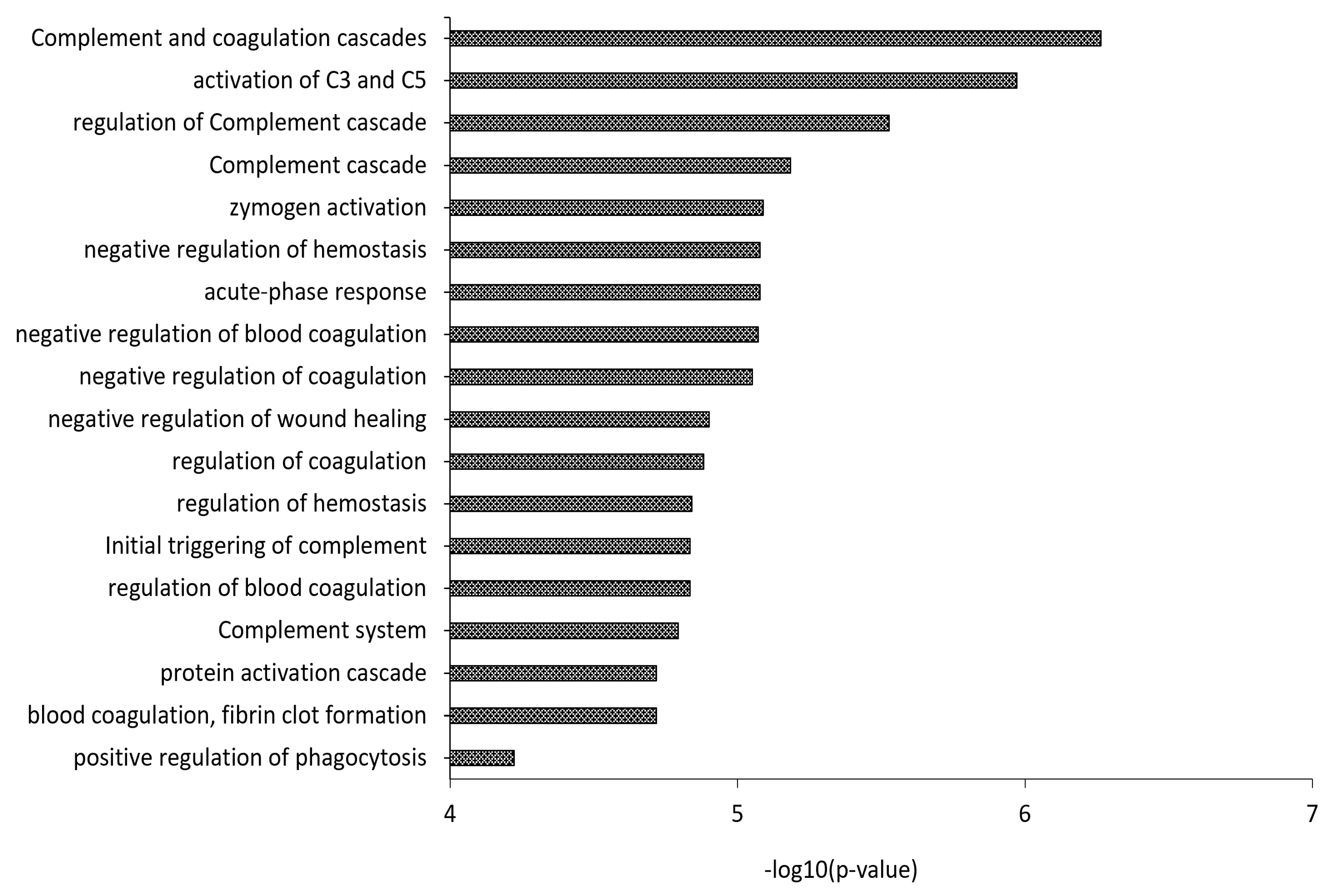
3.2. Proteomics Characterization of HDL and LDL
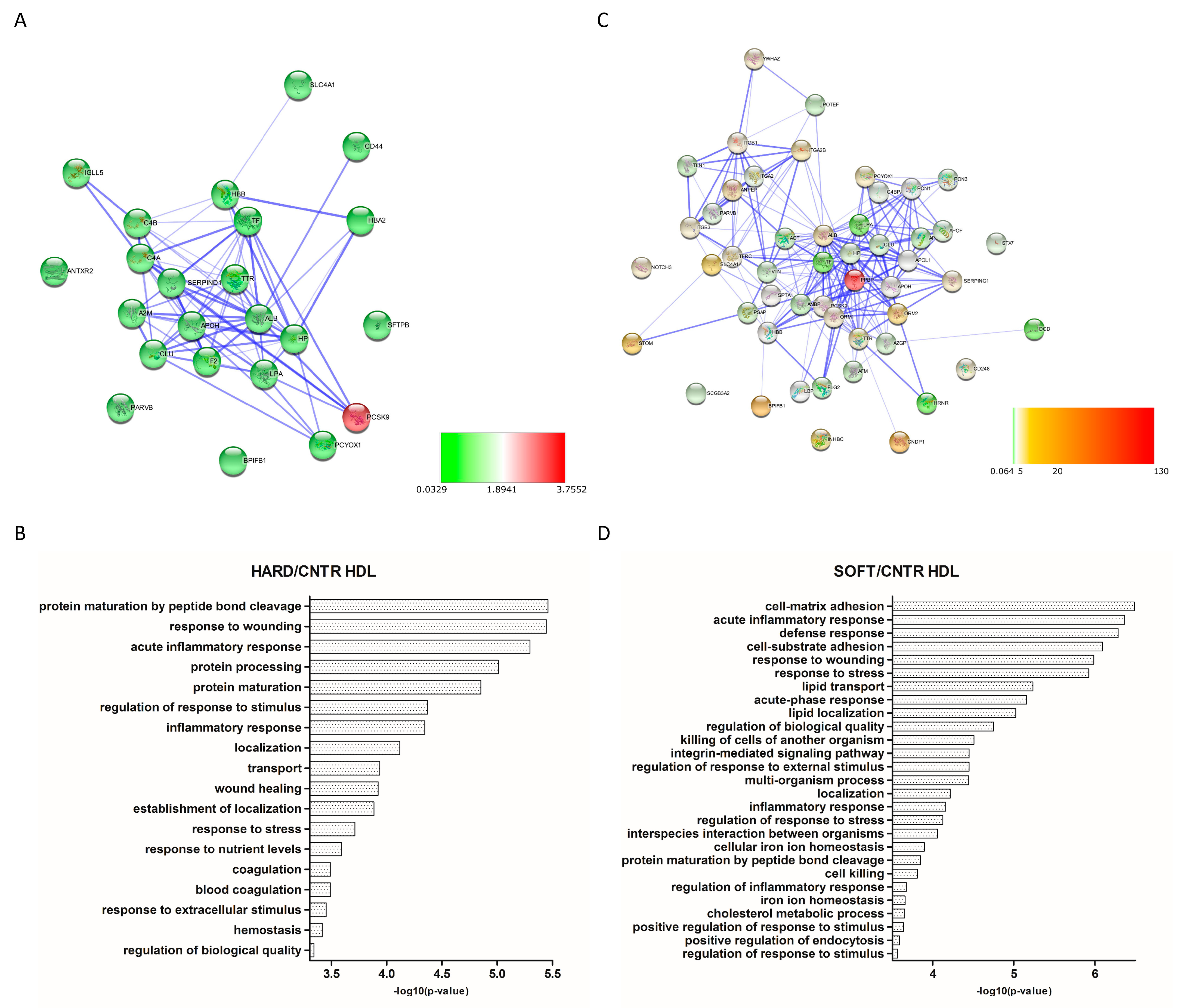
3.3. Differential Expression Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hansson, G.K.; Libby, P.; Tabas, I. Inflammation and plaque vulnerability. J. Intern. Med. 2015, 278, 483–493. [Google Scholar] [CrossRef]
- Libby, P.; Buring, J.E.; Badimon, L.; Hansson, G.K.; Deanfield, J.; Bittencourt, M.S.; Tokgözoğlu, L.; Lewis, E.F. Atherosclerosis. Nat. Rev. Dis. Prim. 2019, 5, 56. [Google Scholar] [CrossRef] [PubMed]
- Puig, N.; Jiménez-Xarrié, E.; Camps-Renom, P.; Benitez, S. Search for Reliable Circulating Biomarkers to Predict Carotid Plaque Vulnerability. Int. J. Mol. Sci. 2020, 21, 8236. [Google Scholar] [CrossRef]
- Zhu, G.; Hom, J.; Li, Y.; Jiang, B.; Rodriguez, F.; Fleischmann, D.; Saloner, D.; Porcu, M.; Zhang, Y.; Saba, L.; et al. Carotid plaque imaging and the risk of atherosclerotic cardiovascular disease. Cardiovasc. Diagn. Ther. 2020, 10, 1048–1067. [Google Scholar] [CrossRef]
- Tuñón, J.; Martín-Ventura, J.L.; Blanco-Colio, L.M.; Lorenzo, O.; López, J.A.; Egido, J. Proteomic strategies in the search of new biomarkers in atherothrombosis. J. Am. Coll. Cardiol. 2010, 55, 2009–2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rocchiccioli, S.; Pelosi, G.; Rosini, S.; Marconi, M.; Viglione, F.; Citti, L.; Ferrari, M.; Trivella, M.G.; Cecchettini, A. Secreted proteins from carotid endarterectomy: An untargeted approach to disclose molecular clues of plaque progression. J. Transl. Med. 2013, 11, 260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eslava-Alcon, S.; Extremera-García, M.J.; González-Rovira, A.; Rosal-Vela, A.; Rojas-Torres, M.; Beltran-Camacho, L.; Sanchez-Gomar, I.; Jiménez-Palomares, M.; Alonso-Piñero, J.A.; Conejero, R.; et al. Molecular signatures of atherosclerotic plaques: An up-dated panel of protein related markers. J. Proteom. 2020, 221, 103757. [Google Scholar] [CrossRef]
- Lygirou, V.; Latosinska, A.; Makridakis, M.; Mullen, W.; Delles, C.; Schanstra, J.P.; Zoidakis, J.; Pieske, B.; Mischak, H.; Vlahou, A. Plasma proteomic analysis reveals altered protein abundances in cardiovascular disease. J. Transl. Med. 2018, 16, 104. [Google Scholar] [CrossRef]
- Delles, C.; Diez, J.; Dominiczak, A.F. Urinary proteomics in cardiovascular disease: Achievements, limits and hopes. Proteom. Clin. Appl. 2011, 5, 222–232. [Google Scholar] [CrossRef]
- Hoofnagle, A.N.; Heinecke, J.W. Lipoproteomics: Using mass spectrometry-based proteomics to explore the assembly, structure, and function of lipoproteins. J. Lipid Res. 2009, 50, 1967–1975. [Google Scholar] [CrossRef] [Green Version]
- Shah, A.S.; Tan, L.; Long, J.L.; Davidson, W.S. Proteomic diversity of high density lipoproteins: Our emerging understanding of its importance in lipid transport and beyond. J. Lipid Res. 2013, 54, 2575–2585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lepedda, A.J.; Cigliano, A.; Cherchi, G.M.; Spirito, R.; Maggioni, M.; Carta, F.; Turrini, F.; Edelstein, C.; Scanu, A.M.; Formato, M. A proteomic approach to differentiate histologically classified stable and unstable plaques from human carotid arteries. Atherosclerosis 2009, 203, 112–118. [Google Scholar] [CrossRef] [Green Version]
- Lepedda, A.J.; Lobina, O.; Rocchiccioli, S.; Nieddu, G.; Ucciferri, N.; De Muro, P.; Idini, M.; Nguyen, H.Q.; Guarino, A.; Spirito, R.; et al. Identification of differentially expressed plasma proteins in atherosclerotic patients with type 2 diabetes. J. Diabetes Complicat. 2016, 30, 880–886. [Google Scholar] [CrossRef] [PubMed]
- Lepedda, A.J.; Zinellu, A.; Nieddu, G.; Zinellu, E.; Carru, C.; Spirito, R.; Guarino, A.; De Muro, P.; Formato, M. Protein sulfhydryl group oxidation and mixed-disulfide modifications in stable and unstable human carotid plaques. Oxid. Med. Cell. Longev. 2013, 2013, 403973. [Google Scholar] [CrossRef]
- Lepedda, A.J.; Formato, M. Oxidative Modifications in Advanced Atherosclerotic Plaques: A Focus on In Situ Protein Sulfhydryl Group Oxidation. Oxid. Med. Cell. Longev. 2020, 2020, 6169825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakashima, Y.; Wight, T.N.; Sueishi, K. Early atherosclerosis in humans: Role of diffuse intimal thickening and extracellular matrix proteoglycans. Cardiovasc. Res. 2008, 79, 14–23. [Google Scholar] [CrossRef] [Green Version]
- Ouimet, M.; Barrett, T.J.; Fisher, E.A. HDL and Reverse Cholesterol Transport. Circ. Res. 2019, 124, 1505–1518. [Google Scholar] [CrossRef]
- Kajani, S.; Curley, S.; McGillicuddy, F.C. Unravelling HDL-Looking beyond the Cholesterol Surface to the Quality Within. Int. J. Mol. Sci. 2018, 19, 1971. [Google Scholar] [CrossRef] [Green Version]
- Vaisar, T.; Pennathur, S.; Green, P.S.; Gharib, S.A.; Hoofnagle, A.N.; Cheung, M.C.; Byun, J.; Vuletic, S.; Kassim, S.; Singh, P.; et al. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J. Clin. Investig. 2007, 117, 746–756. [Google Scholar] [CrossRef] [PubMed]
- Heinecke, J.W. The protein cargo of HDL: Implications for vascular wall biology and therapeutics. J. Clin. Lipidol. 2010, 4, 371–375. [Google Scholar] [CrossRef] [Green Version]
- Alwaili, K.; Bailey, D.; Awan, Z.; Bailey, S.D.; Ruel, I.; Hafiane, A.; Krimbou, L.; Laboissiere, S.; Genest, J. The HDL proteome in acute coronary syndromes shifts to an inflammfatory profile. Biochim. Biophys. Acta 2012, 1821, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.R.; Wang, D.X.; Liu, H.; Zhang, X.X.; Zhao, H.; Hua, L.; Xu, P.; Li, Y.S. A pro-atherogenic HDL profile in coronary heart disease patients: An iTRAQ labelling-based proteomic approach. PLoS ONE 2014, 9, e98368. [Google Scholar] [CrossRef]
- Lepedda, A.J.; Nieddu, G.; Zinellu, E.; De Muro, P.; Piredda, F.; Guarino, A.; Spirito, R.; Carta, F.; Turrini, F.; Formato, M. Proteomic analysis of plasma-purified VLDL, LDL, and HDL fractions from atherosclerotic patients undergoing carotid endarterectomy: Identification of serum amyloid A as a potential marker. Oxid. Med. Cell. Longev. 2013, 2013, 385214. [Google Scholar] [CrossRef]
- Green, P.S.; Vaisar, T.; Pennathur, S.; Kulstad, J.J.; Moore, A.B.; Marcovina, S.; Brunzell, J.; Knopp, R.H.; Zhao, X.Q.; Heinecke, J.W. Combined statin and niacin therapy remodels the high-density lipoprotein proteome. Circulation 2008, 118, 1259–1267. [Google Scholar] [CrossRef] [Green Version]
- Dashty, M.; Motazacker, M.M.; Levels, J.; de Vries, M.; Mahmoudi, M.; Peppelenbosch, M.P.; Rezaee, F. Proteome of human plasma very low-density lipoprotein and low-density lipoprotein exhibits a link with coagulation and lipid metabolism. Thromb. Haemost. 2014, 111, 518–530. [Google Scholar] [CrossRef]
- Vaisar, T.; Mayer, P.; Nilsson, E.; Zhao, X.Q.; Knopp, R.; Prazen, B.J. HDL in humans with cardiovascular disease exhibits a proteomic signature. Clin. Chim. Acta. 2010, 411, 972–979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, Y.; Liu, T.R.; Hu, S.W.; Tian, D.; Li, C.; Zhong, J.K.; Sun, H.G.; Luo, T.T.; Lai, W.Y.; Guo, Z.G. Acute coronary syndrome remodels the protein cargo and functions of high-density lipoprotein subfractions. PLoS ONE 2014, 9, e94264. [Google Scholar] [CrossRef] [PubMed]
- Cubedo, J.; Padró, T.; García-Moll, X.; Pintó, X.; Cinca, J.; Badimon, L. Proteomic signature of Apolipoprotein J in the early phase of new-onset myocardial infarction. J. Proteome Res. 2011, 10, 211–220. [Google Scholar] [CrossRef]
- Rubinow, K.B.; Henderson, C.M.; Robinson-Cohen, C.; Himmelfarb, J.; de Boer, I.H.; Vaisar, T.; Kestenbaum, B.; Hoofnagle, A.N. Kidney function is associated with an altered protein composition of high-density lipoprotein. Kidney Int. 2017, 92, 1526–1535. [Google Scholar] [CrossRef]
- Wang, K.; Zelnick, L.R.; Hoofnagle, A.N.; Vaisar, T.; Henderson, C.M.; Imrey, P.B.; Robinson-Cohen, C.; de Boer, I.H.; Shiu, Y.T.; Himmelfarb, J.; et al. Alteration of HDL Protein Composition with Hemodialysis Initiation. Clin. J. Am. Soc. Nephrol. 2018, 13, 1225–1233. [Google Scholar] [CrossRef] [Green Version]
- Shao, B.; de Boer, I.; Tang, C.; Mayer, P.S.; Zelnick, L.; Afkarian, M.; Heinecke, J.W.; Himmelfarb, J. A Cluster of Proteins Implicated in Kidney Disease Is Increased in High-Density Lipoprotein Isolated from Hemodialysis Subjects. J. Proteome Res. 2015, 14, 2792–2806. [Google Scholar] [CrossRef] [Green Version]
- Shao, B.; Zelnick, L.R.; Wimberger, J.; Himmelfarb, J.; Brunzell, J.; Davidson, W.S.; Snell-Bergeon, J.K.; Bornfeldt, K.E.; de Boer, I.H.; Heinecke, J.W. Albuminuria, the High-Density Lipoprotein Proteome, and Coronary Artery Calcification in Type 1 Diabetes Mellitus. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 1483–1491. [Google Scholar] [CrossRef]
- Gourgari, E.; Ma, J.; Playford, M.P.; Mehta, N.N.; Goldman, R.; Remaley, A.T.; Gordon, S.M. Proteomic alterations of HDL in youth with type 1 diabetes and their associations with glycemic control: A case-control study. Cardiovasc. Diabetol. 2019, 18, 43. [Google Scholar] [CrossRef] [PubMed]
- Kheniser, K.G.; Osme, A.; Kim, C.; Ilchenko, S.; Kasumov, T.; Kashyap, S.R. Temporal Dynamics of High-Density Lipoprotein Proteome in Diet-Controlled Subjects with Type 2 Diabetes. Biomolecules 2020, 10, 520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Reilly, M.; Dillon, E.; Guo, W.; Finucane, O.; McMorrow, A.; Murphy, A.; Lyons, C.; Jones, D.; Ryan, M.; Gibney, M.; et al. High-Density Lipoprotein Proteomic Composition, and not Efflux Capacity, Reflects Differential Modulation of Reverse Cholesterol Transport by Saturated and Monounsaturated Fat Diets. Circulation 2016, 133, 1838–1850. [Google Scholar] [CrossRef]
- Gordon, S.M.; Li, H.; Zhu, X.; Tso, P.; Reardon, C.A.; Shah, A.S.; Lu, L.J.; Davidson, W.S. Impact of genetic deletion of platform apolipoproteins on the size distribution of the murine lipoproteome. J. Proteom. 2016, 146, 184–194. [Google Scholar] [CrossRef] [Green Version]
- Brott, T.G.; Halperin, J.L.; Abbara, S.; Bacharach, J.M.; Barr, J.D.; Bush, R.L.; Cates, C.U.; Creager, M.A.; Fowler, S.B.; Friday, G.; et al. 2011 ASA/ACCF/AHA/AANN/AANS/ACR/ASNR/CNS/SAIP/SCAI/SIR/SNIS/SVM/SVS guideline on the management of patients with extracranial carotid and vertebral artery disease: Executive summary: A report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines, and the American Stroke Association, American Association of Neuroscience Nurses, American Association of Neurological Surgeons, American College of Radiology, American Society of Neuroradiology, Congress of Neurological Surgeons, Society of Atherosclerosis Imaging and Prevention, Society for Cardiovascular Angiography and Interventions, Society of Interventional Radiology, Society of NeuroInterventional Surgery, Society for Vascular Medicine, and Society for Vascular Surgery. Developed in collaboration with the American Academy of Neurology and Society of Cardiovascular Computed Tomography. Catheter. Cardiovasc. Interv. 2013, 81, E76–E123. [Google Scholar] [CrossRef]
- Gray-Weale, A.C.; Graham, J.C.; Burnett, J.R.; Byrne, K.; Lusby, R.J. Carotid artery atheroma: Comparison of preoperative B-mode ultrasound appearance with carotid endarterectomy specimen pathology. J. Cardiovasc. Surg. 1988, 29, 676–681. [Google Scholar]
- Huibers, A.; de Borst, G.J.; Bulbulia, R.; Pan, H.; Halliday, A.; ACST-1 Collaborative Group. Plaque Echolucency and the Risk of Ischaemic Stroke in Patients with Asymptomatic Carotid Stenosis Within the First Asymptomatic Carotid Surgery Trial (ACST-1). Eur. J. Vasc. Endovasc. Surg. 2016, 51, 616–621. [Google Scholar] [CrossRef]
- Knuuti, J.; Wijns, W.; Saraste, A.; Capodanno, D.; Barbato, E.; Funck-Brentano, C.; Prescott, E.; Storey, R.F.; Deaton, C.; Cuisset, T.; et al. 2019 ESC Guidelines for the diagnosis and management of chronic coronary syndromes. Eur. Heart J. 2020, 41, 407–477. [Google Scholar] [CrossRef]
- Nezu, T.; Hosomi, N. Usefulness of Carotid Ultrasonography for Risk Stratification of Cerebral and Cardiovascular Disease. J. Atheroscler. Thromb. 2020, 27, 1023–1035. [Google Scholar] [CrossRef]
- Cismaru, G.; Serban, T.; Tirpe, A. Ultrasound Methods in the Evaluation of Atherosclerosis: From Pathophysiology to Clinic. Biomedicines 2021, 9, 418. [Google Scholar] [CrossRef]
- Gupta, A.; Kesavabhotla, K.; Baradaran, H.; Kamel, H.; Pandya, A.; Giambrone, A.E.; Wright, D.; Pain, K.J.; Mtui, E.E.; Suri, J.S.; et al. Plaque echolucency and stroke risk in asymptomatic carotid stenosis: A systematic review and meta-analysis. Stroke 2015, 46, 91–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Idini, M.; Wieringa, P.; Rocchiccioli, S.; Nieddu, G.; Ucciferri, N.; Formato, M.; Lepedda, A.; Moroni, L. Glycosaminoglycan functionalization of electrospun scaffolds enhances Schwann cell activity. Acta Biomater. 2019, 96, 188–202. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, E.W.; Mendoza, L.; Shteynberg, D.; Slagel, J.; Sun, Z.; Moritz, R.L. Trans-Proteomic Pipeline, a standardized data processing pipeline for large-scale reproducible proteomics informatics. Proteom. Clin. Appl. 2015, 9, 745–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reiter, L.; Claassen, M.; Schrimpf, S.P.; Jovanovic, M.; Schmidt, A.; Buhmann, J.M.; Hengartner, M.O.; Aebersold, R. Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Mol. Cell. Proteom. 2009, 8, 2405–2417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schilling, B.; Rardin, M.J.; MacLean, B.X.; Zawadzka, A.M.; Frewen, B.E.; Cusack, M.P.; Sorensen, D.J.; Bereman, M.S.; Jing, E.; Wu, C.C.; et al. Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: Application to protein acetylation and phosphorylation. Mol. Cell. Proteom. 2012, 11, 202–214. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [Green Version]
- Bancells, C.; Canals, F.; Benítez, S.; Colomé, N.; Julve, J.; Ordóñez-Llanos, J.; Sánchez-Quesada, J.L. Proteomic analysis of electronegative low-density lipoprotein. J. Lipid Res. 2010, 51, 3508–3515. [Google Scholar] [CrossRef] [Green Version]
- Hermus, L.; Lefrandt, J.D.; Tio, R.A.; Breek, J.C.; Zeebregts, C.J. Carotid plaque formation and serum biomarkers. Atherosclerosis 2010, 213, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Formato, M.; Farina, M.; Spirito, R.; Maggioni, M.; Guarino, A.; Cherchi, G.M.; Biglioli, P.; Edelstein, C.; Scanu, A.M. Evidence for a proinflammatory and proteolytic environment in plaques from endarterectomy segments of human carotid arteries. Arter. Thromb. Vasc. Biol. 2004, 24, 129–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Havel, R.J.; Eder, H.A.; Bragdon, J.H. The distribution and chemical composition of ultracentrifugally separated lipoproteins in human serum. J. Clin. Investig. 1955, 34, 1345–1353. [Google Scholar] [CrossRef] [Green Version]
- Ståhlman, M.; Davidsson, P.; Kanmert, I.; Rosengren, B.; Borén, J.; Fagerberg, B.; Camejo, G. Proteomics and lipids of lipoproteins isolated at low salt concentrations in D2O/sucrose or in KBr. J. Lipid Res. 2008, 49, 481–490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Böttcher, A.; Schlosser, J.; Kronenberg, F.; Dieplinger, H.; Knipping, G.; Lackner, K.J.; Schmitz, G. Preparative free-solution isotachophoresis for separation of human plasma lipoproteins: Apolipoprotein and lipid composition of HDL subfractions. J. Lipid Res. 2000, 41, 905–915. [Google Scholar] [CrossRef]
- Collins, L.A.; Olivier, M. Quantitative comparison of lipoprotein fractions derived from human plasma and serum by liquid chromatography-tandem mass spectrometry. Proteome Sci. 2010, 8, 42. [Google Scholar] [CrossRef] [Green Version]
- Gordon, S.M.; Deng, J.; Lu, L.J.; Davidson, W.S. Proteomic characterization of human plasma high density lipoprotein fractionated by gel filtration chromatography. J. Proteome Res. 2010, 9, 5239–5249. [Google Scholar] [CrossRef] [Green Version]
- Klop, B.; van der Pol, P.; van Bruggen, R.; Wang, Y.; de Vries, M.A.; van Santen, S.; O’Flynn, J.; van de Geijn, G.J.; Njo, T.L.; Janssen, H.W.; et al. Differential complement activation pathways promote C3b deposition on native and acetylated LDL thereby inducing lipoprotein binding to the complement receptor 1. J. Biol. Chem. 2014, 289, 35421–35430. [Google Scholar] [CrossRef] [Green Version]
- Speidl, W.S.; Kastl, S.P.; Huber, K.; Wojta, J. Complement in atherosclerosis: Friend or foe? J. Thromb. Haemost. 2011, 9, 428–440. [Google Scholar] [CrossRef] [PubMed]
- Badimon, L.; Vilahur, G. Thrombosis formation on atherosclerotic lesions and plaque rupture. J. Intern. Med. 2014, 276, 618–632. [Google Scholar] [CrossRef] [PubMed]
- Van Tuijl, J.; Joosten, L.A.B.; Netea, M.G.; Bekkering, S.; Riksen, N.P. Immunometabolism orchestrates training of innate immunity in atherosclerosis. Cardiovasc. Res. 2019, 115, 1416–1424. [Google Scholar] [CrossRef] [PubMed]
- Geovanini, G.R.; Libby, P. Atherosclerosis and inflammation: Overview and updates. Clin. Sci. 2018, 132, 1243–1252. [Google Scholar] [CrossRef]
- Hertle, E.; van Greevenbroek, M.M.; Arts, I.C.; van der Kallen, C.J.; Geijselaers, S.L.; Feskens, E.J.; Jansen, E.H.; Schalkwijk, C.G.; Stehouwer, C.D. Distinct associations of complement C3a and its precursor C3 with atherosclerosis and cardiovascular disease. The CODAM study. Thromb. Haemost. 2014, 111, 1102–1111. [Google Scholar] [CrossRef] [PubMed]
- Sotiriou, S.N.; Orlova, V.V.; Al-Fakhri, N.; Ihanus, E.; Economopoulou, M.; Isermann, B.; Bdeir, K.; Nawroth, P.P.; Preissner, K.T.; Gahmberg, C.G.; et al. Lipoprotein(a) in atherosclerotic plaques recruits inflammatory cells through interaction with Mac-1 integrin. FASEB J. 2006, 20, 559–561. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Marcos, L.V.; Lou-Bonafonte, J.M.; Martinez-Gracia, M.V.; Arnal, C.; Navarro, M.A.; Osada, J. Prenylcysteine oxidase 1, a pro-oxidant enzyme of low density lipoproteins. Front. Biosci. 2018, 23, 1020–1037. [Google Scholar] [CrossRef] [Green Version]
- Wieczorek, E.; Ożyhar, A. Transthyretin: From Structural Stability to Osteoarticular and Cardiovascular Diseases. Cells 2021, 10, 1768. [Google Scholar] [CrossRef]
- Serrano, M.; Moreno-Navarrete, J.M.; Puig, J.; Moreno, M.; Guerra, E.; Ortega, F.; Xifra, G.; Ricart, W.; Fernández-Real, J.M. Serum lipopolysaccharide-binding protein as a marker of atherosclerosis. Atherosclerosis 2013, 230, 223–227. [Google Scholar] [CrossRef]
- Lepper, P.M.; Kleber, M.E.; Grammer, T.B.; Hoffmann, K.; Dietz, S.; Winkelmann, B.R.; Boehm, B.O.; März, W. Lipopolysaccharide-binding protein (LBP) is associated with total and cardiovascular mortality in individuals with or without stable coronary artery disease—Results from the Ludwigshafen Risk and Cardiovascular Health Study (LURIC). Atherosclerosis 2011, 219, 291–297. [Google Scholar] [CrossRef]
- Thompson, J.C.; Jayne, C.; Thompson, J.; Wilson, P.G.; Yoder, M.H.; Webb, N.; Tannock, L.R. A brief elevation of serum amyloid A is sufficient to increase atherosclerosis. J. Lipid Res. 2015, 56, 286–293. [Google Scholar] [CrossRef] [Green Version]
- Getz, G.S.; Krishack, P.A.; Reardon, C.A. Serum amyloid A and atherosclerosis. Curr. Opin. Lipidol. 2016, 27, 531–535. [Google Scholar] [CrossRef]
- Schuchardt, M.; Prüfer, N.; Tu, Y.; Herrmann, J.; Hu, X.P.; Chebli, S.; Dahlke, K.; Zidek, W.; van der Giet, M.; Tölle, M. Dysfunctional high-density lipoprotein activates toll-like receptors via serum amyloid A in vascular smooth muscle cells. Sci. Rep. 2019, 9, 3421. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Parameters | “Hard” (n = 31) | “Soft” (n = 44) |
|---|---|---|
| Age (years) * | 67.1 ± 6.9 | 72.6 ± 8.0 |
| Sex ratio, m/f | 22/9 | 34/10 |
| BMI * | 25.3 ± 3.4 | 25.9 ± 3.5 |
| Triglycerides (mg/dL) * | 116.8 ± 48.2 | 124.1 ± 44.6 |
| Total Cholesterol (mg/dL) * | 166.6 ± 41.4 | 181.3 ± 50.5 |
| HDL Cholesterol (mg/dL) * | 47.6 ± 14.1 | 42.3 ± 15.8 |
| LDL Cholesterol (mg/dL) * | 96.8 ± 29.8 | 112.0 ± 45.0 |
| Cholesterol lowering therapy (%) | 23/31 (74.2) | 41/44 (93.2) |
| Glycemia (mg/dL) * | 114.4 ± 35.8 | 107.3 ± 20.9 |
| HbA1C (%) * | 6.6 ± 1.0 | 6.5 ± 1.0 |
| Type 2 Diabetes (%) | 9/31 (29.0) | 19/44 (43.2) |
| Glucose lowering Therapy (%) | 9/9 (100) | 12/19 (63.2) |
| Systolic blood pressure (mmHg) * | 138.8 ± 8.7 | 129.1 ± 12.6 |
| Diastolic blood pressure (mmHg) * | 79.9 ± 8.9 | 72.4 ± 7.3 |
| Anti-hypertensive Therapy (%) | 28/31 (90.3) | 35/44 (79.5) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Finamore, F.; Nieddu, G.; Rocchiccioli, S.; Spirito, R.; Guarino, A.; Formato, M.; Lepedda, A.J. Apolipoprotein Signature of HDL and LDL from Atherosclerotic Patients in Relation with Carotid Plaque Typology: A Preliminary Report. Biomedicines 2021, 9, 1156. https://doi.org/10.3390/biomedicines9091156
Finamore F, Nieddu G, Rocchiccioli S, Spirito R, Guarino A, Formato M, Lepedda AJ. Apolipoprotein Signature of HDL and LDL from Atherosclerotic Patients in Relation with Carotid Plaque Typology: A Preliminary Report. Biomedicines. 2021; 9(9):1156. https://doi.org/10.3390/biomedicines9091156
Chicago/Turabian StyleFinamore, Francesco, Gabriele Nieddu, Silvia Rocchiccioli, Rita Spirito, Anna Guarino, Marilena Formato, and Antonio Junior Lepedda. 2021. "Apolipoprotein Signature of HDL and LDL from Atherosclerotic Patients in Relation with Carotid Plaque Typology: A Preliminary Report" Biomedicines 9, no. 9: 1156. https://doi.org/10.3390/biomedicines9091156
APA StyleFinamore, F., Nieddu, G., Rocchiccioli, S., Spirito, R., Guarino, A., Formato, M., & Lepedda, A. J. (2021). Apolipoprotein Signature of HDL and LDL from Atherosclerotic Patients in Relation with Carotid Plaque Typology: A Preliminary Report. Biomedicines, 9(9), 1156. https://doi.org/10.3390/biomedicines9091156









